Summary
The trehalose biosynthesis pathway is critical for virulence in human and plant fungal pathogens. In this study, we tested the hypothesis that trehalose-6-phosphate phosphatase (T6PP) is required for Aspergillus fumigatus virulence. A mutant of the A. fumigatus T6PP, OrlA, displayed severe morphological defects related to asexual reproduction when grown on glucose (1%) minimal media. These defects could be rescued by addition of osmotic stabilizers, reduction in incubation temperature, or increase in glucose levels (>4%). Subsequent examination of the mutant with cell wall perturbing agents revealed a link between cell wall biosynthesis and trehalose-6-phosphate (T6P) levels. As expected, high levels of T6P accumulated in the absence of OrlA resulting in depletion of free inorganic phosphate (Pi) and inhibition of hexokinase activity. Surprisingly, trehalose production persisted in the absence of OrlA. Further analyses revealed that A. fumigatus contains two trehalose phosphorylases that may be responsible for trehalose production in the absence of OrlA. Despite a normal growth rate under in vitro growth conditions, the orlA mutant was virtually avirulent in two distinct murine models of invasive pulmonary aspergillosis. Our results suggest that further study of this pathway will lead to new insights into regulation of fungal cell wall biosynthesis and virulence.
Keywords: Aspergillus fumigatus, Trehalose, virulence, OrlA, cell wall, trehalose phosphorylase
Introduction
Trehalose (α-D-glucopyranosyle α-D-glucopyranoside) is synthesized and stored in a wide variety of organisms such as bacteria, fungi, plants, insects, and some invertebrates (Crowe et al., 1992, Thevelein, 1984, Singer & Lindquist, 1998). Unlike these other organisms, humans are unable to produce trehalose but can utilize trehalose through a trehalase enzyme (Gancedo & Flores, 2004, Ishihara et al., 2000, Murray et al., 2000). Importantly, fungal mutants of enzymes at different key steps in the trehalose biosynthesis pathway are required for human and plant fungal pathogenesis (Zaragoza et al., 2002, Zaragoza et al., 1998, Van Dijck et al., 2002, Petzold et al., 2006, Foster et al., 2003). Thus, the lack of human orthologs and the importance of trehalose biosynthesis in human and plant fungal pathogenesis has led to the hypothesis that this pathway is a promising antifungal drug target.
Mechanisms of trehalose biosynthesis in fungi have been extensively studied in the model yeast Saccharomyces cerevisiae. In S. cerevisiae, trehalose is synthesized in a two-step reaction (TPS/TPP) that is carried out by a four-subunit protein complex (Elliott et al., 1996, Vuorio et al., 1993, Gancedo & Flores, 2004, Cabib & Leloir, 1958). A glucosyl residue from uridine-diphospho-glucose (UDP-glucose) is transferred to glucose-6-phosphate (G6P) by trehalose-6-phosphate synthase (Tps1p) resulting in production of trehalose-6-phosphate (T6P). T6P is subsequently dephosphorylated by trehalose-6-phosphate phosphatase (Tps2p) to obtain trehalose (reviewed in (Gancedo & Flores, 2004). Additionally, there are two regulatory subunits in the protein complex (Tsl1p and Tps3p) that participate in these reactions by interacting with the Tps1p and Tps2p complex (Bell et al., 1998, Reinders et al., 1997). All subunits are required for optimal enzymatic activity.
Five routes of trehalose biosynthesis have now been described in various organisms (Avonce et al., 2006). With regard to the pathogenic fungi, it seems that the TPS/TPP pathway is the primary route utilized, though mechanistic details have yet to be carried out to the extent that they have in S. cerevisiae. Some fungi have been reported to utilize the trehalose phosphorylase (TP) pathway for trehalose biosynthesis. For example, the basidiomycete mushrooms Grifola frendosa and Pleurotus ostreatus use TP to produce trehalose from G1P and glucose (Saito et al., 1998a, Kitamoto, 2000). TP activity also appears to play a key role in trehalose metabolism in Agaricus bisporus due to a lack of trehalose synthase and trehalase activity (Wannet et al., 1998). Importantly, this pathway of trehalose biosynthesis is negatively regulated by free inorganic phosphate (Pi) (Wannet et al., 1998).
Trehalose has been primarily thought to serve as a storage carbohydrate in fungi due to the presence of a trehalase enzyme, which hydrolyzes trehalose into two molecules of glucose (Nwaka et al., 1995, Thevelein, 1984). Trehalose also functions as an important stress protectant against a variety of stress conditions including dehydration, heat and cold shock, oxidative stress, and antimicrobial drug treatments (Thevelein, 1984, Crowe et al., 1992, Elbein et al., 2003, Sano et al., 1999, Hottiger et al., 1994, Elliott et al., 1996, Kandror et al., 2002, Cao et al., 2008). Moreover, trehalose may be an important carbohydrate source for reproductive propagules of fungi. Trehalose is abundant in mature ascospores in yeast, and Aspergillus nidulans conidia have high concentrations of trehalose that is rapidly degraded upon induction of conidial germination (Kane & Roth, 1974, Fillinger et al., 2001). Recently, a regulator of conidiogenesis, VosA, was identified in A. nidulans and also found to regulate genes involved in trehalose biosynthesis in conidia (Ni & Yu, 2007). Perhaps most intriguingly, there is a growing understanding of the role that genes, proteins, and intermediate metabolites involved in trehalose biosynthesis are playing as key regulators of basic carbon metabolism in fungi (Wilson et al., 2007, Thevelein & Hohmann, 1995, Elliott et al., 1996, Blazquez et al., 1993).
With regard to fungal pathogenesis, the trehalose pathway has been characterized in three human pathogenic yeast (Cryptococcus neoformans, Cryptococcus gattii, and Candida albicans) and one filamentous fungal plant pathogen, Magnaporthe grisea (Zaragoza et al., 2002, Zaragoza et al., 1998, Wilson et al., 2007, Van Dijck et al., 2002, Petzold et al., 2006, Ngamskulrungroj et al., 2009, Foster et al., 2003). However, studies on the trehalose pathway of the most common cause of human invasive mold infections, Aspergillus fumigatus, have not been undertaken. In this study, we have begun a comprehensive examination of the trehalose biosynthesis pathway in this important human pathogenic fungus by identifying and functionally characterizing the T6P phosphatase OrlA, (S. cerevisiae Tps2p ortholog). Our results confirm previous suggestions that the trehalose biosynthesis pathway is a promising antifungal drug target, and also suggest a critical role for this pathway in regulating key aspects of A. fumigatus biology including glycolytic flux, cell wall integrity, and virulence.
Results
Construction of Trehalose-6-Phosphate Phosphatase mutant and reconstituted strains
To identify the putative trehalose-6-phosphate phosphatase gene involved in trehalose biosynthesis in A. fumigatus, we searched the genome sequence of A. fumigatus strain A1163 with the A. nidulans OrlA (AN3441) and S. cerevisiae Tps2p protein sequences using BLAST algorithms. Searches with both OrlA and Tps2p revealed a putative single ortholog in A. fumigatus, AFUB_043350, which we subsequently designated OrlA consistent with previous nomenclature in A. nidulans (Borgia et al., 1996). OrlA protein sequences from A. nidulans and A. fumigatus displayed 72% amino acid identity and 82% protein sequence similarity while Sc Tps2p and Af OrlA displayed 44% amino acid identity and 59% protein sequence similarity. Analysis of the Af OrlA protein sequence with InterPro revealed 3 highly conserved domains: a glycosyl transferase family 20 domain (amino acids 281-663), a trehalose phosphatase domain (amino acids 695-931), and a HAD-superfamily hydrolase subfamily IIB domain (amino acids 693-905). These results suggest that AFUB_043350 encodes the A. fumigatus trehalose-6-phosphate phosphatase, OrlA.
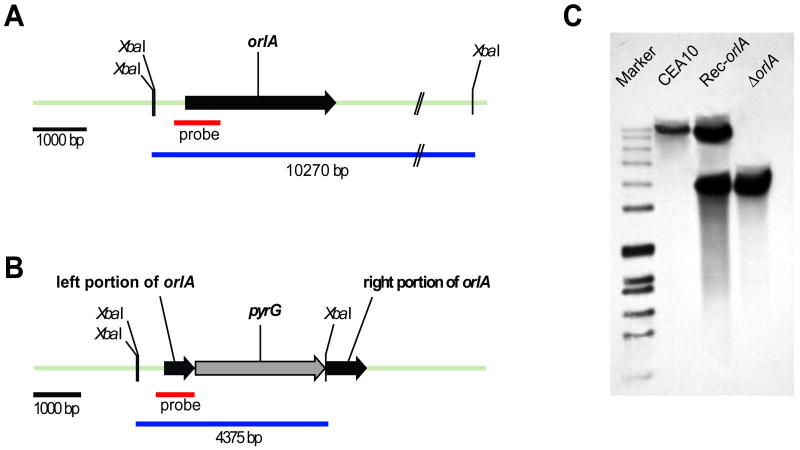
In order to determine the role of OrlA in A. fumigatus biology, we generated a null mutant of orlA by disruption of the coding sequence in the uracil/uridine auxotrophic A. fumigatus strain (CEA17) with the selectable marker pyrG from Aspergillus parasiticus as we have previously described (Willger et al., 2008) (Fig. 1). To confirm that phenotypes of the orlA mutant were solely due to loss of OrlA, we generated a reconstituted strain by ectopic reintroduction of the wild-type orlA gene into the orlA mutant strain (Rec-orlA strain). All strains were rigorously confirmed by both PCR and Southern blot analyses (Fig. 1, and data not shown). All examined strains produced the expected hybridization signals when XbaI digested genomic DNA was hybridized with a DNA probe of the 5′ (upstream) orlA ORF flanking sequence (Fig. 1A-C). The orlA mutant has a single band at 4,375 bp as expected for a clean disruption of the orlA open reading frame by the A. parasiticus pyrG marker (Fig. 1B-C). Subsequently, the reconstituted strain (Rec-orlA) contained hybridization bands corresponding to both the orlA mutant and wild-type alleles indicating successful ectopic reconstitution of the orlA gene in the orlA mutant background (Fig. 1A, 1B, and 1C). mRNA abundance of the orlA gene was determined in all strains by qRT-PCR and expression of orlA was equivalent between the wild-type and Rec-orlA strains while absent in the orlA mutant (data not shown).
Fig. 1.
Generation of strains used in this study. (A) Locus of orlA gene in wild-type strain (CEA10) and (B) locus of orlA after successful introduction of gene replacement construct (C) Southern blot analysis of wild-type, reconstituted strain (Rec-orlA) and ΔorlA strains. The XbaI digested genomic DNA of all strains were separated on a 1% agarose gel, blotted, and hybridized with a 900 bp genomic DNA probe from the orlA upstream sequence. The expected fragment sizes of the orlA locus in the wild-type and orlA mutant were observed and are 10,270 bp and 4,375 bp respectively. The reconstituted strain (Rec-orlA) contains the expected hybridization signals for a single ectopic insertion of the wild-type allele of orlA (top) and maintenance of the disrupted orlA locus (bottom).
Abolished asexual reproduction and abnormal hyphal morphology of the orlA mutant
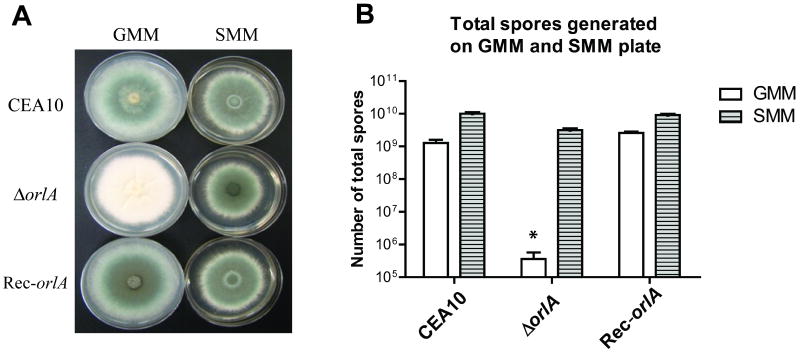
Although the orlA mutant has a wild-type like hyphal growth rate on glucose minimal media (GMM) at 37°C, we observed an almost complete loss of conidiation in the absence of orlA (Fig.2A). When quantified, total conidia production of the orlA mutant on GMM at 37°C was significantly less than the wild-type and reconstituted strains (P= 0.02 and <0.01 respectively) (Fig. 2B). The conidiation defect could be partially rescued by growth at temperatures at or below 30°C and fully rescued on media containing an osmotic stabilizer (1.2 M sorbitol or 1% glycerol) (Fig. 2A, 2B and data not shown). We confirmed that temperature stress is a factor in the conidiation defect by allowing growth of the mutant at 37°C for 2 days followed by a subsequent shift to lower temperatures (25°C and 30°C). We found that the orlA mutant was able to produce conidia on new tissue generated after the temperature shift to lower temperatures (data not shown).
Fig. 2.
Colony morphology and conidia production in the absence of OrlA. (A) Normal growth rate but abolished asexual conidiation of the orlA mutant is observed on glucose minimal media (GMM) at 37°C. This defect could be recovered on 1.2M sorbitol minimal media (SMM) at 37°C. (B) Conidia production of wild-type, orlA mutant and orlA reconstituted strain on two different media GMM and SMM incubated at 37°C for 4 days prior to harvesting and counting. Data is the mean and standard deviation of three biological replicates. *Total conidia production of the mutant cultured on GMM is significantly less than the wild-type and reconstituted strains (P = 0.020 and 0.003 respectively).
Besides the effect of temperature on asexual reproduction, a temperature sensitive phenotype was observed at temperatures above 42°C. When subjected to heat stress (45°C) the orlA mutant displayed a significant decrease in growth in comparison with the wild-type and reconstituted strain when grown on both GMM and SMM media (data not shown). All strains grew better under high temperature stress when the osmotic stabilizing agent sorbitol was added to the media indicating that significant cell wall stress is placed on the fungus during high temperature growth. In the orlA mutant of A. nidulans, thermotolerance was completely suppressed as conidia were unable to germinate at high temperature (42°C) due to cell lysis (Borgia et al., 1996, Borgia & Dodge, 1992). However, conidia of the orlA mutant of A. fumigatus were able to survive and germinate at 45°C and we did not observe any cell lysis at these elevated temperatures. Taken together, these results strongly suggest that loss of trehalose-6-phosphate phosphatase activity severely impacted the ability of A. fumigatus to undergo asexual reproduction and tolerate temperature stress. We hypothesized that the ability of temperature and osmotic stabilizers to rescue these phenotypes suggested a possible defect in the cell wall of the orlA mutant.
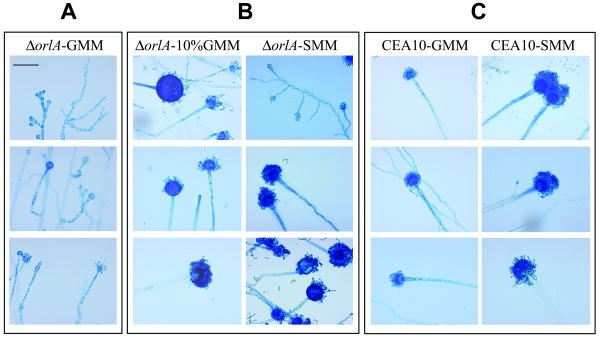
To test our hypothesis we first examined the orlA mutant hyphal morphology using traditional slide cultures and lactophenol cotton blue stains (Fig. 3). Strikingly, asexual reproduction was abolished in the absence of orlA as we found only conidiophores and vesicles produced on GMM with limited and abnormal phialide and conidia production (Fig. 3A). Abnormal hyphal morphology was also apparent when the orlA mutant was grown on GMM at 37°C. Mutant hyphae are clearly dysmorphic with shorter length compartments between septa and truncated hyphal tips with knobby, abnormal branching (Fig. 3A). When placed on high GMM (>4% glucose) or osmotic stabilizing media (Fig. 3B), these abnormal morphological phenotypes largely, but not completely, disappeared consistent with these culture conditions being able to restore conidiation. Interestingly, the ability of glucose to restore the conidiation defect of the mutant was dose dependent as restoration of conidiation started at around a concentration of 4% glucose and increased proportionately to the amount of glucose in the media (data not shown).
Fig. 3.
Hyphal morphology of the orlA mutant is altered. (A) Slide cultures and lactophenol cotton blue staining reveal dysmorphic hyphae and malformed asexual reproductive structures which formed vesicles lacking phialides in the orlA mutant grown on GMM at 37°C. (B) The ability to form normal hyphae, asexual reproductive structures, and conidia is restored in the orlA mutant grown on GMM containing 10% glucose or minimal media containing 1.2 M sorbitol (SMM) at 37°C. (C) Wild-type CEA10 generates normal hyphae and asexual reproductive structures when grown on GMM and SMM at 37°C. All representative pictures were captured under light microscopy at 400× magnification (reference bar = 50 μm).
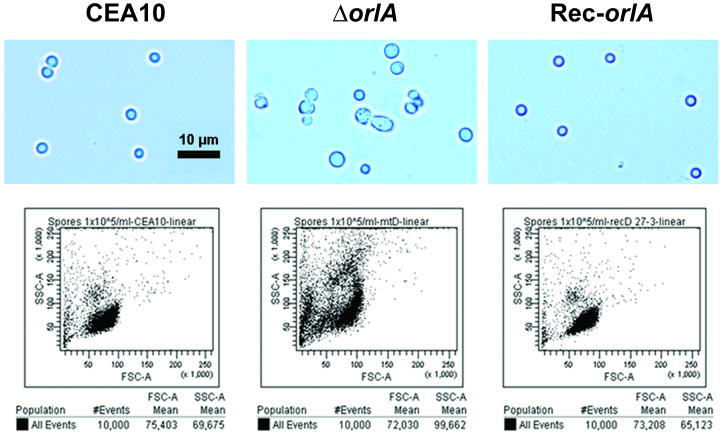
However, we observed that restoration of conidia production on high glucose media (10% glucose) or SMM resulted in conidia with abnormal morphology (Fig. 3B). The morphology of restored conidia of the orlA mutant grown on SMM media at 37°C are aberrant in shape and size as observed using light microscopy and confirmed by forward and side scatter measurements of conidia size and density using FACS analysis (Fig. 4).
Fig. 4.
Flow cytometry analysis reveals a heterogeneous population of orlA mutant conidia from SMM when cultured at 37°C while wild-type (CEA10) and reconstituted strains (Rec-orlA) display a homogenous population of conidia. FACS analysis via forward and side scatter parameters of equal amounts (10,000 events) of conidia. Differences in conidia size and shape can be observed via light microscopic observation at 400× magnification. A reference bar is 10 μm length.
Based on the FACS analysis, it is clear that a wide range of conidial morphologies is produced in the absence of orlA. This led us to question the viability and germination ability of the orlA mutant conidia. Conidia viability of all strains grown on SMM after incubation at 37°C did not differ after 10 days (all strains displayed a 60-70% survival rate). However, after one month of incubation the orlA mutant conidia displayed an approximate 45% decrease in survival compared to day 3, whereas approximately 20% and 21% decreases in viability were observed in the wild-type and reconstituted strains respectively (data not shown).
When the ability of orlA mutant conidia from SMM plates at 37°C were examined for their ability to germinate in liquid GMM, we observed similar germination rates for all three strains with a slight but statistically insignificant trend for increased germination rates in the absence of orlA (data not shown). Thus, although dysmorphic, orlA mutant conidia are viable and have virtual wild-type levels of germination. Taken together, these morphological analyses of the orlA mutant suggest that loss of trehalose-6-phosphate phosphatase activity results in a defect in the cell wall that affects the ability of the fungus to form normal hyphae, asexual reproduction structures, and conidia.
Loss of orlA results in increased sensitivity to cell wall perturbing agents
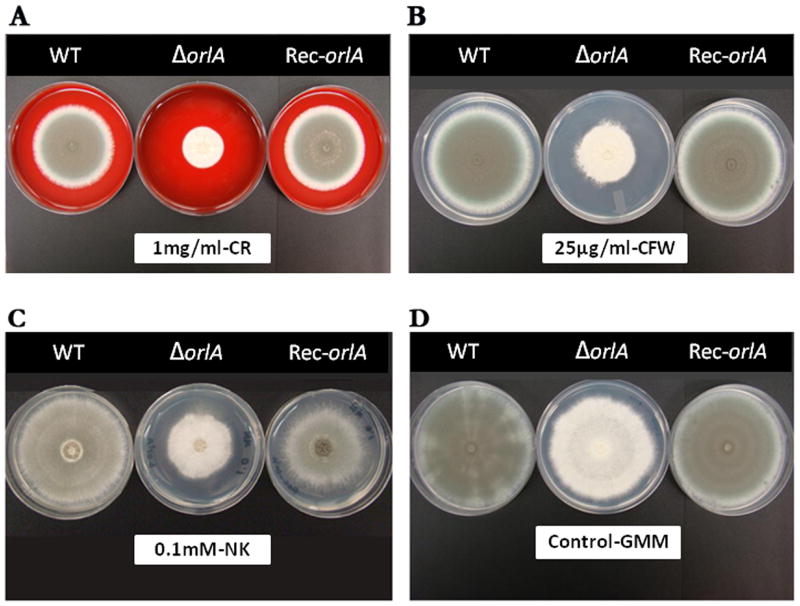
We next tested the hypothesis that the orlA mutant had a defect in the cell wall by utilizing several known cell wall perturbation agents. The mutant displayed wild-type levels of susceptibility to the echinocandin caspofungin, a specific β-1,3-glucan inhibitor, suggesting that β-1,3-glucan synthesis or content is not altered in the absence of OrlA. Susceptibility of the mutant to triazoles and amphotericin B was also tested and found to be the same as the wild-type and reconstituted strains (data not shown). Next, we tested the mutant's response to calcofluor white (CFW), congo red (CR), and Nikkomycin Z (NK). As observed in Figure 5, loss of orlA resulted in increased susceptibility to all three cell wall perturbing agents compared to the wild-type and reconstituted strains (Fig. 5A-C). The growth rate (radius growth/day) of the reconstituted strain, but not the orlA mutant, was similar to the wild-type on all cell wall perturbing agents.
Fig. 5.
The orlA mutant is sensitive to cell wall perturbing agents. Cell wall defects were observed in the orlA null mutant cultured on GMM at 37°C containing various cell wall inhibitors. (A) Congo Red (CR) 1 mg/ml. (B) Calcofluor White (CFW) 25 μg/ml. (C) Nikkomycin Z (NK) 0.1mM. (D) GMM media without inhibitors was used as a control that represents the normal growth of all strains incubated at 37°C for 3 days. Concentrations presented are the optimum concentrations used to determine the growth defect of the orlA mutant (ΔorlA) compared to the wild-type (CEA10) and reconstituted strains (Rec-orlA). The experiment was repeated in biological triplicates with identical results.
On CFW containing media, the growth rate of Rec-orlA (1.73±0.01) was comparable to CEA10 (1.65±0.03), while a decreased rate was observed in the orlA mutant (0.95±0.11). On NK, the orlA mutant (0.68±0.03) was again more susceptible than Rec-orlA (0.94±0.02) and CEA10 (1.09±0.03). Congo red had the strongest effect on the orlA mutant (Fig. 5A). Growth rates on CR containing media were similar in Rec-orlA (1.38±0.01) and CEA10 (1.27±0.03), but a drastic effect was observed on the orlA mutant (0.59±0.01). CR is thought to inhibit cell wall assembly enzymes that connect chitin to β-1,3-glucan and β-1,6-glucan. However, the mutant also displayed increased sensitivity to CFW and NK (Fig. 5B and 5C). CFW is an inhibitor of chitin polymer assembly while Nikkomycin Z is a chitin synthase inhibitor (Ram & Klis, 2006, Kaur et al., 2007, Brun et al., 2007). Taken together, these results suggest that β-1,3-glucan and β-1,6-glucan synthesis is likely unaffected, or at least not reduced, in the absence of OrlA. However, a significant defect either in the formation of the chitin-glucan matrix, which is critical for cell wall integrity, or actual chitin biosynthesis seems to be a consequence of OrlA loss.
We reasoned that the apparent cell wall defect in the orlA mutant could be due to alteration in the transcription of cell wall biosynthesis genes. To test this hypothesis, we examined the mRNA abundance of cell wall biosynthesis genes in the orlA mutant in comparison with the wild-type at two defined time points utilizing qRT-PCR (Table 1). 6.5 hours after conidial germination in liquid GMM at 37°C germ tubes had formed in both the orlA mutant and wild-type strains. mRNA abundance of all 7 chitin synthase genes at 6.5 hours was not significantly altered in the orlA mutant compared to the wild-type. After 24 hours of growth in liquid GMM at 37°C, when both strains had formed masses of mycelia, chitin synthase mRNA abundance generally increased in the orlA mutant strain in comparison with the wild-type. However, it is unclear if the observed differences are biologically relevant. As expected from caspofungin's lack of an effect on the orlA mutant, no difference was observed between the wild-type and orlA mutant in the expression of the β-1,3- glucan synthase fksA at either time point. The most apparent differences in mRNA abundance at both 6.5 hours and 24 hours were observed in the three α-1,3-glucan synthases agsA, agsB, and agsC. In particular, the largest change in mRNA abundance was observed for agsC, which was increased over 16.35 (±7.6) fold in the absence of orlA at 24 hours. mRNA abundance changes in the cell wall remodeling enzymes gelA, gelB, and gelC (1,3-β - glucanosyltransferase) were also observed at 24 hours in the absence of orlA. However, inconsistencies between the replicates with these 3 genes prevents definitive conclusions from being drawn about the effect of OrlA loss on the mRNA abundance of these important cell wall remodeling enzyme encoding genes. Overall, we hypothesize that the general observed increases in mRNA abundance of the cell wall biosynthesis genes in the absence of orlA likely reflects the occurrence of alternations in the cell wall composition that results in positive transcriptional feedback of the required biosynthesis genes. Yet, the relatively modest increases in mRNA abundance may also suggest that the observed alterations in the orlA mutant cell wall occur due to changes in non-transcriptional based mechanisms.
Table 1.
Quantitative Real-Time PCR analysis of cell wall biosynthesis genes in the orlA mutant and wild-type. mRNA abundance for each gene was normalized to β-tubulin in each strain and normalized mRNA abundance levels are from the orlA mutant relative to the wild-type strain at the respective time points (6.5 hours and 24 hours growth in glucose minimal media at 37°C). Data represents the average value of two biological replicates, each repeated in duplicate, and standard deviation.
| Gene | Predicted Function | Location | Fold Change | Fold Change |
|---|---|---|---|---|
| 6.5 hours | 24 hours | |||
| agsA | α-1,3-glucan synthase | Afu3g00910 | 0.25 (±1.9) | 4.75 (±1.6) |
| agsB | α-1,3-glucan synthase | Afu2g11270 | 2.65 (±0.2) | 1.45 (±0.1) |
| agsC | α-1,3-glucan synthase | Afu1g15440 | 3.95 (±0.2) | 16.35 (±7.6) |
| chsA | chitin synthase A | Afu2g01870 | -0.10 (±2.0) | 3.35 (±0.2) |
| chsB | chitin synthase B | Afu4g04180 | -0.20 (±1.8) | 2.20 (±0.1) |
| chsC | chitin synthase C | Afu5g00760 | 0.05 (±1.8) | 1.65 (±0.4) |
| chsD | chitin synthase D | Afu1g12600 | -0.15 (±2.5) | 1.30 (±0) |
| chsE | chitin synthase E | Afu2g13440 | 0.25 (±1.9) | 2.85 (±1.5) |
| chsF | chitin synthase F | Afu8g05630 | -1.95 (±5.3) | 1.45 (±0.6) |
| chsG | chitin synthase G | Afu3g14420 | -1.80 (±0.7) | 1.70 (±0.8) |
| fksA | β-1,3-glucan synthase catalytic subunit | Afu6g12400 | 2.70 (±2.3) | 1.65 (±0.8) |
| gelA | 1,3-β-glucanosyltransferase | Afu2g01170 | 1.35 (±0.2) | 8.10 (±3.4) |
| gelB | 1,3-β-glucanosyltransferase | Afu6g11390 | 8.20 (±10.0) | 9.45 (±11.1) |
| gelC | 1,3-β-glucanosyltransferase | Afu2g12850 | 1.30 (±5.1) | 5.65 (±6.6) |
Trehalose and Trehalose-6-phosphate production in the orlA mutant is altered
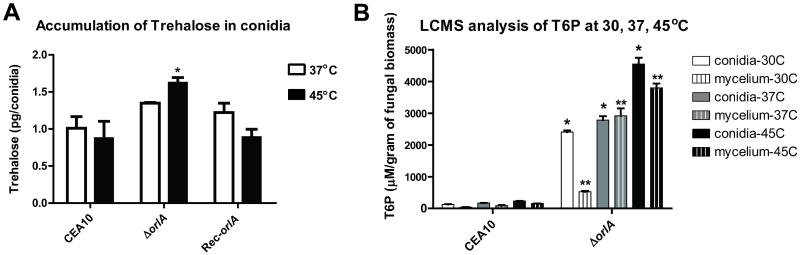
Based on knowledge of the S. cerevisiae trehalose biosynthesis pathway, the most likely hypothesis to explain the observed phenotypes of the orlA mutant is that loss of OrlA activity results in a significant increase in trehalose-6-phosphate (T6P) and subsequent loss of trehalose production. To test this hypothesis, we measured trehalose and T6P production in the wild-type, reconstituted, and orlA mutant strains. Surprisingly, we observed that orlA mutant conidia contained wild-type levels of trehalose when cultured at 37°C, and an approximate ∼2 fold increase in trehalose content when exposed to high temperature stress (P value < 0.05) (Fig. 6A). This unexpected result raises the question whether OrlA is the trehalose-6-phosphate phosphatase in A. fumigatus.
Fig. 6.
Production of trehalose and trehalose-6-phosphate (T6P). (A) Conidia for trehalose accumulation measurements were cultured at 37°C and 45°C. At 37°C, trehalose amounts found in the mutant were not significantly different from the wild-type and reconstituted strains; however at 45°C the mutant accumulated greater amounts of trehalose compared to the wild-type and reconstituted strains (*P =0.04 and 0.02 respectively. (B) T6P accumulation in conidia and mycelia were measured with LCMS from cell free culture extracts grown at 30°C, 37°C and 45°C. T6P concentration was back calculated from a T6P standard curve and normalized to the input weight of fungal biomass. In all conditions, T6P accumulated in conidia and mycelium of the orlA mutant and was significantly higher than the wild-type (CEA10) (*P<0.05 in conidia, **P<0.05 in mycelia).
However, liquid chromatography mass spectrometry (LCMS) analyses revealed that, as expected for a trehalose-6-phosphate phosphatase mutant, T6P accumulated in the orlA mutant to significant levels (2,400-4,700μM/gram conidia and 500-3,900 μM/gram mycelia) in both conidia and mycelia cultured at 30°C, 37°C, and 45°C (Fig. 6B) (Lunn et al., 2006). T6P levels were also found to increase with temperature suggesting that trehalose synthase activity encoded by TPS1 orthologs is increased with temperature stress. Taken together, these results suggest that an alternate mechanism of trehalose biosynthesis exists in A. fumigatus in the absence of OrlA. Though unexpected, this result is not unprecedented as a similar persistence of trehalose biosynthesis was found in the A. nidulans orlA mutant (Borgia et al., 1996) and a C. albicans tps2/tps2 mutant (Van Dijck et al., 2002).
A. fumigatus contains two trehalose phosphorylase encoding genes
Currently, the mechanism by which trehalose-6-phosphate phosphatase mutants in Aspergillus spp. and C. albicans generate trehalose is unknown. A preliminary microarray analysis of the orlA mutant revealed a significant increase in the mRNA abundance of a gene annotated as a trehalose synthase, CCG-9 (AFUB_062450) (data not shown). This gene is not part of the well-studied trehalose biosynthesis pathway in S. cerevisiae, but is part of a trehalose synthesizing enzyme family known as trehalose phosphorylases (TP) (Saito et al., 1998a, Saito et al., 1998b, Han et al., 2003, Eis & Nidetzky, 1999, Belocopitow & Marechal, 1970, Aisaka & Masuda, 1995, Aisaka et al., 1998). TPs are capable of producing trehalose from α or β-glucose-1-phosphate and have been found in a variety of organisms including bacteria, yeasts, green algae, and basidiomycete mushrooms (Saito et al., 1998a, Kitamoto, 2000). A. fumigatus contains two putative TP encoding genes, the aforementioned AFUB_062450 and another gene, AFUB_037080. A third gene with putative sequence similarity to TPs was found, but appears to be a pseudogene based on current genome annotation. The predicted TP protein sequences have 36% similarity and both contain a highly conserved glycosyltransferase GTB type superfamily protein domain. In filamentous fungi, a TP encoding gene has been partially characterized in Neurospora crassa and named CCG-9 (clock controlled gene 9) (Shinohara et al., 2002). AFUB_062450 has 32% sequence similarity while AFUB_037080 has 49% sequence similarity with N. crassa CCG-9.
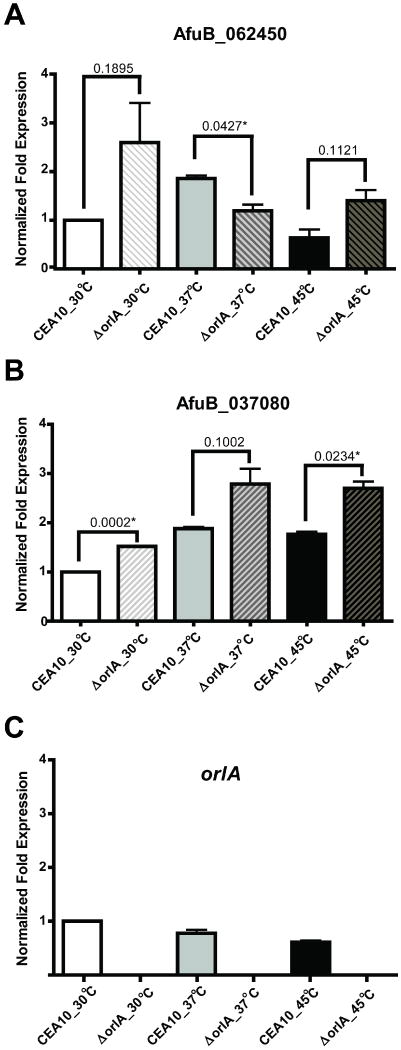
We hypothesize that the persistence of trehalose biosynthesis in the orlA mutant is due to TP activity. Consequently, we examined the mRNA abundance of the two putative TP genes in conidia of both the orlA mutant and wild-type harvested from cultures grown on SMM at 30°C, 37°C, and 45°C (conditions in which the mutant produces wild-type levels of trehalose). In comparison to wild-type conidia from cultures incubated at 30°C, conidia of both the wild-type and orlA mutant had mRNA abundance levels of AfuB_037080 with cultures from 30°C and 45°C having a significant increase in the orlA mutant (Fig. 7B). On the other hand, AFUB_062450 is the most abundant TP in the mutant conidia cultured at 30°C (Fig. 7A). In addition, AFUB_037080 mRNA was present at higher levels in the orlA mutant conidia under thermal stress conditions (37°C and 45°C) (Fig. 7B). mRNA abundance of orlA was essentially unchanged between the different temperature conditions, and no orlA mRNA was detected in the orlA mutant (Fig. 7C). These results suggest that increased TP mRNA levels could potentially explain the production of trehalose in the orlA mutant. Additional fungal genome analyses revealed putative TP orthologs in A. nidulans and other filamentous ascomycetes, but failed to identify a clear TP ortholog in C. albicans.
Fig. 7.
mRNA abundance of trehalose phosphorylase genes (A) AFUB_062450 (B) AFUB_037080 and (C) orlA was measured in conidia from the wild-type (CEA10) and orlA mutant. RNA samples were extracted from conidia cultured at 30°C, 37°C and 45°C. mRNA abundance was normalized to actin and expression values are relative to the CEA10 sample at low temperature (30°C). Results are the mean and standard deviation of two biological replicates and were calculated using the 2-ΔΔCt method (Livak & Schmittgen, 2001).
Potential activation of TP enzymes by depleted free Pi levels in the orlA mutant
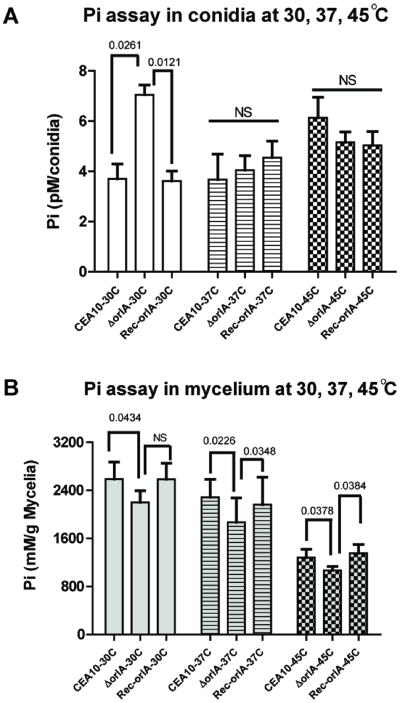
While the qRT-PCR data suggested slight increases in the mRNA levels of TP encoding genes in the orlA mutant, TP activity is also inhibited by high levels of free inorganic phosphate (Pi) (Wannet et al., 1998). We hypothesized that the accumulation of high T6P levels in the orlA mutant would result in sequestration of free Pi that could lead to induction of TP enzyme activity. To test this hypothesis, we measured levels of Pi in cell free extracts of the wild-type, orlA mutant and reconstituted strains incubated at 30°C, 37°C, and 45°C. We observed that in the mycelia, the orlA mutant contains less Pi than the wild type and reconstituted strains (P value <0.05 at all incubation conditions except at 30°C) (Fig. 8B). However, in conidia there was not a statistically significant difference between the fungal strains under thermal stress conditions with the exception of a higher Pi level in the orlA mutant at 30°C (Fig. 8A). The higher amount of Pi levels in the orlA mutant conidia may be a result of the difference in conidia size that we observed at 30°C (data not shown). Taken together, at similar conditions in which we observed the accumulation of T6P in the orlA mutant (Fig. 6B), associated Pi levels were consequently decreased in the absence of OrlA (Fig. 8B). This result suggests that Pi is sequestered by T6P which could induce the activation of TP enzymes. Confirmation of these results await further experimentation.
Fig. 8.
Free inorganic phosphate (Pi) is sequestrated by T6P in the absence of OrlA. Conidia (A) and mycelia (B) biomass for Pi assays were cultured at 30°C, 37°C and 45°C. At all conditions, Pi levels found in mycelia of the orlA mutant were significantly decreased from the wild-type and reconstituted strains (P value < 0.05 as indicated) where as an insignificant difference (NS; P value >0.05) was found in conidia cultured at 37°C and 45°C. However, at 30°C, a significant increase in Pi levels was found in the orlA mutant relative to wild type and reconstituted strains.
Hexokinase activity is decreased in the orlA mutant but addition of GlcNAc to growth medium does not rescue cell wall defects
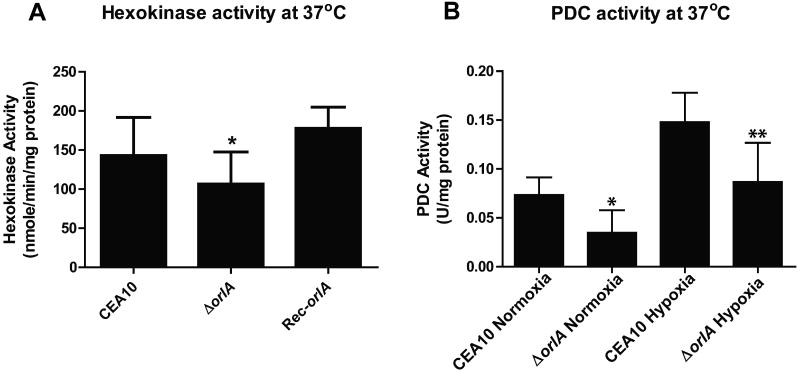
T6P has been observed to inhibit hexokinase activity in vitro in several fungi. Thus, we hypothesized that the large accumulation of T6P in the orlA mutant would lead to decreased hexokinase activity. Cell free extracts from the orlA mutant displayed an approximate ∼30% decrease in hexokinase activity in comparison with the wild-type and reconstituted strains (Fig. 9A). Importantly, this assay cannot distinguish between hexokinase and glucokinase activity. However, while glucose entry into glycolysis can be regulated by both hexokinase and glucokinase, fructose entry into glycolysis is solely regulated by hexokinase. Thus, we next tested the growth of the orlA mutant on fructose minimal media (FMM, 1% fructose). The orlA mutant grows slower than the wild-type and reconstituted strains when cultured on FMM. The radial growth rate (centrimeter diameter/day) of the orlA mutant was 1.27±0.00,the wild-type 2.20±0.03, and 2.16±0.05 in the reconstituted strain. This result confirms that hexokinase activity is likely significantly reduced in the orlA mutant.
Fig. 9.
Key steps in glycolysis are altered in the absence of OrlA. (A) Hexokinase activity of the wild-type, the orlA mutant and the reconstituted strain indicates a decrease in hexokinase activity in the absence of orlA (P value = 0.04) relative to wild-type. Results presented are the mean and standard deviations from three independent experiments. (B) Pyruvate decarboxylase activity is decreased in the absence of OrlA also suggesting a potential decrease in glycolytic flux. Respective strains were grown in GMM at 37°C in normoxic (20% O2) or hypoxic (1% O2) conditions. Results presented are the mean and standard deviations from two independent experiments. *P = 0.03 **P = 0.05 (wild-type vs. mutant respectively for normoxia and hypoxia).
To further examine whether glycolytic flux was potentially inhibited in the orlA mutant, we measured pyruvate decarboxylase (PDC) activity of the orlA mutant in comparison with the wild-type and reconstituted strains grown in liquid GMM at 37°C (Fig. 9B). The orlA mutant displayed a significant decrease in PDC activity suggesting decreased flux through the lower part of glycolysis occurs in the absence of OrlA. This result may be due to decreased levels of the glycolytic intermediates due to inhibition of hexokinase activity, or sequestration of free Pi in the absence of OrlA.
Since aminosugar biosynthesis is linked to intermediates of glycolysis, we next tested whether addition of N-acetylglucosamine (GlcNAc) to growth media could suppress the conidiation or hyphal morphology defect of the orlA mutant on GMM (Borgia et al., 1996). We tested GlcNAc at two concentrations (100 μg/ml and 1 mg/ml) in GMM and observed no change in the asexual conidiation or hyphal morphology defects of the orlA mutant at either concentration (data not shown). While a decrease in hexokinase activity in the orlA mutant is likely to result in a decrease in the needed precursors (Fructose-6-phosphate) for aminosugar biosynthesis, and therefore could explain the subsequent observed cell wall defects, this remains to be conclusively determined.
Loss of OrlA function results in virulence attenuation in murine models of invasive pulmonary aspergillosis
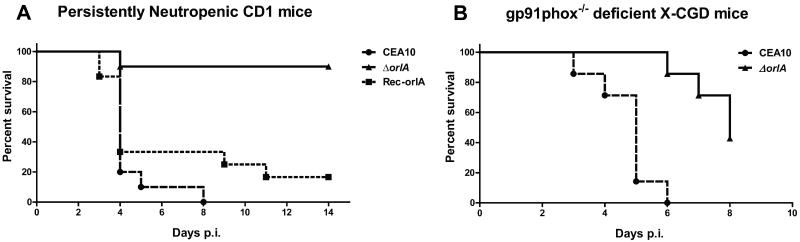
Given the previous associations between the trehalose biosynthesis pathway and fungal virulence, we examined the virulence of the A. fumigatus orlA mutant, reconstituted and wild-type strains in a chemotherapeutic murine model of invasive pulmonary aspergillosis. Despite the normal growth rate of the mutant at mammalian body temperature on glucose minimal media, the orlA mutant displayed a severe virulence attenuation in immunocompromised CD1 mice (Fig. 10A) (P value <0.0005 versus wild type and P=0.0007 versus reconstituted strain). Next, we examined the virulence of the orlA mutant in mice deficient in the gp91 Phox subunit of the NADPH oxidase (X-CGD mice) (Pollock et al., 1995, Morgenstern et al., 1997). In this murine model, the orlA mutant also displayed a statistically significant decrease in virulence (P = 0.0005) (Fig. 10B). Importantly, X-CGD mice infected with the orlA mutant do eventually succumb to the infection. Culture of lung homogenates from X-CGD mice infected with the orlA mutant on day 10 of the infection revealed the persistence of the fungus in these animals. These striking results suggest that OrlA is part of the virulence arsenal of A. fumigatus and strongly supports our hypothesis that the trehalose pathway in A. fumigatus and other fungal pathogens is a potential global antifungal drug target.
Fig. 10.
OrlA is a critical component of the Aspergillus fumigatus virulence arsenal. (A) Outbred CD-1 male mice (n = 10 each fungal strain or mock control) were immunosuppressed as described in the experimental procedures to generate persistently neutropenic mice. Mice were inoculated intranasally with 106 conidia/25 μl of wild-type CEA10, ΔorlA, or the orlA reconstituted strain Rec-orlA. A cohort of 10 mice was also mock infected with 0.01% Tween80 in sterile phosphate buffered saline. No mock infected animals perished in either experiment. Infection experiments were repeated two times with similar results. Log-Rank tests revealed that the survival curves between the mutant and wild-type CEA10 and mutant and reconstituted strain were statistically significant (P < 0.0001 and P = 0.0007, respectively). No significant difference was observed between the wild-type and reconstituted strains (P = 0.2659). (B) X-CGD murine model of IPA, mice were inoculated intranasally with 105 conidia/25 μl of wild type and orlA mutant. The delay in death of mice infected with the orlA mutant is statistically significant (P value = 0.0005).
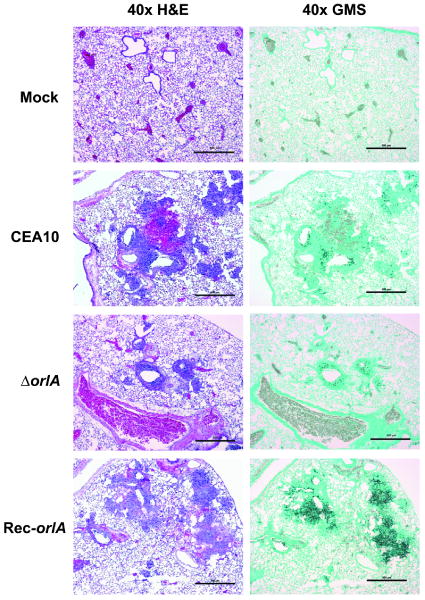
To better understand the mechanism behind the virulence defect in the orlA mutant, we utilized histopathology analyses to visualize the host response and in vivo fungal burden during infection in the immunocompromised CD1 mouse model. On day 3-post infection, whole lung histopathology of mice infected with all 3 strains of fungus showed multiple sites of infection surrounded by leukocytes. At this time point, all mice exhibited symptoms of invasive pulmonary aspergillosis including ruffled fur, lethargy, and slightly labored breathing. No clear differences between the mutant and wild-type infected animals were observed in terms of lesion numbers (data not shown). However, a closer examination of hematoxylin and Eosin (H&E) stained lungs revealed inflammation, drastic tissue damage, and hemorrhaging in the wild-type and reconstituted strains whereas orlA mutant infected mice had less inflammation and damage (Fig. 11 H&E staining). It is clear from the histopathology that the orlA mutant was able to germinate and develop hyphae in vivo in the lung, but it appears that the mutant stimulated a less aggressive inflammatory response which corresponded with slight decreases in fungal burden within given lesions (Fig. 11 GMS staining).
Fig. 11.
Histopathology from immunosuppressed CD1 mouse model observed on day 3 after infection. Mice were intranasally inoculated with 106 conidia and euthanized on day 3 post infection. Lung sections of representative infections stained with hematoxylin and eosin (H&E) or Gomori methenamine silver stain (GMS) are presented. Mock infected animals display normal healthy lung architecture with lack of necrosis, inflammation, and fungal elements. Wild-type (CEA10) and reconstituted strain infected lungs displayed significant numbers of inflammatory and necrotic lesions (H&E stains) surrounding sites infiltrated by fungal growth (black stained fungal hyphae as observed via GMS staining). Interestingly, the orlA mutant infected mice consistently contained less necrosis, inflammation, and fungal growth. Bar = 500 μm for 40× magnification.
By day 7 post infection, all animals in the wild-type and reconstituted strain infected groups had perished and only orlA mutant infected mice survived. H&E and GMS staining revealed that the orlA mutant infected mice were able to partially clear the fungal infection as evidenced by contained, localized inflammatory lesions with minimal fungal growth (data not shown). The localized lesions surrounding the airways observed in orlA mutant infected mice were reminiscent of histopathology observed with mice infected with the A. fumigatus SREBP mutant, which cannot grow in low oxygen environments (Willger et al., 2008). However, hypoxic growth of the orlA mutant was unaffected, and thus the mechanism behind the inability of the orlA mutant to proliferate throughout the host tissue and cause mortality is currently unknown.
Discussion
Continued advances in medical technologies that impair the host immune system and increased incidences of immunosuppressive diseases has resulted in substantial increases in invasive fungal infections over the last three decades (Erjavec et al., 2009, Varkey & Perfect, 2008). Due to the genetic similarity between humans and fungi, the antifungal drug arsenal available to treat fungal infections is relatively limited when compared to other infectious diseases. Moreover, as the use of available antifungal drugs has increased, resistant strains have and will continue to emerge (Perlin & Mellado, 2008, Verweij et al., 2007, White et al., 1998, Snelders et al., 2008). Thus, in order to maintain our ability to thwart these often lethal infections, new antifungal agents are urgently needed. Biochemical pathways that are unique to fungi and absent in humans present ideal targets for antifungal drug development. The trehalose biosynthesis pathway in fungi is such a target.
Previous studies on the trehalose biosynthesis pathway in the human pathogenic fungi C. neoformans, C. gattii, and C. albicans have revealed the importance of this pathway in human fungal pathogenesis (Petzold et al., 2006, Van Dijck et al., 2002, Ngamskulrungroj et al., 2009, Zaragoza et al., 1998, Zaragoza et al., 2002). Broad spectrum antifungal agents that are effective against the majority of human pathogenic fungi are clearly desirable, and thus in this study we have started an examination of the trehalose biosynthesis pathway in the most common cause of invasive mold infections in humans, A. fumigatus.
Surprisingly, our bioinformatic analyses of genes encoding proteins predicted to be involved in trehalose biosynthesis in A. fumigatus revealed the presence of multiple functional copies of the trehalose-6-phosphate synthase (TPS1), which is different from findings in A. nidulans, C. neoformans, C. albicans, and the plant pathogenic fungus Magnaporthe grisea (Puttikamonkul et al. unpublished data). However, our analyses of the A. fumigatus genome revealed the presence of one putative trehalose-6-phosphate phophatase (T6PP) (TPS2 ortholog). Consequently, we began our examination of the A. fumigatus trehalose pathway by generation of a tps2 null mutant. Previously, a TPS2 ortholog was characterized in A. nidulans and named orlA (Borgia et al., 1996). Though the phenotype of our A. fumigatus T6PP mutant has different phenotypes than the A. nidulans mutant, most notably a continued ability to grow under high temperature without cell lysis, we elected to maintain the historical name of this gene and protein as orlA.
The first clear phenotype exhibited by the Af orlA mutant was a severe loss of conidiation on glucose minimal media (1% glucose). As observed in Fig. 2A, the mutant colony appears white in contrast to the blue-green pigmentation normally observed for A. fumigatus colonies. Microscopic examination of the mutant strain revealed that this lack of pigmentation was not due to loss of pigment production, but rather the inability to produce functional asexual reproductive structures. A similar, but less apparent, defect in conidiation was also reported for the A. nidulans orlA mutant (Borgia et al., 1996).
Importantly, several environmental conditions could partially rescue the conidiation defect of the Af orlA mutant. First, we observed that conidiation could partially be restored during growth at temperatures at or below 30°C. Thus, while the Af orlA mutant does not display a lethal growth phenotype on glucose containing media at 37°C such as C. neoformans, C. gattii, and S. cerevisiae mutants, it does display a temperature sensitive phenotype. Second, we observed that conidiation could be almost fully restored on media that contained an osmotic stabilizer (either sorbitol or glycerol). Both of these phenotypes are consistent with previously reported phenotypes of the An orlA mutant (Borgia et al., 1996). In addition, the temperature sensitive phenotypes of the C. neoformans and C. gattii mutants could also be rescued with sorbitol. Third, addition of high levels of glucose (>4%) could restore conidiation and morphological defects in a dose dependent manner. Taken together these results are consistent with the presence of a defect at the level of the cell wall in the Af orlA mutant, and also possibly suggest a defect in glycolysis at the point of glucose entry.
Further evidence for a major cell wall defect in the orlA mutant was observed upon microscopic examination of the orlA mutant hyphae, which displayed severe morphological defects when grown on GMM that could partially be rescued by growth on osmotic stabilizing media. When we examined the sensitivity of the mutant to agents known to target the fungal cell wall, we found that the mutant did not display an increased sensitivity to the β-1,3-glucan synthesis inhibitor caspofungin. However, exposure of the mutant to cell wall perturbing agents that either target chitin biosynthesis or the linkage of chitin polymers to β-1,3-glucan and β-1,6-glucan resulted in significant increases in susceptibility. Together, these results suggest that loss of OrlA results in a significant alteration in either chitin biosynthesis itself or the ability to properly assemble the cell wall matrix. This conclusion is also supported by decreased chitin levels found in the An orlA mutant when exposed to high temperature stress (Borgia et al., 1996). Interestingly, C. albicans tps2 mutants also display a significant defect in cell integrity, which has been attributed to a defect in cell wall biosynthesis (Zaragoza et al., 2002). The C. neoformans tps2 mutant displays a severe temperature sensitive growth phenotype (TS) in glucose media at 37°C that can be rescued with addition of sorbitol also implying that a defect in cell wall biosynthesis is responsible for the TS phenotype of this mutant (Petzold et al., 2006).
A potential hypothesis to explain the orlA mutant phenotypes is that the predicted loss of trehalose biosynthesis due to blockage of T6PP results in the observed phenotypes as trehalose is a membrane and cell wall stabilizing agent. Yet several tps2 mutants in fungi, including the An and Af orlA mutants, surprisingly still produce substantial amount of trehalose. Thus, it seems unlikely that defects in the production of trehalose itself are responsible for the observed phenotypes in the Af orlA mutant. It has been hypothesized that residual, non-specific phosphatase activity in the cell can dephosphorylate T6P into trehalose (Van Dijck et al., 2002). Given that C. albicans tps2 mutants produce lower levels of trehalose than wild-type cells, this hypothesis seems plausible in this organism. However, in the Af orlA mutant we observed an increase in trehalose levels in response to heat shock in comparison with the wild-type strain.
Bioinformatic analyses of the A. fumigatus genome revealed that the number of genes involved in trehalose biosynthesis is expanded compared to S. cerevisiae and the pathogenic yeast studied to date (Puttikamonkul et al. unpublished data). Of great interest is the presence of at least two putative trehalose phosphorylase encoding genes in the A. fumigatus genome. Trehalose phosphorylase has been found to produce trehalose from glucose or α or β-glucose-1-phosphate in algae, bacteria, some yeasts, and basidiomycete mushrooms (Saito et al., 1998a, Kitamoto, 2000, Aisaka & Masuda, 1995, Belocopitow & Marechal, 1970, Belocopitow & Marechal, 1974, Eis & Nidetzky, 1999). Interestingly, trehalose phosphorylase from the mushroom Pleurotus sajor-caju could complement the glucose growth and trehalose biosynthesis defect of a S. cerevisiae tps1/tps2 double mutant (Han et al., 2003). In our study, we observed an increase in the mRNA abundance of both trehalose phosphorylase genes in the absence of orlA possibly suggesting that persistent TP activity explains the ability of the orlA mutant to make wild-type levels of trehalose. It is also known that TP activity is inhibited by free inorganic phosphate. In theory, the accumulation of T6P in the absence of OrlA should lead to a significant decrease in free Pi levels in the cell. This could have two affects. First, it could lead to activation of the TP enzymes, and thus explain the persistence of trehalose in the orlA mutant. Second, Pi is a critical component of the lower portion of glycolysis, and thus the decreased PDC activity observed in the olrA mutant could also be explained by the decrease in available Pi. These alternatives, which may not be mutually exclusive, are being tested in our laboratory by generation of TP null mutants in the wild-type and orlA mutant backgrounds.
The relationship between TP encoding genes and genes of the better studied trehalose biosynthesis pathway (TPS1-TPS2-TPS3) remains to be determined. To date, a TP encoding gene has been partially characterized in N. crassa (CCG-9) where it was observed to be critical for clock control of fungal development (Shinohara et al., 2002). However, the ability of the ccg-9 null mutant (with an apparently intact Tps1p ortholog) to make trehalose was not examined and the role of TP enzymes in filamentous fungal biology remains to be explored. In Tps1 double mutants of A. fumigatus (tpsA/tpsB), trehalose production is completely absent (Puttikamonkul et al. unpublished data). However, in this tps1 null mutant T6P levels are also removed and thus high free Pi levels, which would inhibit TP activity, are present. These results illustrate the complexity, differences, and importance of trehalose biosynthesis mechanisms among the fungi.
Since production of trehalose itself is not significantly affected in the orlA mutant, an alternative hypothesis to explain the orlA mutant phenotypes is that T6P inhibits/alters cell wall biosynthesis. In S. cerevisiae, accumulation of T6P in the tps2 mutant resulted in a temperature sensitive phenotype and inability of the mutant to grow on glucose. This led to the hypothesis that accumulation of T6P is toxic to yeast cells (De Virgilio et al., 1993, Sur et al., 1994). This toxicity is hypothesized to manifest itself through inhibition of hexokinase activity and subsequent mis-regulation of glycolysis or by sequestration of intracellular phosphate and subsequent intracellular acidification (Thevelein & Hohmann, 1995, Blazquez et al., 1994, Blazquez et al., 1993). In the An orlA mutant, it was observed that glutamine:fructose-6-phosphate amidotransferase (GFAT) activity was reduced. GFAT is the first enzyme unique to aminosugar biosynthesis and thus N-acetylglucosamine (GlcNAc) could partially rescue some, but not all, of the mutant phenotypes (Borgia et al., 1996). Thus, it seems that T6P in A. nidulans may either directly or indirectly inhibit key enzymes involved in chitin biosynthesis.
A. fumigatus contains seven chitin synthase genes (chsA – chsG) which are split into 6 classes based on amino acid sequences (Bernard & Latge, 2001, Munro & Gow, 2001). Null mutants of chsE and chsG share the morphological and conidiation defect of the Af orlA mutant that can be rescued with an osmotic stabilizer (Mellado et al., 1996, Mellado et al., 2003, Aufauvre-Brown et al., 1997). Transcriptional profiling of cell wall biosynthesis genes in the Af orlA mutant background does not reveal a significant decrease in mRNA abundance in all known chitin synthases in the absence of orlA. Nevertheless, the chitin biosynthesis defects could occur at the post-transcriptional level and be due to the consequence of high T6P levels and/or depletion of free inorganic phosphate (Pi). Our data suggests that the accumulation of T6P in the Af orlA mutant may result in deregulation of glycolysis that could alter biosynthesis of GlcNAc, which is required for aminosugar biosynthesis.
Two key glycolysis intermediates are required to produce the UDP-GlcNAc required for chitin biosynthesis: glucose-6-phosphate and fructose-6-phosphate. Alteration in the levels or flux of these important carbohydrate metabolites could negatively impact the ability of the cell to produce appropriate levels of cell wall material. It has been shown that T6P can inhibit hexokinase activity in vitro in many fungal species (Blazquez et al., 1993, Panneman et al., 1998, Gancedo & Flores, 2004). In our study, we observed significant decreases in hexokinase activity and a growth defect on fructose minimal media, which suggests a possible alteration of flux through glycolysis in the absence of OrlA. This conclusion is also supported by the finding that high levels of glucose can suppress the orlA mutant phenotypes. Rescue by high glucose could be due to simple osmotic stabilization by the high glucose levels or, alternatively, high levels of glucose may overcome the inhibition of hexokinase activity and/or activation of glucokinase activity resulting in glucose-6-phosphate levels that restore aminosugar biosynthesis. A simple model is thereby proposed whereby accumulation of T6P in the absence of OrlA not only depletes available Pi levels, but also inhibits hexokinase activity thereby reducing the production of glucose-6-phosphate and fructose-6-phosphate. Regulation of glucose flux into glycolysis in A. fumigatus appears to be a highly regulated process as evidenced by the presence of no less than 6 putative hexokinase orthologs found in this filamentous fungus (Flipphi et al., 2009). Future studies are focusing on the effect of T6P on glycolysis and cell wall biosynthesis in A. fumigatus.
Since the cell wall is a critical pathogen associated molecular pattern (PAMP), it is not surprising that the orlA mutant is attenuated in virulence in two distinct murine models of invasive pulmonary aspergillosis. Given the normal growth rate of the mutant strain under standard in vitro conditions, the virulence defect of the mutant was surprising. Yet, this result is consistent with the attenuation in virulence of C. albicans, C. gattii, and C. neoformans tps2 mutants (Zaragoza et al., 2002, Van Dijck et al., 2002, Petzold et al., 2006, Ngamskulrungroj et al., 2009). In particular, C. neoformans tps2 mutants have a severe TS growth phenotype due to build up of T6P and are unable to grow at 37°C, which would suggest avirulence in mammals (Petzold et al., 2006). Importantly, our results close the proverbial loop by showing that inhibition of tps2 orthologs in the three most common human fungal pathogens results in virulence attenuation. Thus, development of a strategy to inhibit tps2 function would likely have global applicability in treating invasive fungal infections.
Yet, questions remain with regard to the mechanism behind the virulence attenuation in the Af orlA mutant. Histopathological examinations of mice infected with the orlA mutant strain suggest that the mutant does not induce as strong an inflammatory response as the wild-type and reconstituted strains. This would be consistent with an alteration in the cell wall of the mutant strain. Moreover, while growth of the mutant strain in vivo was clearly observed throughout the infection, tissue proliferation seemed to be slightly decreased in comparison with the wild-type strain. This could be due to a simple inability of the mutant to grow in vivo in microenvironments with multiple stresses (possibly the result of glycolysis deregulation). Alternatively, the apparent alteration in cell wall composition of the mutant strain may influence how the host immune system is responding to (or “seeing”) the mutant. It is important to note that the virulence attenuation was also observed in mice deficient in the ability to generate an ROS burst (X-CGD) but otherwise “normal” immunologically. In the X-CGD mice, the mutant was eventually able to cause mortality pointing to a delayed growth defect in the mutant either due to inhibition of fungal growth as a consequence of OrlA loss, or due to an alteration in the host response to the mutant strain. Studies are ongoing in our laboratory to assess the mechanism behind the virulence attenuation of the orlA mutant strain.
In conclusion, our results confirm previous suggestions that trehalose-6-phosphate phosphatase is a promising target for antifungal drug development. Given that tps2 orthologs are now known to be required for fungal virulence in the three most frequently encountered causal agents of human mycoses, it seems logical to pursue research to better understand how this pathway affects fungal virulence. Since inhibition of orlA in A. fumigatus is not fungicidal per say, it may be that inhibition of this pathway in A. fumigatus would best be utilized in some form of combination therapy, as also previously suggested for C. albicans (Van Dijck et al., 2002). Yet, the association between the trehalose pathway and cell wall biosynthesis is intriguing given the clinical importance of the fungal cell wall both as a virulence factor and as a target of current antifungal drugs. Thus, it is likely that further in-depth analysis of the trehalose pathway in A. fumigatus and other pathogenic fungi will yield new insights into fungal biology and virulence that may prove fruitful for designing new therapeutic strategies to treat human mycoses.
Experimental procedures
Strains and media
Aspergillus fumigatus strain CEA17 (a uracil auxotroph strain lacking pyrG) was used to generate the orlA mutant strain (ΔorlA; orlA∷A. parasiticus pyrG) (d'Enfert, 1996). The corresponding wild-type strain CEA10 (CBS144.89) was utilized throughout this study (kind gift of Dr. Jean-Paul Latge). All strains were routinely grown in glucose minimal medium (GMM) containing a final concentration of 1% glucose, if not stated otherwise, with or without uridine and uracil at 37°C (Shimizu & Keller, 2001). Conidia were harvested after growth on sorbitol minimal media (SMM, 1.2 M sorbitol, 1% glucose) for 3 days, using SMM for standard media in all experiments (with the exception of the virulence studies) due to the restricted ability of the orlA mutant to generate conidia on GMM. For the virulence studies, wild-type and reconstituted strain conidia were obtained from GMM. An examination of conidia from the wild-type and reconstituted strains grown on SMM did not reveal any significant differences in morphology, viability or germination rates compared with GMM (data not shown). FMM is a minimal media that contain 1% fructose in substitution of glucose to determine the ability of fungal strains to utilize fructose as a sole carbon source.
Strain construction
Mutant and reconstituted strains were generated by standard fungal protoplast transformation of A. fumigatus uracil auxotroph strain CEA17. PCR generated gene replacement or reconstitution constructs were utilized. BLAST searches of the A1163 genome sequence at The Central Aspergillus Data REpository (CADRE) database retrieved the orlA open reading frame (AFUB_043350) (Mabey et al., 2004). The 2,999 bp orlA gene was replaced by a gene replacement construct of ∼5kb. The replacement construct was generated by cloning homologous sequences (900 bp upstream and 951 bp downstream) of the orlA locus into plasmid pJW24 (gift from Dr. Nancy Keller) flanking the A. parasiticus pyrG gene (d'Enfert, 1996). This plasmid was then used as a template to amplify the replacement cassette and 10 μg of this PCR product was used for protoplast transformation. Polyethylene glycol mediated protoplast transformation was essentially performed according to the procedure described by Yelton et al. (Yelton et al., 1984). To reconstitute the wild-type orlA gene back into the orlA mutant background, a reconstitution construct was generated by cloning a ∼5.3 kb fragment of orlA from the wild-type strain CEA10 into fungal transformation vector pBC-Hygro (gift from Kevin McCluskey, Fungal Genetics Stock Center), which contains the hygromycin B resistance gene as a selectable marker. 1,187bp of sequence upstream of the orlA coding sequence and 775bp downsteam of the coding sequence were utilized. Subsequent transformants were selected on hygromycin B (150 μg/ml) containing media.
Transformants were initially screened with PCR to identify potential transformants with homologous recombination events at the orlA locus. PCR positive transformants were single spore isolated to eliminate any potential heterokaryons. Lastly, all possible candidates were then verified by Southern blot analysis and a strain carrying a single insertion of the replacement construct at the orlA gene locus was selected for further characterization. For reconstitution transformations, hygromycin resistant colonies were selected and screened with PCR and Southern blot analysis to identify a transformant with a single ectopic insertion of the reconstitution construct. Real time reverse transcriptase PCR was used to confirm expression of the re-introduced gene (Cramer et al., 2006).
Conidia production and morphology
Equal amounts of conidia from the respective strains (1×103) were plated on two different media, GMM and SMM, and incubated at 37°C for 4 days prior to harvesting and counting. Total conidia production was assessed in triplicate and repeated on three separate occasions. Average value and standard deviation are reported with a two-tailed Student's t-test analysis (P<0.05) by GraphPad Prism software version 5 (San Diego, CA).
Flow cytometry analysis (FACS) was utilized to evaluate the morphology of conidia in the orlA mutant compared to wild-type and reconstituted strains. Freshly harvested conidia in 0.01% Tween80 were counted and diluted to 1×105 conidia/ml (Ejzykowicz et al., 2009), samples were flowed through the BD FACS Diva Version 6.0 without other additional preparation steps. Side scatter (SSC) and forward scatter (FSC) analyses were performed on the sorted conidia.
Trehalose measurement
A. fumigatus strains were grown on SMM plates at 37°C for 3 days. A total of 2×108 conidia were used for the trehalose assay as described by d'Enfert C. and Fontaine (d'Enfert & Fontaine, 1997). Briefly, the conidia solution was centrifuged and resuspended in 500 μl of distilled water and incubated at 100°C for 20 min. The conidial extract containing soluble trehalose was separated from cell debris by centrifugation at 11,000×g for 10 min and used as the cell free extract.
Cell free extracts were then tested for trehalose levels according to the Glucose Assay Kit protocols (Sigma Aldrich). Samples (100 μl) were incubated with 100 μl of 0.2M sodium citrate pH 5.5 (0.1M final concentration) and incubated at 37°C overnight with 3mU of porcine kidney trehalase. The same reaction without trehalase was used as a control and utilized to determine the basal level of glucose in each reaction. The concentration of trehalose in the assay reaction (presence of trehalase) was subtracted by levels of glucose found in the control reaction (without trehalase). This ensures that the response measured is due to presence of trehalose and not glucose in the extracts. Concentration was normalized to starting input. Results from triplicate experiments were averaged, standard deviation calculated, and statistical significance determined (P≤0.05) with a two-tailed Student's t-test.
Trehalose-6-phosphate (T6P) measurement
Liquid Chromatography Mass Spectrometry (LCMS) was used for measurement of T6P. Conidia and mycelia extractions were performed by the method modified from Wilson et al. (Wilson et al., 2007). A total of 3×1010 conidia from 3 day old cultures grown on SMM were used for T6P measurement. Briefly, the frozen conidia pellet is ground to a fine powder using a mortar and pestle and dichloromethane was used for extraction of metabolites. Mycelia samples were prepared from a 75 ml liquid GMM culture of 1×107 conidia/ml grown overnight at 37°C. After overnight incubation, the cultures were filtered, lyophilized, ground with a mortar and pestle, and the subsequent powder weighed. For extraction, fungal powder was added to 10 ml of a Methanol: Dichloromethane: Water (6 : 2.5 : 1.5) solution and incubated at 4°C for 30 min. The suspension was centrifuged at 1,240×g for 20 min and the supernatant transferred to a new tube. Re-extraction of the sample pellet in 5 ml of fresh extraction solution was performed and the supernatant added to the previous supernatant. The pooled supernatant was then mixed with 7.5 ml of dichloromethane and 7.5 ml of water and centrifuged at 3,440×g for 20 min. The aqueous phase was collected and lyophilized overnight until completely dry. Resuspension of the dried samples was performed in 1 ml of deionized water and stored at -20°C until performing LCMS analysis.
LCMS was performed using an Agilent 1100 series LC system (Agilent Technologies, CA, USA) consisting of a vacuum degasser, autosampler, and a binary pump equipped with two tandem Luna Cartridges, Luna HILIC, 3μ, 200A°, 4×2.0mm (Phenomenex Inc.). The mobile phases were 100% water (solvent A) and 100% acetonitrile (solvent B). A stepwise-gradient from 95% B to 10% B (25°C) at a flow rate of 0.2ml/min was used with a total run time of 9 min. The LC was directly interfaced to the ESI source of a microTOF (Bruker Daltonics, Bremen, Germany) mass spectrometer without splitting, operated in negative mode. Quantitation was achieved by analyzing samples in triplicate and using T6P-EIC (extracted ion chromatograms). Standard curves were constructed using a Trehalose 6-phosphate (Sigma) standard. The concentration of T6P was normalized to the input weight of fungal tissue for each sample. Results from triplicate experiments were averaged, standard deviation calculated, and statistical significance determined (P≤0.05) with a two-tailed Student's t-test.
Microscopic analysis
A modified slide culture method adapted from Johnson (Johnson, 1946) and Harris (Harris, 1986) was used to visualize the conidia and conidiophores of the orlA mutant compared to the wild-type and reconstituted strains with light microscopy. Briefly, a thin layer of solid media was cut in blocks approximately 10 mm2, aseptically removed and placed on the glass slide which was placed on a support in a sterile 60 mm plastic petri dish. Each side of an agar block was inoculated with conidia of the respective strains. A sterile cover glass slip was placed on the inoculated agar and sterile water was added into the slide culture chamber to maintain humidity. This apparatus was incubated at 37°C until adequate growth and conidiogenesis had occurred (approximately 2-3 days). Both the cover slip and glass slide were carefully taken from the agar block and a drop of fungal mounting medium (lactophenol cotton blue, Sigma) was applied to each specimen. The slides were subsequently observed utilizing a light microscope.
Cell wall perturbation agents
Several cell wall inhibitors were utilized for cell wall integrity tests: congo red (CR, Sigma Aldrich), calcofluor white (CFW, Sigma Aldrich), and Nikkomycin Z (NK, Sigma Aldrich) (Brun et al., 2007, Kaur et al., 2007, Ram & Klis, 2006). CR, CFW or NK were added into GMM at final concentrations of 1mg/ml, 25 μg/ml and 0.1 mM respectively. 1×106 conidia (in a 5μl drop) of each test strain were placed on the center of each plate. The radial growth rates at 37°C of all strains were measured every 24 hours for a period of 5 days for each fungal strain in triplicate.
Quantitative Real-Time PCR
Respective A. fumigatus strains were cultured in liquid GMM for 6.5 and 24 hrs. At the respective time point, the germlings and mycelia were collected with vacuum filtration and lyophilized with a freeze drier (Labconco) prior to homogenization with 0.1 mm glass beads. Total RNA was extracted using the TRIsure reagent (Bioline) according to the manufacturer's protocol and subsequently further purified with the Qiagen RNeasy Plant Mini Kit (Qiagen). Genomic DNA contamination was further removed with Turbo DNase I treatment (Ambion). RNA integrity was confirmed with an Agilent Bioanalyzer. Quantitative real-time PCR analysis of cell wall biosynthesis genes was performed as previously described (Cramer et al., 2008). Primers for the individual cell wall biosynthesis genes are also as listed in Cramer et al., 2008. qRT-PCR experiments were performed in duplicate and the fold expression data presented in Table 1 represents the mean and standard deviation of two biological replicates.
For mRNA abundance of the orlA and trehalose phosphorylase genes, conidia were produced on SMM plates (30 °C, 37 °C, and 45°C) and harvested with 0.01% Tween80. Total conidia were centrifuged and lyophilized overnight prior to homogenization with 0.1 mm glass beads. Total RNA extraction and subsequent processing were as described above. 500 ng of DNase treated total RNA was reverse transcribed with the Quantitect Reverse Transcriptase kit (Qiagen). qRT-PCR was subsequently performed as we have previously described (Cramer et al., 2006). A no DNA template control was used in each analysis. Each sample was tested in triplicate and data was normalized to the actin reference gene and relative to the wild-type control grown at low temperature (30°C) (Livak & Schmittgen, 2001). The results presented are from biological duplicate experiments and statistical significance was determined with an unpaired two-tailed student T-test.
Hexokinase and Pyruvate Decarboxylase Assays
1×107 conidia/ml of the respective test strains were cultured in liquid GMM at 37°C for approximately 20 hr (which is the culture time that T6P accumulated in the orlA mutant). Mycelia was harvested by vacuum filtration, immediately frozen in liquid nitrogen and homogenized with 0.1 mm glass beads in 50mM Triethanolamine buffer pH 7.6 (Anderson et al., 1984). All procedures were conducted at 4°C. The homogenate was centrifuged at 13,000 rpm for 20 min at 4°C. The supernatant was transferred to a new pre-chilled tube and 25 μl of supernatant was used for the hexokinase assay following the manufacturer's protocol (Sigma Aldrich) (Bergmeyer et al., 1983). Hexokinase activity was determined by measuring NADPH production at an absorption intensity of 340 nm with equilibration to 25°C. The ΔA340nm/minute was obtained from the maximum linear rate of kinetic measurements for approximately 5 minutes (Wilson et al., 2007). Protein concentration in the extracted supernatant was quantified with the Bradford Reagent and used to normalize the data. Data presented are the mean and standard deviation of three biological replicates.
For pyruvate decarboxylase activity assays, the protocol of Lockington et al. was essentially followed (Lockington et al., 1997). Briefly, respective test strains were cultured overnight (∼12 hours) at 37°C in liquid GMM (LGMM). Equal amounts of germlings were then incubated in fresh liquid GMM either under normoxic (20% O2) or hypoxic (1% O2) conditions for 48 hours at 37°C in LGMM. Cell free extracts from mycelium were collected as described for the hexokinase assay with the exception that the buffer for extraction was 100 mM KH2PO4, 2 mM MgCl2, and 1 mM DTT. Pyruvate decarboxylase activity was determined by measuring the reduction of NADH at A340nm and equilibrated to 37°C. Kinetic measurements were performed for 5 minutes and the maximum linear rate was calculated for each unit of enzyme. Data presented are the mean and standard deviation of two biological replicates.
Pi assay
Conidia and mycelia of wild type, the orlA mutant and reconstituted strains were grown at 30, 37 and 45°C. All strains were grown on SMM plates and a pellet of 2×109 conidia was lyophilized overnight prior to homogenization with 0.1 mm glass beads. The mycelia was cultured in 50ml liquid GMM at inoculums of 1×107conidia/ml overnight. Mycelia were then harvested, washed with sterile water, and collected with vacuum filtration prior to lyophilization and homogenization. Either 50mg of powdered mycelia or powder of 2×109 conidia was then reconstituted in 1 ml and 0.5 ml of sterile water respectively. The cell free extracts were prepared by vortexing and the clear supernatant was collected after centrifugation at 13,000 rpm for 10 minutes. Pi assay was measured by utilizing malachite green dye and molybdate, which forms a stable colored complex specifically with inorganic phosphate following the manufacturer's protocol; QuantiChrom Phosphate Assay Kit (BioAssay Systems). The cell free extracts were diluted to fall within the standard curve limits; 1:100 and 1:400 dilutions were optimized with conidia and mycelia samples respectively.
Murine virulence tests
Male CD1 mice (Charles River Laboratory, Raleign, NC) (6-8 weeks of age) were housed and supplied with food and antibiotic water ad libitum in the Animal Resources Center at Montana State University. Mice were immunosuppressed with intraperitoneal (IP) injections of cyclophosphamide (150 mg/kg) and subcutaneously (SC) with triamcinolone acetonide (Kenalog) (40 mg/kg) on days -2 and -1 respectively prior to infection. On days +3 and +6 post infection, the same dose of cyclophosphamide and Kenalog were repeated respectively. On day 0, groups of 10 mice were infected via the intranasal route with 1×106 conidia of each A. fumigatus strain: orlA mutant, reconstituted (Rec-orlA), wild-type (CEA10), and a mock control group without conidia (the diluent; 0.01% Tween 80). For experiments with the X-CGD mice, 8-10 week old sex matched mice were utilized. Since the reconstitution strain was shown to have wild-type levels of virulence in the neutropenic model, only the wild-type and orlA mutant were compared in these animals (N = 10 each group). On day 0, mice were infected with 1×105 conidia of the respective fungal strains.
Each mouse was observed twice daily for 14 days after A. fumigatus challenge. Mice were observed for standard signs of IPA including ruffled fur, hunched posture, difficulty breathing and weight loss accounting for more than 20% of body mass. Critically affected mice were humanely euthanized when it is clear recovery was not possible using the above criteria. A log rank test was used for pair wise comparisons of survival levels among the experimental groups, P ≤ 0.05. Mouse experiments for each model were repeated on two separate occasions with similar results. Results presented are for one representative experiment for each model. All animal procedures and protocols were approved by the MSU Institutional Animal Care and Use Committee.
Histopathology
Additional murine experiments were conducted as described for histopathology analysis. In contrast to the survival study, 12 mice per group were used. At specific time points (days 1, 3, 5, 7, and 14 post-infections), two mice in each group were humanely euthanized. One lung was harvested from each mouse and fixed in 10% formalin prior to embedding in paraffin, 5 μm thick sections taken, and stained with either H&E (Hematoxylin and Eosin) or GMS (Gomori's Methenamine Silver) (Huppert et al., 1978). The other lung was kept at -80°C for additional analyses. Microscopic examination was performed on a Zeiss Axioscope2-plus microscope and engaged imaging system. Pictures were captured at 40× object-magnification and a reference bar is included in each image.
Acknowledgments
This work was supported by funding from the National Institutes of Health-NCRR COBRE grants 1P20RR020185 and 2P20RR020185 and the Montana State University Agricultural Experiment Station. Srisombat Puttikamonkul is funded through a Ph.D. fellowship from the Royal Thai Government. Dr. John Perfect is supported by Public Health Service Grant AI 73897. The authors would like to thank Dr. Kelly Craven, The Samuel Roberts Noble Foundation, Kevin Fuller and Dr. Judith Rhodes, University of Cincinnati, and members of the Cramer Laboratory for insightful discussions. Thanks to Dr. Allen Harmsen and laboratory members and Dr. Mark Quinn for technical advice and use of equipment at Montana State University, and the staff at the Animal Resources Center at Montana State University for their assistance and care of the animals used in this study.
Footnotes
During the revision of this manuscript, a study was published by Nadia Al-Bader and colleagues confirming that a tpsA/tpsB mutant is unable to produce trehalose during development and heat shock (Al-Bader et al., 2010).
References
- Aisaka K, Masuda T. Production of trehalose phosphorylase by Catellatospora ferruginea. FEMS Microbiol Lett. 1995;131:47–51. doi: 10.1016/0378-1097(95)00233-u. [DOI] [PubMed] [Google Scholar]
- Aisaka K, Masuda T, Chikamune T, Kamitori K. Purification and characterization of trehalose phosphorylase from Catellatospora ferruginea. Biosci Biotechnol Biochem. 1998;62:782–787. doi: 10.1271/bbb.62.782. [DOI] [PubMed] [Google Scholar]
- Al-Bader N, Vanier G, Liu H, Gravelat FN, Urb M, Hoareau CM, Campoli P, Chabot J, Filler SG, Sheppard DC. The role of trehalose biosynthesis in Aspergillus fumigatus development, stress response and virulence. Infect Immun. 2010 doi: 10.1128/IAI.00813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PJ, Karageuzian LN, Cheng HM, Epstein DL. Hexokinase of calf trabecular meshwork. Invest Ophthalmol Vis Sci. 1984;25:1258–1261. [PubMed] [Google Scholar]
- Aufauvre-Brown A, Mellado E, Gow NAR, Holden DW. Aspergillus fumigatus chsE: A Gene Related to CHS3 of Saccharomyces cerevisiae and Important for Hyphal Growth and Conidiophore Development but Not Pathogenicity. Fungal Genet Biol. 1997;21:141–152. doi: 10.1006/fgbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- Avonce N, Mendoza-Vargas A, Morett E, Iturriaga G. Insights on the evolution of trehalose biosynthesis. BMC Evol Biol. 2006;6:109. doi: 10.1186/1471-2148-6-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell W, Sun W, Hohmann S, Wera S, Reinders A, De Virgilio C, Wiemken A, Thevelein JM. Composition and functional analysis of the Saccharomyces cerevisiae trehalose synthase complex. J Biol Chem. 1998;273:33311–33319. doi: 10.1074/jbc.273.50.33311. [DOI] [PubMed] [Google Scholar]
- Belocopitow E, Marechal LR. Trehalose phosphorylase from Euglena gracilis. Biochim Biophys Acta. 1970;198:151–154. doi: 10.1016/0005-2744(70)90045-8. [DOI] [PubMed] [Google Scholar]
- Belocopitow E, Marechal LR. Metabolism of trehalose in Euglena gracilis. Partial purification and some properties of phosphoglucomutase acting on beta-glucose 1-phosphate. Eur J Biochem. 1974;46:631–637. doi: 10.1111/j.1432-1033.1974.tb03659.x. [DOI] [PubMed] [Google Scholar]
- Bergmeyer HU, Grassl M, Walter HE. In: Methods of Enzymatic Analysis. 3rd. Bergmeyer HU, editor. II. Deerfield Beach, FL: Verlag Chemie; 1983. pp. 222–223. [Google Scholar]
- Bernard M, Latge JP. Aspergillus fumigatus cell wall: composition and biosynthesis. Med Mycol. 2001;39(1):9–17. [PubMed] [Google Scholar]
- Blazquez MA, Lagunas R, Gancedo C, Gancedo JM. Trehalose-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett. 1993;329:51–54. doi: 10.1016/0014-5793(93)80191-v. [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Stucka R, Feldmann H, Gancedo C. Trehalose-6-P synthase is dispensable for growth on glucose but not for spore germination in Schizosaccharomyces pombe. J Bacteriol. 1994;176:3895–3902. doi: 10.1128/jb.176.13.3895-3902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgia PT, Dodge CL. Characterization of Aspergillus nidulans mutants deficient in cell wall chitin or glucan. J Bacteriol. 1992;174:377–383. doi: 10.1128/jb.174.2.377-383.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgia PT, Miao Y, Dodge CL. The orlA gene from Aspergillus nidulans encodes a trehalose-6-phosphate phosphatase necessary for normal growth and chitin synthesis at elevated temperatures. Mol Microbiol. 1996;20:1287–1296. doi: 10.1111/j.1365-2958.1996.tb02647.x. [DOI] [PubMed] [Google Scholar]
- Brun YF, Dennis CG, Greco WR, Bernacki RJ, Pera PJ, Bushey JJ, Youn RC, White DB, Segal BH. Modeling the combination of amphotericin B, micafungin, and nikkomycin Z against Aspergillus fumigatus in vitro using a novel response surface paradigm. Antimicrob Agents Chemother. 2007;51:1804–1812. doi: 10.1128/AAC.01007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E, Leloir LF. The biosynthesis of trehalose phosphate. J Biol Chem. 1958;231:259–275. [PubMed] [Google Scholar]
- Cao Y, Wang Y, Dai B, Wang B, Zhang H, Zhu Z, Xu Y, Jiang Y, Zhang G. Trehalose is an important mediator of Cap1p oxidative stress response in Candida albicans. Biol Pharm Bull. 2008;31:421–425. doi: 10.1248/bpb.31.421. [DOI] [PubMed] [Google Scholar]
- Cramer RA, Jr, Gamcsik MP, Brooking RM, Najvar LK, Kirkpatrick WR, Patterson TF, Balibar CJ, Graybill JR, Perfect JR, Abraham SN, Steinbach WJ. Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot Cell. 2006;5:972–980. doi: 10.1128/EC.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer RA, Jr, Perfect BZ, Pinchai N, Park S, Perlin DS, Asfaw YG, Heitman J, Perfect JR, Steinbach WJ. Calcineurin target CrzA regulates conidial germination, hyphal growth, and pathogenesis of Aspergillus fumigatus. Eukaryot Cell. 2008;7:1085–1097. doi: 10.1128/EC.00086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JH, Hoekstra FA, Crowe LM. Anhydrobiosis. Annu Rev Physiol. 1992;54:579–599. doi: 10.1146/annurev.ph.54.030192.003051. [DOI] [PubMed] [Google Scholar]
- d'Enfert C. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr Genet. 1996;30:76–82. doi: 10.1007/s002940050103. [DOI] [PubMed] [Google Scholar]
- d'Enfert C, Fontaine T. Molecular characterization of the Aspergillus nidulans treA gene encoding an acid trehalase required for growth on trehalose. Mol Microbiol. 1997;24:203–216. doi: 10.1046/j.1365-2958.1997.3131693.x. [DOI] [PubMed] [Google Scholar]
- De Virgilio C, Burckert N, Bell W, Jeno P, Boller T, Wiemken A. Disruption of TPS2, the gene encoding the 100-kDa subunit of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae, causes accumulation of trehalose-6-phosphate and loss of trehalose-6-phosphate phosphatase activity. Eur J Biochem. 1993;212:315–323. doi: 10.1111/j.1432-1033.1993.tb17664.x. [DOI] [PubMed] [Google Scholar]
- Eis C, Nidetzky B. Characterization of trehalose phosphorylase from Schizophyllum commune. Biochem J. 1999;341(Pt 2):385–393. [PMC free article] [PubMed] [Google Scholar]
- Ejzykowicz DE, Cunha MM, Rozental S, Solis NV, Gravelat FN, Sheppard DC, Filler SG. The Aspergillus fumigatus transcription factor Ace2 governs pigment production, conidiation and virulence. Mol Microbiol. 2009;72:155–169. doi: 10.1111/j.1365-2958.2009.06631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein AD, Pan YT, Pastuszak I, Carroll D. New insights on trehalose: a multifunctional molecule. Glycobiology. 2003;13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- Elliott B, Haltiwanger RS, Futcher B. Synergy between trehalose and Hsp104 for thermotolerance in Saccharomyces cerevisiae. Genetics. 1996;144:923–933. doi: 10.1093/genetics/144.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec Z, Kluin-Nelemans H, Verweij PE. Trends in invasive fungal infections, with emphasis on invasive aspergillosis. Clin Microbiol Infect. 2009;15:625–633. doi: 10.1111/j.1469-0691.2009.02929.x. [DOI] [PubMed] [Google Scholar]
- Fillinger S, Chaveroche MK, van Dijck P, de Vries R, Ruijter G, Thevelein J, d'Enfert C. Trehalose is required for the acquisition of tolerance to a variety of stresses in the filamentous fungus Aspergillus nidulans. Microbiology. 2001;147:1851–1862. doi: 10.1099/00221287-147-7-1851. [DOI] [PubMed] [Google Scholar]
- Flipphi M, Sun J, Robellet X, Karaffa L, Fekete E, Zeng AP, Kubiecek CP. Biodiversity and evolution of primary carbon metabolism in Aspergillus nidulans and other Aspergillus spp. Fungal Genet Biol. 2009;46(1):S19–S44. doi: 10.1016/j.fgb.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Foster AJ, Jenkinson JM, Talbot NJ. Trehalose synthesis and metabolism are required at different stages of plant infection by Magnaporthe grisea. EMBO J. 2003;22:225–235. doi: 10.1093/emboj/cdg018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo C, Flores CL. The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res. 2004;4:351–359. doi: 10.1016/S1567-1356(03)00222-8. [DOI] [PubMed] [Google Scholar]
- Han SE, Kwon HB, Lee SB, Yi BY, Murayama I, Kitamoto Y, Byun MO. Cloning and characterization of a gene encoding trehalose phosphorylase (TP) from Pleurotus sajor-caju. Protein Expr Purif. 2003;30:194–202. doi: 10.1016/s1046-5928(03)00104-9. [DOI] [PubMed] [Google Scholar]
- Harris JL. Modified method for fungal slide culture. J Clin Microbiol. 1986;24:460–461. doi: 10.1128/jcm.24.3.460-461.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger T, De Virgilio C, Hall MN, Boller T, Wiemken A. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. II. Physiological concentrations of trehalose increase the thermal stability of proteins in vitro. Eur J Biochem. 1994;219:187–193. doi: 10.1111/j.1432-1033.1994.tb19929.x. [DOI] [PubMed] [Google Scholar]
- Huppert M, Oliver DJ, Sun SH. Combined methenamine-silver nitrate and hematoxylin & eosin stain for fungi in tissues. J Clin Microbiol. 1978;8:598–603. doi: 10.1128/jcm.8.5.598-603.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara R, Taketani S, Sasai-Takedatsu M, Adachi Y, Kino M, Furuya A, Hanai N, Tokunaga R, Kobayashi Y. ELISA for urinary trehalase with monoclonal antibodies: a technique for assessment of renal tubular damage. Clin Chem. 2000;46:636–643. [PubMed] [Google Scholar]
- Johnson EA. An Improved Slide Culture Technique for the Study and Identification of Pathogenic Fungi. J Bacteriol. 1946;51:689–694. doi: 10.1128/jb.51.6.689-694.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandror O, DeLeon A, Goldberg AL. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc Natl Acad Sci U S A. 2002;99:9727–9732. doi: 10.1073/pnas.142314099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane SM, Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R, Ma B, Cormack BP. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc Natl Acad Sci U S A. 2007;104:7628–7633. doi: 10.1073/pnas.0611195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto Y, Osaki N, Tanaka H, Sasaki H, Mori N. Purification and properties of a-glucose 1-phosphate-forming trehalose phosphorylase from basidiomycete, Pleurotus ostreatus. Mycoscience. 2000;41:607–613. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lockington RA, Borlace GN, Kelly JM. Pyruvate decarboxylase and anaerobic survival in Aspergillus nidulans. Gene. 1997;191:61–67. doi: 10.1016/s0378-1119(97)00032-2. [DOI] [PubMed] [Google Scholar]
- Lunn JE, Feil R, Hendriks JH, Gibon Y, Morcuende R, Osuna D, Scheible WR, Carillo P, Hajirezaei MR, Stitt M. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J. 2006;397:139–148. doi: 10.1042/BJ20060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabey JE, Anderson MJ, Giles PF, Miller CJ, Attwood TK, Paton NW, Bornberg-Bauer E, Robson GD, Oliver SG, Denning DW. CADRE: the Central Aspergillus Data REpository. Nucleic Acids Res. 2004;32:D401–405. doi: 10.1093/nar/gkh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado E, Aufauvre-Brown A, Gow NA, Holden DW. The Aspergillus fumigatus chsC and chsG genes encode class III chitin synthases with different functions. Mol Microbiol. 1996;20:667–679. doi: 10.1046/j.1365-2958.1996.5571084.x. [DOI] [PubMed] [Google Scholar]
- Mellado E, Dubreucq G, Mol P, Sarfati J, Paris S, Diaquin M, Holden DW, Rodriguez-Tudela JL, Latge JP. Cell wall biogenesis in a double chitin synthase mutant (chsG-/chsE-) of Aspergillus fumigatus. Fungal Genet Biol. 2003;38:98–109. doi: 10.1016/s1087-1845(02)00516-9. [DOI] [PubMed] [Google Scholar]
- Morgenstern DE, Gifford MA, Li LL, Doerschuk CM, Dinauer MC. Absence of respiratory burst in X-linked chronic granulomatous disease mice leads to abnormalities in both host defense and inflammatory response to Aspergillus fumigatus. J Exp Med. 1997;185:207–218. doi: 10.1084/jem.185.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro CA, Gow NA. Chitin synthesis in human pathogenic fungi. Med Mycol. 2001;39(1):41–53. [PubMed] [Google Scholar]
- Murray IA, Coupland K, Smith JA, Ansell ID, Long RG. Intestinal trehalase activity in a UK population: establishing a normal range and the effect of disease. Br J Nutr. 2000;83:241–245. doi: 10.1017/s0007114500000313. [DOI] [PubMed] [Google Scholar]
- Ngamskulrungroj P, Himmelreich U, Breger JA, Wilson C, Chayakulkeeree M, Krockenberger MB, Malik R, Daniel HM, Toffaletti D, Djordjevic JT, Mylonakis E, Meyer W, Perfect JR. The trehalose synthesis pathway is an integral part of the virulence composite for Cryptococcus gattii. Infect Immun. 2009;77:4584–4596. doi: 10.1128/IAI.00565-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Yu JH. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS One. 2007;2:e970. doi: 10.1371/journal.pone.0000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwaka S, Mechler B, Destruelle M, Holzer H. Phenotypic features of trehalase mutants in Saccharomyces cerevisiae. FEBS Lett. 1995;360:286–290. doi: 10.1016/0014-5793(95)00105-i. [DOI] [PubMed] [Google Scholar]
- Panneman H, Ruijter GJ, van den Broeck HC, Visser J. Cloning and biochemical characterisation of Aspergillus niger hexokinase--the enzyme is strongly inhibited by physiological concentrations of trehalose 6-phosphate. Eur J Biochem. 1998;258:223–232. doi: 10.1046/j.1432-1327.1998.2580223.x. [DOI] [PubMed] [Google Scholar]
- Perlin DS, Mellado E. Antifungal Mechanisms of Action and Resistance. In: Latge JP, Steinbach WJ, editors. Aspergillus fumigatus and Aspergillosis. Washington, DC: ASM Press; 2008. p. 568. [Google Scholar]
- Petzold EW, Himmelreich U, Mylonakis E, Rude T, Toffaletti D, Cox GM, Miller JL, Perfect JR. Characterization and regulation of the trehalose synthesis pathway and its importance in the pathogenicity of Cryptococcus neoformans. Infect Immun. 2006;74:5877–5887. doi: 10.1128/IAI.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock JD, Williams DA, Gifford MA, Li LL, Du X, Fisherman J, Orkin SH, Doerschuk CM, Dinauer MC. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat Genet. 1995;9:202–209. doi: 10.1038/ng0295-202. [DOI] [PubMed] [Google Scholar]
- Ram AF, Klis FM. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat Protoc. 2006;1:2253–2256. doi: 10.1038/nprot.2006.397. [DOI] [PubMed] [Google Scholar]
- Reinders A, Burckert N, Hohmann S, Thevelein JM, Boller T, Wiemken A, De Virgilio C. Structural analysis of the subunits of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae and their function during heat shock. Mol Microbiol. 1997;24:687–695. doi: 10.1046/j.1365-2958.1997.3861749.x. [DOI] [PubMed] [Google Scholar]
- Saito K, Kase T, Takahashi E, Horinouchi S. Purification and characterization of a trehalose synthase from the basidiomycete grifola frondosa. Appl Environ Microbiol. 1998a;64:4340–4345. doi: 10.1128/aem.64.11.4340-4345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Yamazaki H, Ohnishi Y, Fujimoto S, Takahashi E, Horinouchi S. Production of trehalose synthase from a basidiomycete, Grifola frondosa, in Escherichia coli. Appl Microbiol Biotechnol. 1998b;50:193–198. doi: 10.1007/s002530051276. [DOI] [PubMed] [Google Scholar]
- Sano F, Asakawa N, Inoue Y, Sakurai M. A dual role for intracellular trehalose in the resistance of yeast cells to water stress. Cryobiology. 1999;39:80–87. doi: 10.1006/cryo.1999.2188. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Keller NP. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics. 2001;157:591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara ML, Correa A, Bell-Pedersen D, Dunlap JC, Loros JJ. Neurospora clock-controlled gene 9 (ccg-9) encodes trehalose synthase: circadian regulation of stress responses and development. Eukaryot Cell. 2002;1:33–43. doi: 10.1128/EC.1.1.33-43.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MA, Lindquist S. Thermotolerance in Saccharomyces cerevisiae: the Yin and Yang of trehalose. Trends Biotechnol. 1998;16:460–468. doi: 10.1016/s0167-7799(98)01251-7. [DOI] [PubMed] [Google Scholar]
- Snelders E, van der Lee HA, Kuijpers J, Rijs AJ, Varga J, Samson RA, Mellado E, Donders AR, Melchers WJ, Verweij PE. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 2008;5:e219. doi: 10.1371/journal.pmed.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur IP, Lobo Z, Maitra PK. Analysis of PFK3--a gene involved in particulate phosphofructokinase synthesis reveals additional functions of TPS2 in Saccharomyces cerevisiae. Yeast. 1994;10:199–209. doi: 10.1002/yea.320100207. [DOI] [PubMed] [Google Scholar]
- Thevelein JM. Regulation of trehalose mobilization in fungi. Microbiol Rev. 1984;48:42–59. doi: 10.1128/mr.48.1.42-59.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, Hohmann S. Trehalose synthase: guard to the gate of glycolysis in yeast? Trends Biochem Sci. 1995;20:3–10. doi: 10.1016/s0968-0004(00)88938-0. [DOI] [PubMed] [Google Scholar]
- Van Dijck P, De Rop L, Szlufcik K, Van Ael E, Thevelein JM. Disruption of the Candida albicans TPS2 gene encoding trehalose-6-phosphate phosphatase decreases infectivity without affecting hypha formation. Infect Immun. 2002;70:1772–1782. doi: 10.1128/IAI.70.4.1772-1782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkey JB, Perfect JR. Rare and emerging fungal pulmonary infections. Semin Respir Crit Care Med. 2008;29:121–131. doi: 10.1055/s-2008-1063851. [DOI] [PubMed] [Google Scholar]
- Verweij PE, Mellado E, Melchers WJ. Multiple-triazole-resistant aspergillosis. N Engl J Med. 2007;356:1481–1483. doi: 10.1056/NEJMc061720. [DOI] [PubMed] [Google Scholar]
- Vuorio OE, Kalkkinen N, Londesborough J. Cloning of two related genes encoding the 56-kDa and 123-kDa subunits of trehalose synthase from the yeast Saccharomyces cerevisiae. Eur J Biochem. 1993;216:849–861. doi: 10.1111/j.1432-1033.1993.tb18207.x. [DOI] [PubMed] [Google Scholar]
- Wannet WJ, Op den Camp HJ, Wisselink HW, van der Drift C, Van Griensven LJ, Vogels GD. Purification and characterization of trehalose phosphorylase from the commercial mushroom Agaricus bisporus. Biochim Biophys Acta. 1998;1425:177–188. doi: 10.1016/s0304-4165(98)00066-x. [DOI] [PubMed] [Google Scholar]
- White TC, Marr KA, Bowden RA. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA., Jr A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008;4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RA, Jenkinson JM, Gibson RP, Littlechild JA, Wang ZY, Talbot NJ. Tps1 regulates the pentose phosphate pathway, nitrogen metabolism and fungal virulence. EMBO J. 2007;26:3673–3685. doi: 10.1038/sj.emboj.7601795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelton MM, Hamer JE, Timberlake WE. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci U S A. 1984;81:1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Blazquez MA, Gancedo C. Disruption of the Candida albicans TPS1 gene encoding trehalose-6-phosphate synthase impairs formation of hyphae and decreases infectivity. J Bacteriol. 1998;180:3809–3815. doi: 10.1128/jb.180.15.3809-3815.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, de Virgilio C, Ponton J, Gancedo C. Disruption in Candida albicans of the TPS2 gene encoding trehalose-6-phosphate phosphatase affects cell integrity and decreases infectivity. Microbiology. 2002;148:1281–1290. doi: 10.1099/00221287-148-5-1281. [DOI] [PubMed] [Google Scholar]