Summary
Activating transcription factor 2 (ATF2) is regulated by JNK/p38 in response to stress. Here, we demonstrate that the protein kinase ATM phosphorylates ATF2 on serines 490 and 498 following ionizing radiation (IR). Phosphoantibodies to ATF2490/8 reveal dose- and time-dependent phosphorylation of ATF2 by ATM that results in its rapid colocalization with γ-H2AX and MRN components into IR-induced foci (IRIF). Inhibition of ATF2 expression decreased recruitment of Mre11 to IRIF, abrogated S phase checkpoint, reduced activation of ATM, Chk1, and Chk2, and impaired radioresistance. ATF2 requires neither JNK/p38 nor its DNA binding domain for recruitment to IRIF and the S phase checkpoint. Our findings identify a role for ATF2 in the DNA damage response that is uncoupled from its transcriptional activity.
Introduction
Activating transcription factor 2 (ATF2) is a member of the bZIP family of transcription factors, which has been implicated in transcriptional regulation of a wide set of genes, including cytokines (Liu and Green, 1990; Falvo et al., 2000; Tsai et al., 1996), cell cycle control proteins (Shimizu et al., 1998), and apoptosis factors (Bhoumik et al., 2002). ATF2 transcriptional activity requires homodimerization or heterodimerization with other members of the bZIP family, of which c-Jun has been best characterized (van Dam et al., 1993).
Several studies have implicated ATF2 in DNA repair, although mechanisms underlying its possible regulation and function are not well understood. ATF2 was indirectly implicated in regulating repair of ionizing radiation (IR)-induced double strand breaks (DSB; Masson et al. [2001]), as well as in repair of UV-induced lesions (Hayakawa et al., 2003). An additional link between ATF2 and DNA repair comes from its association with TIP49b (Cho et al., 2001), a member of the chromatin remodeling complex (Kanemaki et al., 1999), which is part of the TIP60 histone acetylase complex implicated in DSB repair (Ikura et al., 2000). Studies in S. pombe implicated Atf1/Pcr1 (an ATF2 homolog) in meiotic recombination hot spots (Steiner et al., 2002). Further, atf1 and pcr1 are required for deacetylation of histone H3 (and H4), a prerequisite for subsequent H3 lysine 9 methylation and Swi6-dependent heterochromatin assembly (Jia et al., 2004; Kim et al., 2004). Lysine 9 methylation of histone H3 was also shown to be important for locus-specific stability of polycomb complexes, which have been implicated in transcriptional control (Ringrose et al., 2004). These observations indicate that via its effect on chromatin organization ATF2 may contribute to the DNA damage response.
The ATM protein kinase phosphorylates key factors in various damage response pathways, most notably those that activate cell cycle checkpoints. ATM is considered a primary transducer of cellular responses to DSBs and is missing or inactivated in patients with ataxia telangiectasia (Shiloh, 2003). ATM/ATR substrates include the transcription factor p53 (Banin et al., 1998), p53's E3 ubiquitin ligase Mdm2 (Maya et al., 2001), the BRCA1 protein (Cortez et al., 1999), the checkpoint protein Chk2 (Matsuoka et al., 1998), the cohesin subunit Smc1 (Kim et al., 2002), and the damage response proteins H2AX, Mdc1/NFBD1, Nbs1, and 53BP1 (Paull et al., 2000, Goldberg et al., 2003, Lim et al., 2000, Wang et al., 2002). Here, we identify ATF2 as a previously unrecognized substrate for the ATM family of kinases and demonstrate its role in the DNA damage response.
Results
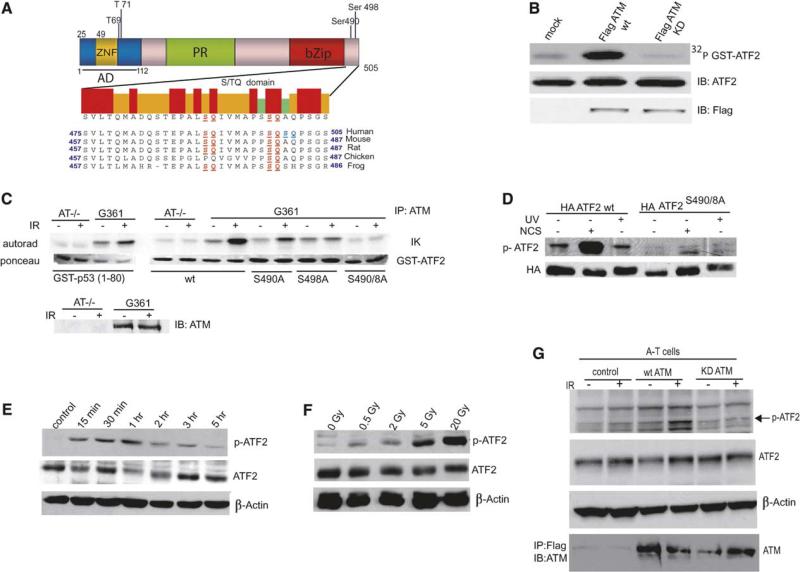
ATM Phosphorylates ATF2 on Amino Acids 490 and 498
Examination of the ATF2 amino acid sequence identified two highly conserved sites within the C-terminal of ATF2 for phosphorylation by phosphatidylinositol 3-kinase-like (PIKK) family members ATM/ATR/DNA-PK (Figure 1A). Immunokinase reactions using Flag-ATM revealed efficient phosphorylation of GST-ATF2 in vitro (Figure 1B). Mutation of either of the two putative sites reduced, and mutating both sites abolished, ATM phosphorylation of ATF2. Phosphorylation of ATF2 was not observed using immune complexes obtained from A-T cells (Figure 1C), similar to what has been observed with the ATM target p53 (Banin et al., 1998). These observations indicated that ATM is capable of phosphorylating ATF2 on serines 490 and 498 in vitro.
Figure. 1. ATM Phosphorylates ATF2 on Amino Acids 490 and 498.
(A) Outline of major functional domains in ATF2 and the amino acid sequence section that harbors the phosphoacceptor sites for ATM in human, mouse, rat, chicken, and frog. ATM phosphor-acceptor sites on ATF2 are highlighted. Domains indicated: PR, proline rich; ZNF, zinc finger; bZip, basic leucine zipper.
(B) In vitro phosphorylation of ATF2 by ATM. Immunokinase reactions were performed using Flag-tagged wild-type (wt) or kinase-dead mutant forms of ATM expressed in 293T cells that had been immunopurified from IR-treated cells. Protein G-bound ATM was incubated with a bacterially expressed and purified spliced form of GST-sATF2 (Δ35–395) using 32PγATP. GST-sATF2 was separated on SDS-PAGE followed by immunoblot (middle panel) and autoradiography (upper panel). Control for the equal amount of ATM used in these reactions is shown in the lower panel.
(C) Immunokinase reactions were carried out as detailed in (B), except that the ATM used for these reactions was immunopurified from G361 melanoma cells, which harbor high levels of ATM, and from A-T cells after mock or 5 Gy IR treatment. GST-ATF2 (wt or mutated on S490A, S498A, or both) was expressed in bacteria and purified for kinase reactions using immunoprecipitated ATM. GST-p53 (aa 1–80) served as positive control. Ponceau staining revealed the amount of substrate used and immunoblot of ATM confirmed the high level of ATM expression in G361 but not in A-T cells.
(D) ATF2 phosphorylation on aa 490 and 498 is induced within 1 hr after NCS but not UV treatment. 293T cells were transfected with wt or mutant (on both aa 490 and 498) forms of HA-ATF2 followed by treatment with UV (254 nm, 24 J/m2) or radiomimetic drug NCS (300 ng/ml). Proteins prepared 1 hr later were used for immunoblot analysis with antibodies raised against the phosphorylated form of ATF2 on aa 490 and 498 (p-ATF2). The membrane was reprobed with antibodies to HA to reveal relative expression of the transfected constructs.
(E) Kinetics of ATF2 phosphorylation by ATM. Melanoma cells (MeWo) were subjected to IR (5 Gy) and proteins were prepared at the indicated time points. Analysis of ATF2 phosphorylation on aa 490 and 498 (p-ATF2), ATF2 expression levels (middle panel), and β-actin are shown.
(F) Dose-dependent phosphorylation of ATF2 by ATM. MeWo cells were subjected to IR at the indicated doses, and proteins prepared after 1 hr were subjected to analysis as indicated in panel (D).
(G) Wild-type, but not kinase-dead, ATM induces phosphorylation of endogenous ATF2 in A-T cells. GM05849 cells derived from an ATM patient treated by IR (5 Gy) and proteins prepared 1 hr later were subjected to immunoblot analysis with p-ATF2 antibodies. The membrane was reprobed with total ATF2 antibodies (second panel). β-actin and ATM expression is shown in the lower panels.
We next generated phosphoantibodies that specifically recognize ATF2 phosphorylated on aa 490 and 498 (p-ATF2). Recognition of ATF2 by these phosphoantibodies was inhibited by phosphorylated but not by nonphosphorylated peptides on these sites (Figure S1A in the Supplemental Data available with this article online), a finding that establishes the specificity of these antibodies. p-ATF2 detected the wt but not ATF2 mutated on the 490 and 498 phosphoacceptor sites expressed in cells within 1 hr after treatment with the radiomimetic drug neocarzinostatin (NCS) but not with UV irradiation (Figure 1D). Phosphorylation of ATF2 was observed as soon as 15 min after IR with a peak after 1hr(Figure 1E). Doses, as low as 0.5 Gy, were sufficient to induce phosphorylation of ATF2, although the level of phosphorylation increased proportionately to the IR dose (Figure 1F). Phosphorylation of endogenous ATF2 on 490 and 498 was also seen in human diploid fibroblasts (IMR90; Figure S1B) but not in A-T cells subjected to IR treatment (Figure 1G). Ectopic expression of wt but not the kinase-dead form of ATM restored endogenous ATF2 phosphorylation on aa 490 and 498 in A-T cells (Figure 1G). These data suggest that following formation of DSB, phosphorylation of ATF2 on aa 490 and 498 is mediated by ATM.
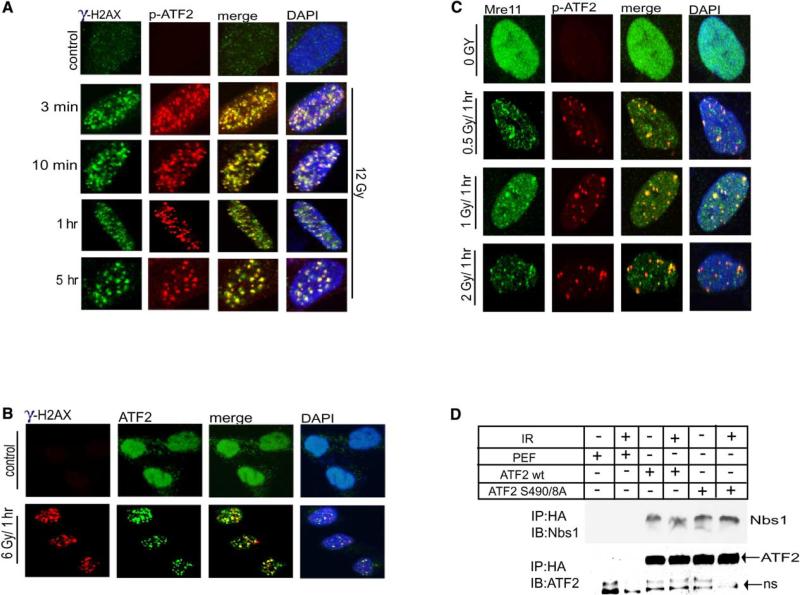
ATF2 Phosphorylated by ATM Is Colocalized with γ-H2AX and Components of the MRN Complex in DNA Repair Foci
To investigate possible changes in localization of ATF2, we performed a series of immunocytochemistry analyses. After IR, ATF2 was identified by antibodies to ATF2 phosphorylated on aa 490 and 498 within foci resembling those formed in response to DSB (Figure 2A). The signal was out-competed using the phosphorylated, but not the nonphosphorylated, peptide, demonstrating the specificity of ATF2 phosphoantibodies in immunocytochemistry analysis (Fig. S1c).
Figure 2. p-ATF2 Is Localized Immediately after IR to DSB Repair Foci.
(A) ATF2 foci colocalize with γ-H2AX foci as early as 3 min after IR. IMR90 cells were subjected to IR (12 Gy) and fixed at the indicated time points, followed by analysis using antibodies to p-ATF2 and to γ-H2AX.
(B) IR-induced ATF2 foci formation. Cells (WM793) were untreated or irradiated (6 Gy), fixed 1 hr after irradiation, and stained with anti-ATF2 antibody (C19).
(C) ATF2 is localized in repair foci following low doses of IR. IMR90 cells were subjected to IR (0.5 Gy, 1 Gy, or 2 Gy) and fixed at the indicated time points, followed by analysis using antibodies to p-ATF2 and to Mre11.
(D) ATF2 associates with Nbs1. 293T cells were transfected with HA-ATF2 wt or mutant on ATM phosphoacceptor sites and were subjected to IR (6 Gy) 24 hr later. Proteins prepared prior to and 1 hr after IR were subjected to immunoprecipitation with antibodies to HA followed by immunoblot analysis using antibodies to Nbs1, as indicated. The membrane was reprobed with antibodies to ATF2 to reveal amounts precipitated (lower panel). NS, nonspecific; PEF, empty vector driven by elongation factor promoter.
To further assess the possible role of ATF2 in the DNA damage response, we determined whether ATF2 foci colocalize with other DNA damage response proteins recruited to DSB sites. Significantly, ATF2 foci were detected as early as 3 min after IR and were colocalized with γ-H2AX (Figure 2A), which have been associated with DSB sites and represent one of the earliest responses to DNA damage (Paull et al., 2000). Over the time period following IR the number of such foci decreased, whereas their size increased (Figure 2A), similar to what was observed with other members of the MRN complex. IRIF detected by p-ATF2 antibodies were also observed using antibodies against the nonphosphorylated form of ATF2, suggesting that a major fraction of ATF2 is recruited to such foci (Figure 2B), similarly to what was seen with Mre11 (Maser et al., 1997). ATF2 foci were formed in response to doses as low as 0.5 Gy (Figure 2C), in line with the finding that minimal degrees of DSB suffice for activation of ATM (Bakkenist and Kastan, 2003). Inhibition of ATF2 expression by specific RNAi efficiently blocked detection of ATF2 phosphorylation by the p-ATF2 antibodies after IR (Figures S2A and S2B) and abolished the localization of ATF2 in IRIF recognized by ATF2 (Figure S2C) and p-ATF2 Ab (Figure S2D).
As early as 10 min after IR, and in a dose-dependent manner, p-ATF2 foci colocalized with Rad50 and Mre11 (Figures S3A and S3B), and at a later time point, also with Nbs1 (Figure S3C). Importantly, p-ATF2 was also found within these foci following detergent-based extraction (Figure S3D), indicating that p-ATF2 is tightly bound to damaged DNA. These observations establish the recruitment of p-ATF2 to IRIF. A series of immunoprecipitations confirmed the association of ATF2 with Nbs1 (Figure 2D) and Mre11 (data not shown), which were not dependent on ATM-phosphorylation of ATF2, similar to what was observed for other MRN components (Stewart et al., 2003).
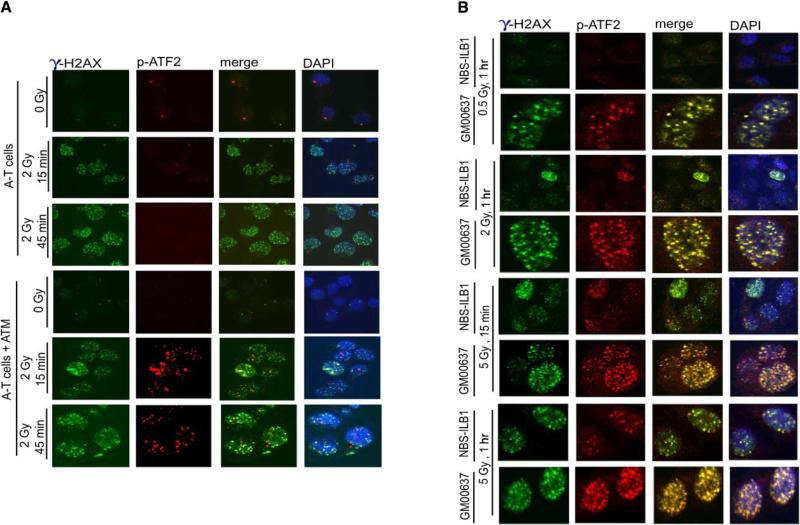
ATF2 Localization into IRIF Is ATM Dependent
To ascertain the role of ATM in ATF2 recruitment to IRIF, we used A-T cells. ATF2 foci were not found in IRIF in A-T cells (up to 5 Gy) and within early time periods (up to 1 hr) after IR. Upon reconstitution of exogenous ATM into the A-T cells, ATF2 was found in IRIF (Figure 3A). These observations provide direct evidence of ATM's role in localization of ATF2 in IRIF.
Figure 3. Localization of ATF2 in IRIF Requires ATM and at Early Time Points, Nbs1.
(A) ATF2 localization in IRIF is ATM dependent. Control or A-T (AT22IJE-T) cells and A-T cells that were reconstituted for ATM by exogenous expression were subjected to IR (2 Gy) and fixed at the indicated time points, followed by analysis as detailed above. γ-H2AX staining is also shown.
(B) Nbs1 is important for immediate localization of ATF2 to repair foci after exposure to low dose IR. Control fibroblasts (GM00637) or NBS (NBS-ILB1) cells were subjected to IR at the indicated doses, and cells were fixed at 15 min or 60 min as indicated. Staining was carried out using antibodies to phospho-ATF2 or γ-H2AX.
Cells from patients with the Nijmegen breakage syndrome (NBS) are deficient in NBS and exhibit impaired recruitment of Mre11-Rad50 into IRIF (Carney et al., 1998). Therefore, we next assessed possible changes in ATF2 localization into IRIF of NBS cells. Neither ATF2 nor γ-H2AX were identified within such foci within 1 hr after exposure of NBS cells to a low dose of IR (0.5 Gy or 2 Gy; Figure 3B). However, ATF2 was seen in IRIF of NBS cells after exposure to a higher dose of IR and in greater numbers at a later time point (5 Gy; 1 hr; Figure 3B). These data suggest that Nbs1 is important for immediate recruitment of ATF2 into IRIF formed upon exposure to low-dose IR, similar to its recruitment of Mre11 and Rad50 (Carney et al., 1998). Upon exposure to high doses of IR, the localization of ATF2 to IRIF is distinct from the MRN complex.
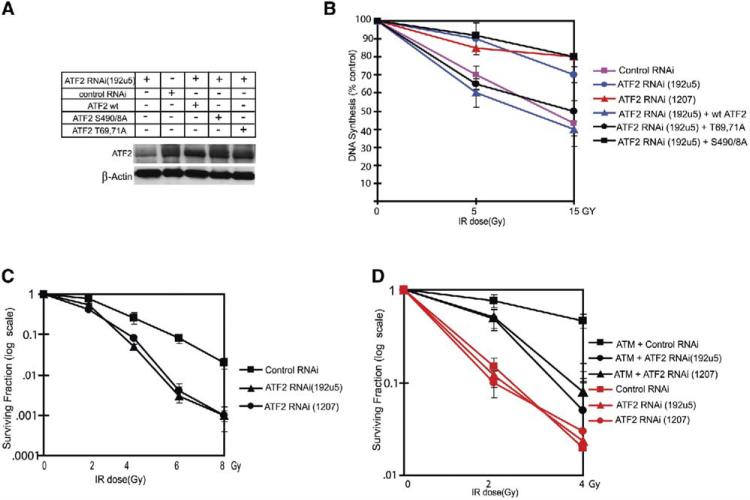
ATF2 Is Required for the IR-Induced S Phase Checkpoint and for Radioresistance
Common to cells obtained from NBS and A-T patients is impaired S or G2/M phase checkpoint control after IR (Shiloh, 2003, Lim et al., 2000). We accordingly monitored changes in DNA synthesis after IR of cells whose ATF2 expression was inhibited by specific RNAi (Figure 4A and S2a). Inhibition of ATF2 expression caused a decrease in the fraction of cells found in the S phase of the cell cycle under normal growth conditions (Figure S4A). Expression of a transcription-inactive form of ATF2 (aa 69 and 71) in cells whose ATF2 expression was inhibited failed to increase the rate of DNA synthesis, in contrast to the expression of either wt or phosphomutant (490 and 498) forms of ATF2 (Figure S4B). This finding is consistent with the ability of ATF2 mutated in its ATM phosphoacceptor sites to activate an ATF2 reporter to the same level as wt ATF2 (Figure S5A). Thus, ATF2 affects the rate of DNA synthesis and cell cycle progression in cells maintained under non-stressed conditions through its ability to activate transcription, of cyclin A and cyclin D (Beier et al., 1999; Shimizu et al., 1998). Given these results, it was important to distinguish between the effects of ATF2 on the cell cycle checkpoint after IR and its role during normal nonstressed growth conditions. To this end we identified melanoma cell lines (MeWo and LU1205) in which ATF2 does not alter cell cycle distribution under normal growth (Figures S4C and S4D); these cells were used for the subsequent analysis.
Figure 4. ATF2 Is Required for Checkpoint Control and Resistance to IR.
(A) Mutant ATF2 forms expressed at levels of endogenous ATF2. Wild-type, transcriptionally inactive (T69, 71A), and ATM phosphomutant (S490 and 498A) forms of ATF2 were transfected into MeWo cells after inhibiting endogenous ATF2 expression with RNAi to achieve expression levels equivalent to that of endogenous ATF2 (transfection efficiency = 65%). Immunoblot analysis using ATF2 antibodies was carried out 36 hr after transfection. These conditions were used throughout the related experiments.
(B) ATF2 is required for inhibition of DNA synthesis after IR. MeWo cells were infected with ATF2-RNAi (192U5, which targets 5# UTR) followed by transfection of empty vector or ATF2 forms. DNA synthesis was monitored using 14C followed by 3H-thymidine labeling. Change in the percent of DNA synthesis is shown based on five independent experiments 45 min after treatment with IR (5 Gy or 15 Gy). Calculation is based on the ratios of [3H]:[14C] and expressed DNA synthesis, which was normalized to control values (0 Gy) for every individual sample (pSuper, ATF2 RNAi, and the like). Data shown reflect triplicate measurements carried out in three experiments, which were used for calculating standard deviation as reflected in error bars.
(C) ATF2 affects cell radiosensitivity. Sensitivity to IR was determined by the colony formation assay in U2OS cells infected with control or two different ATF2 RNAi (Figure S4A; 192U5 and 1207). Surviving fraction is plotted as log of colonies after IR/colonies without IR. Error bars reflect standard deviations of triplicate samples for each point based on three experiments.
(D) ATM is required for ATF2-dependent radioresistance. A-T cells and A-T cells reconstituted with ATM (4000 cells per 60 mm plate) were subjected to IR at the indicated doses, and CFE were counted 18 days later. Cells were also subjected to infection with two different ATF2 or a control RNAi, to determine the role of ATF2 in radioresistance in A-T cells. The surviving fractions were plotted by calculating the log of colonies after IR/colonies without IR. Error bars reflect standard deviation based on triplicate analysis.
Consistent with IR-induced S phase arrest, IR-treated MeWo cells infected with control RNAi exhibited about 40% and 60% inhibition of DNA synthesis following exposure to 5 Gy and 15 Gy, respectively (Figure 4B). Strikingly however, inhibition of ATF2 expression using two different retroviral RNAi constructs (Figures 4B and S2A) markedly attenuated inhibition of DNA synthesis after IR to only 10%–15% (Figure 4B). This finding suggests that ATF2 is required for the intact intra-S phase checkpoint. Cells in which ATF2 expression was inhibited (using ATF2 RNAi) were reconstituted with a transcriptionally inactive form of ATF2 which was sufficient to restore the inhibition of DNA synthesis following IR to the degree seen upon reconstitution with the wt form of ATF2 (Figure 4B). Conversely, reconstitution with ATF2 mutated on both ATM phosphoacceptor sites failed to restore inhibition of the radioresistant DNA synthesis (RDS) phenotype following IR, caused by inhibition of ATF2 expression (Figure 4B). These observations suggest that ATF2 is important for IR-induced S phase checkpoint control, for which it does not require phosphorylation on aa 69 and 71 (required for its transcriptional activities, as noted below).
Since radiosensitivity of cells is also affected by proteins involved in DSB repair, we monitored survival of cells expressing different ATF2-RNAi after exposure to IR. Inhibition of ATF2 expression reduced (up to over 10-fold at a dose of 6 Gy) the number of colonies formed within 12 days following IR (Figure 4C), further substantiating the previously reported role of ATF2 in cell survival following DNA damage (Bhoumik et al., 2002). Additional analysis was carried out in A-T cells reconstituted with different forms of ATF2. As expected, reconstitution of A-T cells with ATM restored resistance to IR (over 15-fold; Figure 4D). Inhibition of ATF2 expression in A-T cells did not alter the level of sensitivity to IR (Figure 4D). Nevertheless, inhibition of ATF2 expression in A-T cells reconstituted with ATM reduced their level of resistance (Figure 4D). These findings suggest that ATM is required for ATF2-mediated radioresistance.
ATF2's Role in the DNA Damage Response Does Not Require Its Transcriptional Activity
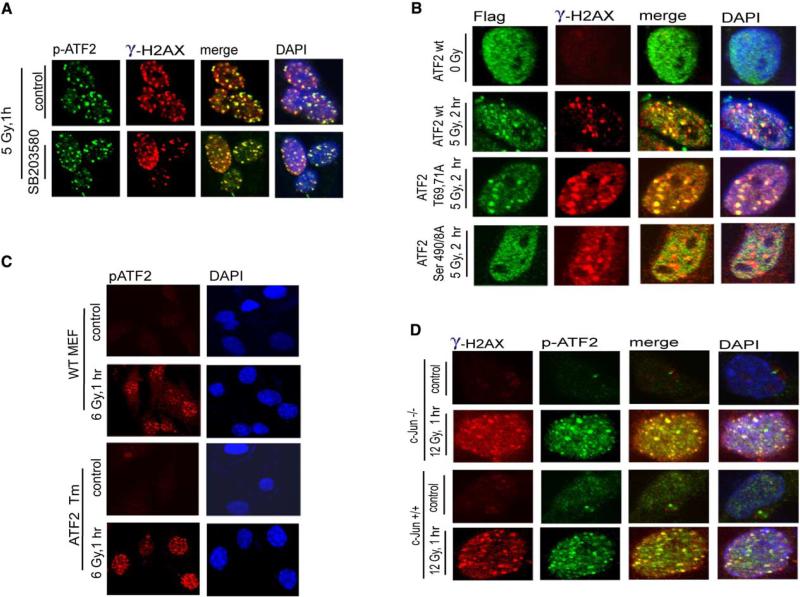
Given ATF2's role as a transcription factor, we next assessed whether its transcriptional activities are required for its DNA damage response. Since phosphorylation of ATF2 by JNK/p38 on Thr69/71 is prerequisite to its transcriptional activities, we first monitored ATF2 localization within IRIF in cells pretreated with a pharmacological inhibitor of p38/JNK. As shown in Figure 5A, localization of ATM-phosphorylated ATF2 in DSB repair foci was not affected by inhibition of p38/JNK activity, suggesting that amino-terminal phosphorylation of ATF2 is not required for its localization in IRIF. The latter is consistent with the notion that IR is a poor inducer of p38/JNK kinases within the time frame and at the doses used in the current study (Chen et al., 1996).
Figure 5. ATF2 Role in the DNA Damage Response Is Uncoupled from Its Transcriptional Activity.
(A) ATF2 phosphorylation by JNK/p38 is not required for recruitment to DSB repair foci after IR. A pharmacological inhibitor of p38/JNK (SB203580, 10 μM, which is sufficient for inhibition of both JNK and p38 kinases) was added to MeWo cells 2 hr before mock treatment or IR (5 Gy). The inhibitor was kept in the medium for 1 hr after treatment, when cells were fixed and subjected to analysis with antibodies to γ-H2AX and p-ATF2.
(B) Colocalization of transcriptionally inactive ATF2 with γ-H2AX in IRIF following IR. ATF2 forms (wt, transcriptionally inactive [T69,71A], and ATM phosphomutants [S490/A] were transfected into MeWo cells that were inhibited for ATF2 expression with corresponding RNAi. Immunostaining of the exogenous forms of ATF2 was carried out 2.5 hr after IR using antibodies to the Flag tag (green). Cells were also analyzed using antibodies to γ-H2AX (red).
(C) ATF2 lacking the DNA binding domain is localized to DSB-induced foci. ATF2 mutant cells expressing a transcriptionally inactive form of ATF2 that lacks DNA binding and part of the leucine zipper domains were subjected to mock or IR treatment (6 Gy) and analyzed to detect ATF2 localization in DSB-induced foci using p-ATF2 antibodies.
(D) ATF2 foci are formed in IR-treated c-Jun−/− cells. Cells lacking c-Jun were irradiated (12 Gy) and fixed 1 hr later for immunostaining using p-ATF2 and γ-H2AX antibodies.
We next used cells whose ATF2 expression was inhibited and reconstituted with wt or transcriptionally inactive (mutated on aa 69 and 71) ATF2 or ATM phosphorylation mutant ATF2 (aa 490 and 498). Expression levels of ectopic ATF2 forms were normalized to that of endogenous ATF2 (Figure 4A). Whereas cells expressing ATF2 490 and 498 no longer exhibited ATF2 within DSB-induced repair foci following IR, ATF2 mutated on aa 69 and 71 was found within such foci, like the wt ATF2 protein (Figure 5B). Thus, this requirement of ATF2 was similar for reconstitution of repair foci and the IR-induced S phase checkpoint (Figure 4B).
To further substantiate that ATF2's role in the DNA damage response is independent of its transcriptional activities, we used mouse embryo fibroblasts (MEFs) obtained from ATF2 mutant mice, in which the complete DNA binding and part of the leucine zipper domains were deleted, thereby rendering ATF2 transcriptionally inactive with impaired dimerization capabilities (Breitwieser et al., unpublished studies). Immunoblot analysis confirmed that the mutant ATF2 protein migrated faster (Figure S5B) and was no longer found on the promoter of target genes, as revealed by CHIP assays (Figure S5C). Importantly, despite the lack of DNA binding and leucine zipper domains, ATF2 was found within DSB repair foci after IR, similar to the wt form (Figure 5C). This finding substantiates our conclusion that transcriptional activities of ATF2 are not required for its function in the DNA damage response.
Since ATF2 transcriptional activities are primarily me diated by its heterodimerization with c-Jun, we assessed possible changes in ATF2 localization within IRIF in c-Jun−/− cells. As shown (Figure 5D), ATF2 localization into DSB-repair foci was normal in c-Jun−/− cells, suggesting that ATF2 does not require c-Jun for its localization into IRIF.
ATF2 Is Important for IR-Mediated Activation of ATM, Chk1, and Chk2
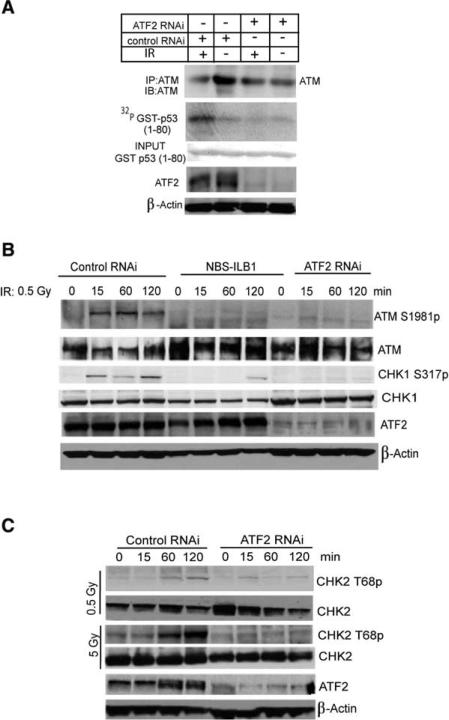
Given the role of ATF2 in the regulation of IR-induced cell cycle checkpoints, we assessed the possible effect of ATF2 on activities of Chk1, Chk2 and its kinase ATM. Significantly, IR-induced activation of ATM, measured by immunokinase assays using p53 as a substrate, was markedly reduced in cells whose ATF2 expression was inhibited (Figure 6A). Subsequent analysis was carried out by monitoring ATM phosphorylation on Ser 1981 (Bakkenist and Kastan, 2003). After exposure to low-dose IR (0.5 Gy) the level of ATM activation was reduced upon inhibition of ATF2 expression, similar to what was observed in NBS cells (Figure 6B). These data suggest that ATF2, somewhat similar to Nbs1, contributes to maintenance of ATM activity in response to the formation of DSB.
Figure 6. ATF2 Is Important for IR-Induced Activation of ATM and Chk2.
(A) ATF2 is important for activation of ATM. U2OS cells were infected with either control or ATF2 RNAi, and 72 hr later, cells were subjected to IR (6 Gy). Proteins prepared 1 hr after IR were subjected to IP with antibodies to ATM. Immunoprecipitated material was used for immunokinase reactions using GST-p531–80 as a substrate. Upper panel depicts the autoradiograph, whereas lower panels show input and level of ATF2 as well as β-actin expression.
(B) ATF2 and Nbs1 are important for ATM and Chk1 activation by IR. GM00637 cells were subjected to infection with control or ATF2 RNAi and were analyzed in parallel to NBL-ILB1 cells at the indicated time points following a low dose of IR (0.5 Gy) for ATM activation using phosphorylation of ATM on Ser1981 (upper panel) and the phosphorylation of Chk1 on Ser317 (third panel). Also shown are total ATM (second panel), total Chk1 (fourth panel), ATF2 expression (fifth panel), and β-actin, used as a loading control (lower panel).
(C) Inhibition of ATF2 expression impairs IR-induced Chk2 phosphorylation. Control or ATF2 RNAi-infected cells were treated by IR (0.5 Gy or 5 Gy) 72 hr postinfection and proteins prepared at the indicated time points. Immunoblot analysis using CHK2 antibodies detects phosphorylated (Thr 68; upper panel) and nonphosphorylated (second panel) forms of Chk2. Controls for the expression level of endogenous ATF2 and β-actin are shown.
Consistent with its effect on ATM, ATF2 was also important in IR-induced activation of Chk1, measured by its phosphorylation on Ser 317 (Figure 6B). Similarly, inhibition of ATF2 expression reduced the level of Chk2 phosphorylation measured by its phosphorylation on Thr 68 in response to IR (Figure 6C). Collectively these data indicate that ATF2 plays an important role in the activation of ATM, and consequently of Chk1 and Chk2.
Localization of Mre11 and Nbs1 in IRIF Foci Is Reduced upon Inhibition of ATF2
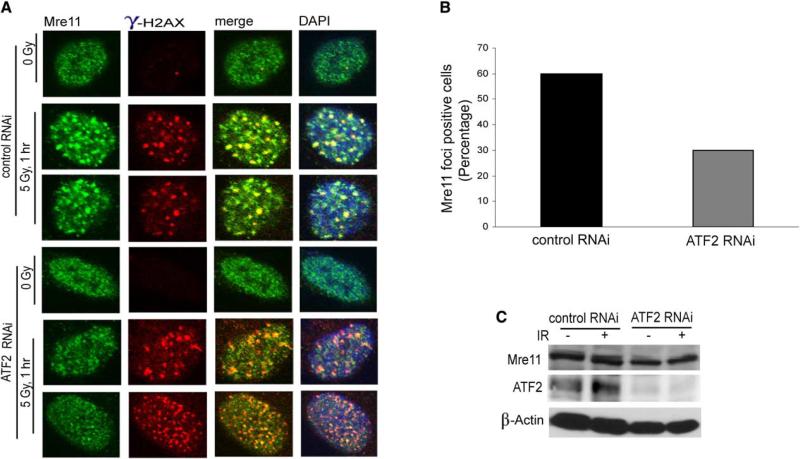
Because Mre11 has been implicated in the upstream signaling required for activation of ATM (Uziel et al., 2003; Carson et al., 2003; Lee and Paull, 2004; Costanzo et al., 2004), we explored possible effects of ATF2 on localization of Mre11 to IRIF. Inhibition of ATF2 expression by RNAi reduced recruitment of Mre11 to IRIF (Figure 7A). Counting the number of IRIF revealed a 50% decrease in cells whose ATF2 expression was inhibited (Figure 7B). The effect of ATF2 on recruitment of Mre11 into IRIF did not affect Mre11 expression (Figure 7C). Inhibition of ATF2 expression also reduced recruitment of Nbs1 into IRIF (Figure S5D). Unlike the effect of ATF2 on recruitment of Mre11 and Nbs1, localization of neither 53BP1 nor Mdc1/NFBD1 into IRIF was altered upon inhibition of ATF2 expression (data not shown). Collectively, these data suggest that ATF2 expression contributes to the selective recruitment of Mre11 and Nbs1 into IRIF.
Figure 7. ATF2 Is Affecting the Recruitment of the Mre11 Complex to DSB Repair Foci.
(A) Inhibition of ATF2 reduces recruitment of Mre11 to IRIF. MeWo cells infected with ATF2 RNAi were subjected to IR (5 Gy) and 1 hr later, fixed for IHC. Shown is staining with antibodies to Mre11 and γ-H2AX.
(B) Quantification of inhibition of Mre11 recruitment upon inhibition of ATF2. Data shown in panel (A) were quantified by multiple counts and plotted.
(C) ATF2 does not alter expression of Mre11. IMR90 cells were infected with either control or ATF2 RNAi, and 72 hr later, cells were subjected to IR (6 Gy). Proteins prepared 1 hr after IR were analyzed for Mre11 levels (upper panel) and ATF2 expression (middle panel). β-actin was used as a loading control (lower panel).
Discussion
The current study identifies ATF2 as a substrate for ATM and reveals its role as a participant in the DNA damage response. Concomitant to IR induced activation of ATM, ATM phosphorylates ATF2, resulting in its rapid recruitment into IRIF and pointing to its possible role as a sensor/adaptor in very early stages of the DNA damage response. Further, our data show that ATM-phosphorylated ATF2 is contributing to the recruitment of Mre11 and Nbs1 to IRIF, a finding that points to its role in coordinating the DNA damage response. ATF2 also affects the S phase checkpoint in response to IR and affects radiosensitivity, similarly to what is seen in cells that harbor mutant NBS1, MRE11, or A-T genes. Our data also demonstrate that ATF2 is important for activation of ATM as well as for concomitant activation of Chk1 and Chk2. Further studies will delineate the mechanism underlying ATF2 activation of ATM, which is likely to contribute to maintenance of active ATM at IRIF. Lastly, ATF2's role in the DNA damage response is distinct from its transcriptional activities, an observation that underscores the significance of our findings and establishes a paradigm for uncoupling transcription from DNA damage control.
Of interest is to address how ATF2 could contribute to ATM activation. Two of the possibilities currently considered relate to ATF2 being part of an upstream signal for ATM or for ATF2 ability to be part of mechanism that serves to maintain active ATM at IRIF. The link between ATF2 and chromatin organization is supported by independent studies from yeast and from mammalian systems. ATF2 associates with TIP49b (Cho et al., 2001), which is part of the TIP60 histone acetylase complex implicated in DNA repair and chromatin organization (Kanemaki et al., 1999; Ikura et al., 2000). Further, binding of TIP60 to phosphorylated H2AX was recently shown to be an important step in subsequent modifications that are part of the DNA damage response (Morrison et al., 2004). Atf1 and pcr1 (pombe homologs of ATF2) were shown to contribute to deacetylation of certain lysines on histone H3 and H4, a prerequisite for heterochromatin assembly (Jia et al., 2004; Kim et al., 2004). The latter changes are consistent with the notion that activation of ATM could be induced upon changes in chromatin organization (Bakkenist and Kastan, 2003). At this point, we equally entertain the second possibility, which would position ATF2 as part of mechanism to maintain active ATM at the damaged sites. Preliminary results revealed that for its ability to activate ATM, ATF2 needs to be phosphorylated on residues 490 and 498, but not on aa 69 and 71 (data not shown). Thus, either initial increase in ATM activity would suffice to promote ATF2 contribution to DNA damage response, and in turn to maintain ATM active at the sites of DSB, or, other PIKK may mediate such activation, which would consequently promote activation of ATM. Regardless of the initial signal, such changes are expected to result in maintaining active ATM at the site of DSB.
Upon its phosphorylation by ATM, ATF2 is recruited to DSB repair foci as one of the immediate early events (3 min). While ATF2 colocalizes with components of the MRN complex and γ-H2AX, it also affects recruitment of Mre11 and Nbs1 into repair foci. Of interest is that ATF2 did not affect localization of 53BP1 into repair foci, suggesting that the latter are subject to a different regulation, somewhat similar to the parallel interacting pathways shown for 53BP1 and Mdc1/NFBD1 in ATM activation (Mochan et al., 2003).
Importantly, the contribution of ATF2 to the DNA damage response is mediated by the transcriptionally inactive form of the protein. This conclusion is supported by the following observations: (1) transcriptionally inactive ATF2 (69/71 mutant) is recruited to IRIF as efficiently as wt ATF2; (2) transcriptionally inactive ATF2 mediates the IR-induced S phase checkpoint as well as wt ATF2; (3) MEFs of ATF2 mutant mice in which the ATF2 DNA binding domain and part of the dimerization domains were deleted exhibit physiologic localization of this transcriptionally dead form of ATF2 into IRIF; (4) p38/JNK inhibitors do not interrupt localization of ATF2 into IRIF; (5) ATF2 is found on promoters of RAD50 and ATM (4 hr after exposure to genotoxic stress) without altering their transcription (Hayakawa et al., 2004); and lastly (6) ATF2 localization into repair foci does not require its primary transcriptional heterodimeric partner c-Jun, as evident from analysis in c-Jun−/− cells. The latter is of further interest in light of the report that c-Jun is also localized into DSB repair foci (MacLaren et al., 2004). Since c-Jun was not shown to be associated with, or phosphorylated by, ATM, its localization in IRIF may be ATF2 dependent.
In all, ATF2 appears to play distinct functions in the cell cycle and the DNA damage response. First is its contribution to the cell cycle under nonstressed conditions. Transcriptional activity of ATF2 is necessary for maintaining physiologic cell cycle control under normal nonstressed growth conditions, probably through its established effects on cyclin A and cyclin D (Beier et al., 1999; Shimizu et al., 1998). Second is the phosphorylation of ATF2 by ATM required for its recruitment to IRIF, as for its recruitment of MRN components and for its role in IR-induced checkpoint control. For this function, ATF2 does not require its transcriptional activities. Third is ATF2's contribution to ATM activation, which is likely to contribute to maintenance of active ATM at sites of DNA damage. These observations raise the possibility that the balance between transcriptionally active and nonactive ATF2 (i.e., phosphorylated on the 69 and 71 sites versus the aa 490 and 498 sites) may affect the cell's ability to respond to DNA damage by means of altered RDS and radioresistance. These findings offer insight into our understanding of the immediate DNA damage response and into the relationship between transcription and DNA damage control. In as much, the present study establishes a paradigm for a function of a transcription factor in the DNA damage response that is independent of its transcriptional activities.
Experimental Procedures
Cells
Cultures of HeLa, 293, IMR90, GM00637, and A-T (GM05849) cells were obtained from Coriell repository or ATCC and maintained according to the supplier's recommendations. NBS-ILB1 cells (Kraakman-van der Zwet et al., 1999) were kindly obtained from T. Halazonetis; MeWo, LU1205, and WM793 melanoma cells were kindly provided by M. Herlyn; and the A-T cells (AT22IJE-T) as well as those that were reconstituted for ATM were obtained from Y. Shiloh (Ziv et al., 1997). ATF2 mutant mice were generated by insertion of loxP sites into genomic sequences flanking exons 8 and 9 of the ATF2 gene (encoding the whole DNA binding and most of the leucine zipper domain) and induction of recombination by transiently expressing Cre recombinase in ES cells (W.B., unpublished data). Primary mouse embryonic fibroblasts (MEFs) were generated from E13.5 embryos derived from crossing heterozygous animals carrying a germline allele of the mutant ATF2 gene.
Expression Vectors
ATF2 wt and mutant forms on aa 69, 71, 490, and 498 (generated with the aid of a QuikChange site-directed mutagenesis kit, Stratagene) were cloned into BamHI and NotI sites within the mammalian expression vector pEF HA or bacterial expression vector pGEX-4T. Purification of GST-ATF2 was performed under standard conditions. ATM constructs were previously described (Bakkenist and Kastan, 2003).
Kinase Reactions
In vitro kinase assays were performed using G361, 293T, or U2OS cells transfected with either FLAG-ATM wt or FLAG-ATM-KD (10 μg). Bacterially expressed and purified GST-ATF2 (full length or the spliced form containing aa 1–48 and aa 310–505) or GST p53 1–80 aa were incubated with immunopurified endogenous or exogenous ATM coupled to protein G beads in the presence of kinase buffer (30 μl) containing 10 μCi of [γ32P]ATP. The reaction mixtures were incubated at 30°C for 20 min before separation on SDS-PAGE, electroblotting, and analysis on a phosphorimager.
Phosphoantibodies to ATF2 Amino Acids 490 and 498
Antibodies to phosphorylated aa 490 and 498 on ATF2 antibody were produced in rabbits immunized with keyhole-limpet hemocyanin-conjugated phosphopeptide (TEPALpSQIVM and APSpSQSQPSG), derived from aa 485–494 and aa 495–502 of ATF2, respectively. The phosphospecific antibodies were affinity purified (Phosphosolutions).
Immunostaining
Cells were plated on cover slips and irradiated, fixed at indicated time points, and processed as described (Maser et al., 1997; Carney et al., 1998). Antibodies used were monoclonal or polyclonal γ-H2AX (1:500), pSmc1, Smc1 (Upstate Biotechnology), Mre11, Nbs, Rad50 (1:500), ATM, 53BP1, Chk2 (Genetex Inc.), ATF2 (Santa Cruz), p-ATF2 (1:500; Phosphosolutions), ATM-pSer1981 (Rockland Immunochemicals), Mdc1/NFBD1 (Bethyl Labs) and pChk1-S317, pChk2-T68 (Cell Signaling).
RNA Interference
For RNAi of ATF2 expression, the p-Super vector system that directs synthesis of siRNAs in mammalian cells was used (Brummelkamp et al., 2002). The two targets chosen for RNAi of ATF2 were as follows: 5#UTR192 TAAAGCTCATGGCCCACCA210 (pRS192ut) and within the coding sequence 1207AATGAAGTGGCACAGCTGA1225 (pRS1207) of the human ATF2 gene (National Center for Biotechnology Information [NCBI] accession number NM_001880).
Reconstitution experiments were carried out by transfection of ATF2 plasmids (1 μg) with lipofectamine in MeWo cells that were infected (48 hr earlier) with ATF2 RNAi.
Cell Cycle Checkpoint Analysis
Cells (293 or MeWo) were infected with RNAi and 72 hr later were labeled (24 hr) with 10 nCi/ml of [methyl-14C]-Thymidine (Perkin Elmer) followed by incubation (8 hr) with nonradioactive medium. The cells were than transfected and 48 hr later were irradiated (0, 5, or 15 Gy) using a 137Cs source (dose rate: 5.1 Gy/min). 45 min after irradiation cells were pulse-labeled (2.5 μCi/ml of [methyl-3H]Thymidine; Perkin Elmer) for 15 min. Inhibition of DNA synthesis was measured as described (Lim et al., 2000). The ratios obtained for 3H (counts/min) over 14C (counts/min) (3H/14C ratio) were corrected for channel crossover. The ratio of DNA synthesis after exposure to ionizing radiation was calculated as 3H/14C ratio in irradiated over the 3H/14C ratio in unirradiated cells.
For FACS analysis cells that were about 50% confluent were treated with thymidine (2 mM in DMEM and FBS) for 16 hr. Cells were washed with PBS three times and incubated for 8 hr in DMEM and FBS in the absence of excess thymidine. Cells were then treated again with thymidine (2 mM) in DMEM and FBS (16 hr) to result in cells that were arrested at the G1/S phase boundary of the cell cycle. Once synchronized in G1/S phase, cells were washed with PBS, thereby allowing their release into the cell cycle. Cells were than harvested at various time points after G1/S phase, which corresponded to G2 and M phase as determined by flow cytometry analysis.
Colony-Forming Assay
U2OS, or early passage A-T cells were infected with control RNAi or ATF2 RNAi . 72 hr later cells were resuspended to reach same density (5 × 104 cells/ml), irradiated, and subsequently plated in triplicates within the indicated range of cell densities. Cells were monitored for colony formation for 12 days (18 days for A-T cells) at 37°C. Colonies (containing 50 or more cells) were stained with 2% crystal violet, 50% ethanol, and counted. Each point represents a triplicate sample in experiments reproduced twice.
CHIP Assays
Wt and mutant MEFs were treated with anisomycin (25 μg/ml for 30 min) and fixed in formaldehyde (1%). Whole cell extracts were sonicated and immunoprecipitated (IP) with ATF2 antibodies C19 (Santa Cruz Biotechnology) or with IgG control antibodies. Afterward, reverse crosslinking DNA was purified using a PCR Purification Kit (Qiagen) and amplified by PCR (35 cycles) using primers GCGAGGAACGCAGGACGCGCCGTG (5′) and GCCCTCGCGTTGGCAGGGAGCCCG (3′) for ATF3, CTCCTCTGCGCAGGCGCGTCCTC (5′) and GTAGAGCCCAGGAGCCGCGAGCTG (3′) for Cyclin A (CcnA) promoter sequences containing ATF binding sites, and GGGAAGCCCATCACCATCTTC (5′) and CACCAGTAGACTCCACGACATACTCA (3′) for the GAPDH control sequence. Control PCR reactions were carried out using whole cell extracts (WCE) as template DNA.
Supplementary Material
Acknowledgments
We thank Barry Rosenstein for assistance with IR settings, Hans Snoeck for advice regarding FACS analysis, Michael Kastan for ATM expression vectors, Toru Ouchi for antibodies, Ron Wisdom for c-Jun−/− cells, M. Herlyn for melanoma cells, and John Petrini and Matthew O'Connell for reagents and most helpful advice. We also thank members of the Ronai Lab and Rolf Jessberger, Tanya Paull, and Bob Abraham for discussions. Support from National Institutes of Health (NIH) grant (CA 51995 to Z.R.) is gratefully acknowledged.
Footnotes
Supplemental Data
Supplemental Data include five additional figures and can be found with this article online at http://www.molecule.org/cgi/content/full/18/5/577/DC1/.
References
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Beier F, Lee RJ, Taylor AC, Pestell RG, LuValle P. Identification of the cyclin D1 gene as a target of activating transcription factor 2 in chondrocytes. Proc. Natl. Acad. Sci. USA. 1999;96:1433–1438. doi: 10.1073/pnas.96.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoumik A, Huang TG, Ivanov V, Gangi L, Qiao RF, Woo SL, Chen SH, Ronai Z. An ATF2-derived peptide sensitizes melanomas to apoptosis and inhibits their growth and metastasis. J. Clin. Invest. 2002;110:643–650. doi: 10.1172/JCI16081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Chen YR, Meyer CF, Tan TH. Persistent activation of c-Jun N-terminal kinase 1 (JNK1) in gamma radiation-induced apoptosis. J. Biol. Chem. 1996;271:631–634. doi: 10.1074/jbc.271.2.631. [DOI] [PubMed] [Google Scholar]
- Cho SG, Bhoumik A, Broday L, Ivanov V, Rosenstein B, Ronai Z. TIP49b, a regulator of activating transcription factor 2 response to stress and DNA damage. Mol. Cell. Biol. 2001;21:8398–8413. doi: 10.1128/MCB.21.24.8398-8413.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Paull T, Gottesman M, Gautier J. Mre11 assembles linear DNA fragments into DNA damage signaling complexes. PLoS Biol. 2004;2:e110. doi: 10.1371/journal.pbio.0020110. 10.1371/journal.pbio.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- Falvo JV, Parekh BS, Lin CH, Fraenkel E, Maniatis T. Assembly of a functional beta interferon enhanceosome is dependent on ATF-2-c-jun heterodimer orientation. Mol. Cell. Biol. 2000;20:4814–4825. doi: 10.1128/mcb.20.13.4814-4825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D'Amours D, Rahman D, Pappin D, Bartek J, Jackson SP. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- Hayakawa J, Depatie C, Ohmichi M, Mercola D. The activation of c-Jun NH2-terminal kinase (JNK) by DNA-damaging agents serves to promote drug resistance via activating transcription factor 2 (ATF2)-dependent enhanced DNA repair. J. Biol. Chem. 2003;278:20582–20592. doi: 10.1074/jbc.M210992200. [DOI] [PubMed] [Google Scholar]
- Hayakawa J, Mittal S, Wang Y, Korkmaz KS, Adamson E, English C, Omichi M, McClelland M, Mercola D. Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation following genotoxic stress. Mol. Cell. 2004;16:521–535. doi: 10.1016/j.molcel.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- Kanemaki M, Kurokawa Y, Matsu-ura T, Makino Y, Masani A, Okazaki K, Morishita T, Tamura TA. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J. Biol. Chem. 1999;274:22437–22444. doi: 10.1074/jbc.274.32.22437. [DOI] [PubMed] [Google Scholar]
- Kim HS, Choi ES, Shin JA, Jang YK, Park SD. Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the MAP kinase pathway and a histone deacetylase Clr6. J. Biol. Chem. 2004;279:42850–42859. doi: 10.1074/jbc.M407259200. [DOI] [PubMed] [Google Scholar]
- Kim ST, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 2002;16:560–570. doi: 10.1101/gad.970602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman-van der Zwet M, Overkamp WJ, Friedl AA, Klein B, Verhaegh GW, Jaspers NG, Midro AT, Eckardt-Schupp F, Lohman PH, Zdzienicka MZ. Immortalization and characterization of Nijmegen Breakage syndrome fibroblasts. Mutat. Res. 1999;434:17–27. doi: 10.1016/s0921-8777(99)00009-9. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- Liu F, Green MR. A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1a protein. Cell. 1990;61:1217–1224. doi: 10.1016/0092-8674(90)90686-9. [DOI] [PubMed] [Google Scholar]
- MacLaren A, Black EJ, Clark W, Gillespie DA. c-Jun-deficient cells undergo premature senescence as a result of spontaneous DNA damage accumulation. Mol. Cell. Biol. 2004;24:9006–9018. doi: 10.1128/MCB.24.20.9006-9018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, Moas M, Buschmann T, Ronai Z, Shiloh Y, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RS, Monsen KJ, Nelms BE, Petrini JH. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson C, Menaa F, Pinon-Lataillade G, Frobert Y, Radicella JP, Angulo JF. Identification of KIN (KIN17), a human gene encoding a nuclear DNA-binding protein, as a novel component of the TP53-independent response to ionizing radiation. Radiat. Res. 2001;156:535–544. doi: 10.1667/0033-7587(2001)156[0535:iokkah]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- Mochan TA, Venere M, DiTullio RA, Jr., Halazonetis TD. 53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage. Cancer Res. 2003;63:8586–8591. [PubMed] [Google Scholar]
- Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. INO80 and gamma-H2AX Interaction Links ATP-Dependent Chromatin Remodeling to DNA Damage Repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–95. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Ehret H, Paro R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol. Cell. 2004;16:641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Steiner WW, Schreckhise RW, Smith GR. Meiotic DNA breaks at the S. pombe recombination hot spot M26. Mol. Cell. 2002;9:847–855. doi: 10.1016/s1097-2765(02)00489-6. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Nomura Y, Suzuki H, Ichikawa E, Takeuchi A, Suzuki M, Nakamura T, Nakajima T, Oda K. Activation of the rat cyclin A promoter by ATF2 and Jun family members and its suppression by ATF4. Exp. Cell Res. 1998;239:93–103. doi: 10.1006/excr.1997.3884. [DOI] [PubMed] [Google Scholar]
- Tsai EY, Jain J, Pesavento PA, Rao A, Goldfeld AE. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol. Cell. Biol. 1996;16:459–467. doi: 10.1128/mcb.16.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam H, Duyndam M, Rottier R, Bosch A, de Vries-Smits L, Herrlich P, Zantema A, Angel P, van der Eb AJ. Heterodimer formation of c-Jun and ATF-2 is responsible for induction of c-jun by the 243 amino acid adenovirus E1A protein. EMBO J. 1993;12:479–487. doi: 10.1002/j.1460-2075.1993.tb05680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- Ziv Y, Bar-Shira A, Pecker I, Russell P, Jorgensen TJ, Tsarfati I, Shiloh Y. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene. 1997;15:159–167. doi: 10.1038/sj.onc.1201319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.