Abstract
In response to an assault by foreign organisms, peripheral B cells can change their antibody affinity and isotype by somatically mutating their genomic DNA. The ability of a cell to modify its DNA is exceptional in light of the potential consequences of genetic alterations to cause human disease and cancer. Thus, as expected, this mechanism of antibody diversity is tightly regulated and coordinated through one protein, activation induced deaminase (AID). AID produces diversity by converting cytosine to uracil within the immunoglobulin loci. The deoxyuracil residue is mutagenic when paired with deoxyguanosine, since it mimics thymidine during DNA replication. Additionally, B cells can manipulate the DNA repair pathways so that deoxyuracils are not faithfully repaired. Therefore, an intricate balance exists which is regulated at multiple stages to promote mutation of immunoglobulin genes, while retaining integrity of the rest of the genome. Here we discuss and summarize the current understanding of how AID functions to cause somatic hypermutation.
1. INTRODUCTION
Diversity in antibodies is produced during two stages in B cell development. In pre-B cells, rearrangement of variable (V), diversity (D), and joining (J) gene segments occurs to produce the primary repertoire of immunoglobulin (Ig) receptors. In mature B cells, Ig receptors undergo affinity maturation (AM) and class switch recombination (CSR) to produce the secondary, or memory, repertoire of antibodies. The latter event occurs after antigen binds to the receptor, which initiates a dynamic cascade of cell signaling events to cause cellular activation (Gauld et al., 2002; Kurosaki, 2002; Niiro and Clark, 2002). The result of this activation is the differentiation of B cells into plasma or memory cells, which now express a large repertoire of antibodies to clear a plethora of different foreign antigens.
Diversity in the secondary repertoire is created by modifying rearranged V(D)J sequences and switching heavy chain constant genes (CH). Alteration of the V gene sequence is achieved by either direct mutagenesis or DNA strand breaks during gene conversion (GC), where strand breaks are repaired using different pseudo-V gene segments in a templated recombination mechanism. In either case, cells containing mutations which increase antibody affinity will be selected to divide and further mutate, while mutations which decrease affinity will be lost through apoptosis. Alteration of the CH gene occurs by DNA strand breaks in the switch (S) regions flanking the different CH gene exons. Breaks in two different S regions are then repaired by non-homologous end joining to remove the intervening introns and exons. This recombination event allows for production of a defined VDJ exon with different CH gene isotypes to regulate antibody function.
A single enzyme is responsible for initiating diversity in V(D)J and CH genes: activation induced deaminase (AID), which is a cytosine deaminase that enzymatically converts cytosine to uracil. Uracil is mutagenic when paired with guanosine in DNA, since dU mimics dT during replication, and the U:G mismatch triggers error-prone DNA repair in B cells. Thus, AID introduces somatic hypermutation (SHM) by converting dC to dU. In this review, the initiating events caused by AID will be referred to as SHM, regardless of whether dU is found in the V or S regions. If dU occurs in V(D)J genes, SHM can produce AM or GC. If dU occurs in S regions, SHM can produce CSR. Furthermore, the proteins that process dU, such as UNG, MSH2, MSH6, and DNA polymerases, have the same activity whether dU is located in the V(D)J or S regions. Therefore, SHM, caused by AID-generated dU, underpins the three mechanisms of AM, GC, and CSR.
One key aspect of AID biology is the balance between mutagenic diversity and genomic integrity. When AID functions at non-Ig loci, both mutation and translocations can promote carcinogenesis (Ramiro et al., 2007). Thus it is imperative to the organism that AID activity be tightly controlled to inhibit possible oncogenic transformation, while still allowing for the production of a wide diversity of antibodies. In this review, we will highlight the intricate aspects of AID biology and regulation.
2. AID, THE MASTER CATALYST
The mechanisms of AM, GC, and CSR were significantly advanced by the ground-breaking discovery of AID (Muramatsu et al., 1999) and its subsequent genetic analysis in humans, mice, and chickens (Arakawa et al., 2002; Rada et al., 2002b; Revy et al., 2000). Broader analysis of AID indicates that an intricate network of regulatory mechanisms controls its expression at the levels of gene transcription, mRNA stability, protein localization, protein phosphorylation, and cell signaling.
2.1. Gene transcription
The Aicda locus, which encodes AID, is comprised of four regions which control transcription (Yadav et al., 2006). Starting at the 5' end of the locus, the first region is located about 8 kb upstream of exon 1 in the mouse, and contains potential motifs for NF-κB, STAT6, C/EBP, and Smad3/4 proteins (Tran et al., 2009; Yadav et al., 2006). This region may respond to stimulation by the mitogen lipopolysaccharide (LPS) and the T-cell mimic anti-CD40 antibody to upregulate expression of AID after stimulation. The second region is located about 1 kb upstream of exon 1, and has sites for NF-κB, Stat 6, Sp transcription factors, HoxC4, and Pax5 (Dedeoglu et al., 2004; Gonda et al., 2003; Park et al., 2009; Yadav et al., 2006). The third region is found in the intron between exon 1 and exon 2, and contains sites for NF-κB, E proteins, Pax5, and several other factors (Gonda et al., 2003; Sayegh et al., 2003; Tran et al., 2009; Yadav et al., 2006). The fourth region is located about 6 kb downstream of exon 5 in the mouse (Tran et al., 2009; Yadav et al., 2006), and appears to function as an enhancer (Crouch et al., 2007). Many of the sites in the first three regions bind to transcription factors that are upregulated after B cell stimulation, and so they likely play a role in inducing AID in vivo.
Conversely, the proteins Id1, Id2, and Id3 reduce CSR (Goldfarb et al., 1996; Quong et al., 1999), potentially by inhibiting AID transcription (Gonda et al., 2003; Sayegh et al., 2003). The Id proteins function by binding to stimulatory factors such as E47 and Pax5, which prevents their binding to DNA. Other factors inhibit transcription by binding to sites in the third region (Tran et al., 2009), and may play a role in restricting AID expression to B cells, and not to other cell types.
Additional proteins bind to sites in the second region that appear to function independently of antigen stimulation. For example, Sp1 and Sp2 proteins bind to sites in vitro (Yadav et al., 2006), but their role in vivo is not known. Recently, the sex hormones estrogen and progesterone have been found to regulate AID expression (Pauklin and Petersen-Mahrt, 2009; Pauklin et al., 2009). Both estrogen and progesterone response elements have been found within the second region, and they could have a potential role in upregulating AID in hormone-based cancers and autoimmunity (Maul and Gearhart, 2009; Petersen-Mahrt et al., 2009). However, as with the Sp-binding sites, further research is required to understand the role these factors play during normal B cell development and activation. Finally, B cells from old mice and humans have less AID and reduced CSR compared to B cells from young mice and humans. This may be due in part to degradation of E47 mRNA, which encodes molecules that stimulate AID transcription (Frasca et al., 2008). In B cells from old mice, tristetraprolin binds to E47 mRNA and degrades it, whereas in B cells from young mice, tristetraprolin is phosphorylated and cannot bind to the mRNA (Frasca et al., 2007). Thus, by defining the factors that limit antibody diversity with age, it may be possible to increase the efficacy of vaccines in the elderly.
2.2. mRNA transcripts
Once the Aicda gene is transcribed, the level of transcripts can be controlled by regulation though microRNA molecules. Specifically, miR-155 binds to the 3’ untranslated region of AID mRNA, and destabilizes the message to reduce SHM and CSR. However, in vivo analysis of miR-155 function in SHM is complicated by the global defects seen in different lymphoid cells, which alters germinal center cell number and function (Kohlhaas et al., 2009; Thai et al., 2007; Vigorito et al., 2007). To overcome the global effect, specific mutants of the 3’ untranslated region of AID, which prevent binding of miR-155, were utilized to examine SHM and CSR in vivo. As predicted, AID expression was increased in splenic and Peyer's patch mutant B cells (Dorsett et al., 2008; Teng et al., 2008), and there was a dramatic increase in chromosomal translocations between Myc and Igh genes (Dorsett et al., 2008). However, the increased AID protein only modestly elevated the level of SHM in the V or S regions and decreased AM, suggesting that excess AID was not specifically targeted to the Ig locus. This is consistent with earlier studies showing that overexpression of AID does not always produce increased SHM or CSR, perhaps due to inactivation of the protein by an unknown mechanism (Muto et al., 2006). Another molecule, miR-181b, regulates AID expression in a similar manner, by binding to the 3' untranslated region and lowering the levels of AID and CSR (de Yebenes et al., 2008). Nonetheless, since both of these miR molecules affect multiple genes as well as Aicda, their biological role in AID expression remains unclear.
2.3. Protein localization
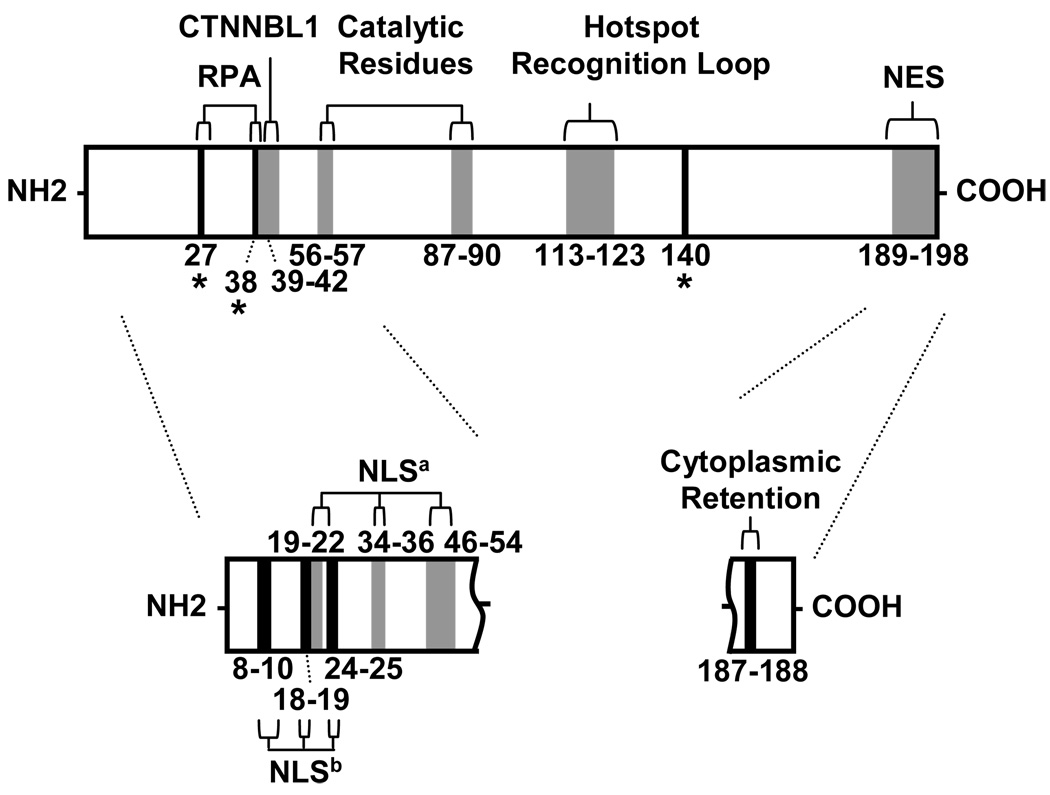
Extensive analyses have identified post-translational mechanisms that coordinate AID sub-cellular localization. Surprisingly, AID protein is far more abundant in the cytoplasm than in the nucleus, as first seen in the Ramos cell line using artificial AID-GFP constructs (Rada et al., 2002a) and in primary B cells looking at endogenous AID (Schrader et al., 2005). Three mechanisms appear to be involved in actively moving AID in and out of the nucleus, and their respective amino acid residues are illustrated in Fig. 1. First, high levels of AID may be retained in the cytoplasm through an anchor sequence in the C-terminal region of AID (Patenaude et al., 2009). This would be advantageous, as the protein is quickly degraded when it is in the nucleus through polyubiquitination (Aoufouchi et al., 2008). Second, AID is actively imported into the nucleus through the use of a N-terminal nuclear localization signal (NLS); however, the exact amino acids which form the NLS are currently unclear. Di Noia and colleagues speculated that a non-classical NLS exists in AID (Fig. 1 - NLSa) (Patenaude et al., 2009), while Honjo and colleagues have identified a classical bipartite NLS (Fig. 1 -; NLSb) (Ito et al., 2004; Shinkura et al., 2004). In addition it has also been proposed that AID may passively diffuse into the nucleus (Brar et al., 2004; McBride et al., 2004). However, the identification of an interaction of AID with importin-α suggests that AID does contain a NLS for active nuclear import (Patenaude et al., 2009). Third, a conserved nuclear export signal (NES) in the C-terminal residues 189–198 transports most of the protein out of the nucleus (Brar et al., 2004; Ito et al., 2004; McBride et al., 2004). Treatment of B cells with leptomycin B, a potent inhibitor of the CRM1 export receptor, increased the abundance of AID in the nucleus. Further dissection of the C-terminal domain showed that export can be abolished by the single point mutation F198A, resulting in increased SHM and CSR activity (McBride et al., 2004). Therefore, these three mechanisms exquisitely regulate the amount of AID in the nucleus, ensuring that there will be only low levels of the mutagenic protein after cell activation. Perturbing any one of these pathways can affect the fine balance between antibody diversity and chromosomal mutagenesis.
FIGURE 1.
Amino acids defining AID activity. Amino acids have been identified that bind co-factors, cause catalysis, recognize the hotspot motif, and shuttle the protein in and out of the nucleus. Confirmed residues that function in these activities are shown in the upper diagram. Amino acids that are phosphorylated are marked by asterisks and black bars; NES, nuclear export signal. Putative residues that may function are shown in the lower diagram. These include nuclear localization signals (NLS) in the N-terminal region identified by two groups, NLSa (Patenaude et al., 2009), in gray bars, and NLSb (Ito et al., 2004) in black bars. Two residues that may cause retention of AID in the cytoplasm are shown in the C-terminal region (Patenaude et al., 2009).
2.4. AID phosphorylation
While sub-cellular localization and degradation coordinate the access of AID to genomic DNA, phosphorylation regulates the activity of the protein. AID phosphorylation was first identified by examining catalytic differences between protein purified from B cells (AIDBcell) or from 293 kidney cells (AID293). Alt and colleagues reported that AID293 was less active than AIDBcell when tested for deamination activity using an in vitro transcription-based assay (Chaudhuri et al., 2004). Subsequent analysis by mass spectroscopy identified four phosphorylation sites in AID: T27, S38, T140, and Y184 (Basu et al., 2005; McBride et al., 2006; McBride et al., 2008; Pasqualucci et al., 2006). Residues T27 and S38 are phosphorylated in a coordinated fashion by protein kinase A (PKA), and they regulate protein-protein interaction between AID and replication protein A (RPA) (Basu et al., 2005; Pasqualucci et al., 2006). Mutation of these residues in B cells stimulated ex vivo or in DT40 cells inhibits SHM, GC, and CSR (Basu et al., 2005; Chatterji et al., 2007; McBride et al., 2006; Pasqualucci et al., 2006; Vuong et al., 2009). Indeed, S38 appears to be the key phosphorylation site in vivo, as mice with a mutation of this residue had reduced SHM and CSR (Cheng et al., 2009; McBride et al., 2008). Recently Nussenzweig and colleagues reported phosphorylation of AID residue T140 in mouse B cells after activation with LPS and IL-4 (McBride et al., 2008). Unlike S38, T140 is not a substrate for PKA phosphorylation but rather for protein kinase C (PKC). Furthermore, mutation of T140 to alanine in mice showed that SHM was affected more significantly than CSR, suggesting differential phosphorylation of S38 and T140 can produce different biological outcomes. In contrast to residues T27, S38, and T140, phosphorylation of Y184 may not play a significant role in AID function, since replacement of the amino acid in B cells did not have an effect on CSR (Basu et al., 2005).
2.5. Cell signaling
As implied in its name, AID is induced after activation by exogenous stimuli, such as bacterial or viral molecules, CD40 ligand, and antigen or anti-Ig. To study the signaling pathways for these activators, murine splenic B cells can be conveniently stimulated ex vivo, and the levels of AID and CSR can be measured. Abundant AID expression and CSR occurs after stimulation with LPS, which binds to toll-like receptor 4, or with anti-CD40 antibody, which binds to the CD40 receptor. In contrast, AID expression is delayed and CSR is ablated when cells are stimulated with anti-IgM, which binds to the Ig receptor (Heltemes-Harris et al., 2008; Jabara et al., 2008). Furthermore, AID expression and CSR are actually suppressed when anti-IgM is added to cells stimulated with LPS or anti-CD40 (Heltemes-Harris et al., 2008; Jabara et al., 2008; Rush et al., 2002). This inhibition has been linked to upregulation of the phosphatidyl inositol 3 kinase (PI3K) pathway (Doi et al., 2008; Heltemes-Harris et al., 2008; Omori et al., 2006). Indeed, balancing the levels of PI3K activation may determine whether CSR is induced or suppressed. B cells treated with LPS and IL-4 activate PI3K signaling, which phosphorylates AKT to a low enough concentration to allow CSR. In contrast, the addition of anti-IgM along with LPS and IL-4 enhanced AKT phosphorylation to a greater extent to inhibit CSR (Heltemes-Harris et al., 2008). A recent report shows that anti-IgM also inhibits AID expression through the calcium-signaling pathway (Hauser et al., 2008). Taken together, these reports suggest that IgM crosslinking ex vivo mimics the end of a germinal center response, where B cells with high affinity receptors stop SHM and are converted into memory and plasma cells. However, the complete story has yet to emerge, as another stimulator, 8-mercaptoguanosine, which binds to toll-like receptor 7 located in endosomes, has the ability to work synergistically with IgM crosslinking to promote AID expression and CSR (Tsukamoto et al., 2009). Perhaps after B cell receptor crosslinking, stimulation from different cellular microenvironments regulates the outcome of B cell differentiation.
3. THE TARGETING ENIGMA
3.1. Targeting to the Ig loci
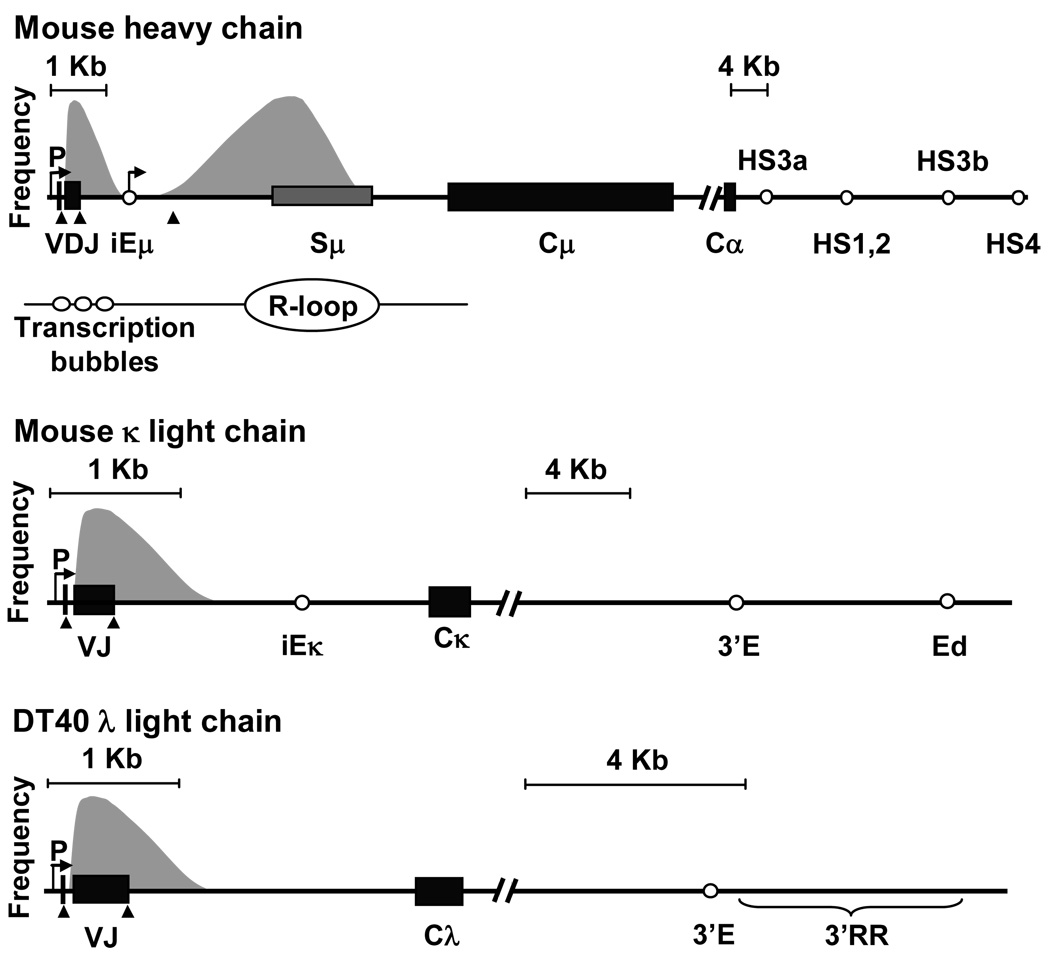
The immunoglobulin loci are mutated in well defined regions encoding rearranged V genes on the heavy and light chain loci, and S regions on the heavy chain locus. Sequence analysis has shown that mutation occurs in a 2 kb region around V(D)J genes (Lebecque and Gearhart, 1990), and in a 4–7 kb region around S regions (Xue et al., 2006). Thus it can be assumed that AID functions on 10−5 to 10−6 of the genome at a given time, suggesting that exquisite levels of regulation target AID to such a small percentage of the genome. Examination of the mutational pattern in both the V(D)J and the S regions shows that mutation occurs in close proximity to either the V gene promoter or S intron promoters, respectively. This has led to the hypothesis that AID is recruited to the Ig region in association with the transcriptional machinery. In fact, mice with transgenes that have been altered to express different amounts of transcripts have mutation frequencies that correlate with the level of transcription (Bachl et al., 2001; Fukita et al., 1998; Sharpe et al., 1991). Transcription of the heavy and light chain loci is coordinated by three well characterized promoters/enhancers shown in Fig. 2: V gene promoter, intronic enhancer (iE), and downstream enhancers (3’E and hypersensitive (HS) sites). While all three elements and the V/S regions are involved in transcription and SHM, the specific role for each has been harder to elucidate.
FIGURE 2.
Targeting elements for SHM in the Ig loci. For all loci, the relative mutation frequency is shown by shaded peaks over the V region or S region. P, promoter; arrows, start of transcription; iE, intronic enhancer; triangles, location of donor splice sites in the leader exon, V(D)J exon, and iEμ intronic exon. AID may be active in the V regions by transcription bubbles, and in the S regions by R-loops. 3’ enhancers downstream of the C genes are shown on a different scale. For the mouse Igh chain locus, 3’ enhancers include 4 hypersensitive (HS) sites. For the mouse Igκ locus, there are two enhancers, 3’E and Ed. For the chicken DT40 Igλ locus, identified enhancers are 3’E and a regulatory region, 3’RR.
The V gene promoter can be replaced with other non-Ig promoters in transgenes and still promote SHM, suggesting that the promoter may only serve to maintain adequate levels of transcription (Betz et al., 1994; Fukita et al., 1998; Shen et al., 2001; Tumas-Brundage and Manser, 1997). However, this may not be the whole story since at least one promoter, human elongation factor 1-α, can induce high levels of transcription without supporting SHM (Yang et al., 2006). Furthermore, substitution of the V gene promoter has not been studied in the endogenous locus with knock-in mice, so its requirement is not fully resolved.
The seeming lack of specificity for promoters would suggest that the V(D)J sequence contains the information required for targeting. However, when the VJ sequence in removed and replaced by different sequences in Igκ transgenes, the new sequence is still subject to SHM (Peters and Storb, 1996; Yelamos et al., 1995). Additionally, when a 750 bp insert is introduced between the leader sequence and the VDJ sequence, the mutation window shifts ~750 bp towards the promoter (Tumas-Brundage and Manser, 1997). Interestingly, when the promoter is removed from the leader sequence by insertion of 2 kb of λ DNA, SHM is lost, indicating that targeting may be linked by the proximity of the promoter to the leader sequence and/or the leader splice site (Winter et al., 1997). However, the caveat still remains that replacement of the V(D)J sequence in the endogenous context has not been studied, and so its requirement is not certain.
Switch regions are also a target for SHM, and sustain as high a frequency of mutation as the V region. Knockout mice with a partial deletion of Sμ tandem repeats had decreased CSR (Luby et al., 2001; Schrader et al., 2007), and when the entire 4.6 kb region containing Sμ tandem repeat sequences was deleted, SHM and CSR were ablated (Khamlichi et al., 2004). Thus the repetitive sequences in Sμ are a magnet for AID activity, and are required.
Intronic enhancers are located in the intron sequence between either the VDJ sequence and the S region for Cμ, or the VJ sequence and C gene in the Igκ locus. A plethora of papers in the 1990's addressed the role of iE for SHM in murine transgenes encoding rearranged V and C genes from the Igh and Igκ loci (Odegard and Schatz, 2006). Interpretation of the varying results was complicated by the random location of transgenes in the genome, which could affect transcription levels. As technology advanced, it became possible to directly delete iE from the endogenous locus in knockout mice. These studies consistently showed that iE had no effect on SHM in the Igh or Igk loci (Inlay et al., 2006; Perlot et al., 2005).
3' enhancers are located downstream of the C genes on the heavy and light chain loci, and are important for transcription of rearranged V genes. Interestingly, it now appears that all three loci have multiple 3' enhancers which play a role in SHM. As in the iE studies, conflicting results were obtained from transgenic mice, whereas more reliable data was found in germline knockout mice. The IgH 3’E is characterized by the presence of four DNaseI HS sites downstream of the Cα gene (Dariavach et al., 1991; Lieberson et al., 1991; Madisen and Groudine, 1994; Matthias and Baltimore, 1993; Pettersson et al., 1990). Partial deletion of the HS sites did not affect SHM (Le Morvan et al., 2003), whereas deletion of the entire four sites in a 230 kb bacterial artificial chromosome mouse model showed reduced transcription and SHM (Dunnick et al., 2009). The Igκ locus has a 3'E (Meyer and Neuberger, 1989) and a downstream enhancer, Ed (Liu et al., 2002). Deletion of 3'E reduced transcription but did not affect SHM (Inlay et al., 2006; van der Stoep et al., 1998), whereas deletion of Ed reduced both transcription and SHM (Xiang and Garrard, 2008). Likewise, the chicken Igλ locus has a defined 3'E (Bulfone-Paus et al., 1995) and another downstream enhancer, 3'RR (Kothapalli et al., 2008). In the DT40 cell line, deletion of the 3'E had no effect on SHM (Yang et al., 2006), but deletion of 3'RR ablated transcription and SHM (Kothapalli et al., 2008). Thus, the presence of two enhancers for both Igκ and Igλ explains why the low levels of SHM, previously seen with single deletions, may be due to residual activity of the additional elements. Furthermore, in DT40 cells, the 3'RR appears to contain a site that recruits AID to the Igλ locus (Blagodatski et al., 2009; Kothapalli et al., 2008).
To summarize, transcription is necessary for SHM. The V gene promoter may not be required, the V(D)J sequence may not be required, the S region sequence is required, the iE is not required, and both the 3' E and downstream enhancers are required. Finally, the downstream enhancer may contain a motif that recruits AID to the Ig loci, although more work is needed to establish this.
In addition to AID being recruited to the Ig loci, it has become increasingly clear that AID is erroneously targeted to non-Ig genes throughout the genome. In the absence of DNA repair proteins uracil DNA glycoslyase (UNG) and mismatch repair protein MSH2, Schatz and colleagues found a high mutation frequency for several non-Ig genes which was only 10-fold lower than in V genes (Liu et al., 2008). The handful of different genes that might be targets for AID suggests that the promoters/enhancers of Ig genes are not the only elements which attract AID. Interestingly, the recent finding that Igλ, Igh, and Myc are spatially contained in close proximity in the nucleus suggests that nuclear organization affects both Ig and non-Ig targeting (Wang et al., 2009a).
Other transcriptional events such as chromatin acetylation may play a role in coordinating AID activity. Examination of the abundance of histone acetylation upon stimulation in vivo or ex vivo, suggests that the V and Sμ regions are maintained in a hyperacetylated (open) state independent of cellular activation, and the C region had a lower level of acetylation (Li et al., 2004a; Odegard et al., 2005; Wang et al., 2006; Wang et al., 2009b). Furthermore, downstream S regions are maintained at a low level of acetylation until stimulated with specific cytokines to promote transcription (Li et al., 2004a; Nambu et al., 2003; Wang et al., 2006; Wang et al., 2009b; Woo et al., 2003). While these events seem to be independent of AID, the difference in the acetylation state might coordinate a functional window for AID activity in the V and S regions, and block activity in the C region.
3.2. Sub-targeting to V and S regions
Once recruited to the Ig loci, AID-generated mutations show a distinctive bell-shaped pattern, suggesting increased activity in particular regions (Fig. 2). This pattern of mutation is advantageous to antibody diversity in that the peak of mutation is either over the V exon or the repetitive core in the S regions. Thus, in the V region, SHM promotes AM and GC, and in the S region, SHM initiates double strand breaks for CSR. This focusing of mutation in a defined region, or sub-targeting, is determined by the nucleotide sequence of the loci which coordinates transcription and AID activity.
Targeting to the V region is similar for rearranged V genes on the Igκ, Igλ, and Igh loci. Mutations start just downstream of the V promoter, proceed for about 2 kb, and then trail off (Lebecque and Gearhart, 1990). The pattern is the same regardless of which J gene segment is being used. For example, if a V gene rearranges to JH1, mutations cease 1550 bp before the intronic enhancer, iEμ, whereas if a V gene rearranges to JH4, mutations end just before iEμ. This indicates that the V gene promoter determines the start of mutation, and the intronic enhancer does not stop mutation. Rather, AID appears to be recruited to the promoter, proceeds for two kb and then may dissociate from the ongoing transcription complex. Indeed, one of the most perplexing questions is, why do mutations start and why do they stop? The frequency of mutations, about 10−2 to 10−3 mutations per bp, is similar in both coding and flanking sequences around the V gene, and is higher in the complementarity-determining regions due to selection during AM for amino acids giving high affinity interactions with the cognate antigen.
Targeting to the intronic S region promotes a high level of deamination events in close proximity to allow for double strand break formation. Mutations start downstream of the intronic exon promoter, accumulate for about 4–7 kb depending on the length of the S region, and then fall off (Xue et al., 2006). The pattern, although longer, is thus similar to that in the V region, in that AID may assemble at the intronic promoter, proceed through the S region, and then dissociate. The S regions in mammals are unique in that they contain clusters of G nucleotides on the nontranscribed strand, and repetitive hotspot motifs for AID deamination, WGCW (W = A or T). The G clusters have been shown to form R-loops, or RNA-DNA hybrids, in vivo (Yu et al., 2003) and in vitro (Roy and Lieber, 2009). The corresponding C clusters on the transcribed strand can stably hybridize to G-rich RNA. One effect of R-loop structure is that the nontranscribed strand becomes single-stranded in regions larger than transcription bubbles to maximize deamination by AID. In contrast to mice, switching in frogs is not as efficient, since the Sμ regions do not have G clusters or R-loops (Zarrin et al., 2004).
A second effect of R-loop structure is to slow down RNA polymerase II molecules as they move through the S region. It has been shown in vitro that R-loops block transcription (Canugovi et al., 2009; Tornaletti et al., 2008), because the polymerases may have difficulty unwinding the stable RNA-DNA hybrid. A recent study (Rajagopal et al., 2009) measured the density of polymerases in vivo by nuclear run-on, and found a 5–10 fold increase in polymerases located in close proximity to the Sμ repetitive core. These polymerases appeared to be piling up because they encountered a road-block ahead caused by the RNA-DNA hybrids. Once the polymerases slowly make it through the repetitive core, they speed up again as the R-loop density is lower. In addition to Sμ, R-loops are present in other S regions (Huang et al., 2006; Yu et al., 2003), and RNA polymerase II has been shown to accumulate in Sγ3 (Wang et al., 2009b), which suggests a conserved mechanism for deamination in S regions. If AID is associated with transcription, this model of paused polymerases may allow AID more opportunity to deaminate DNA, producing more mutations and strand breaks.
Is AID differentially targeted to V or S regions during certain cellular responses? This may seem to occur during stimulation of B cells ex vivo with mitogens, where mutations are found only in S regions and not in V regions (Reina-San-Martin et al., 2003). Conversely, in B cell lines such as Ramos and DT40, mutations accumulate in V regions and apparently not in S regions, as the cells do not undergo switching. In addition, IgM memory cells from humans (Klein et al., 1998; Rosner et al., 2001) and mice (Dogan et al., 2009) have mutations in the V region, but have not switched isotypes. This suggests that specific co-factors may guide AID to the V or S regions; however, other interpretations are possible. Mutations may occur first in S regions in cells stimulated ex vivo because of the formation of R-loops and RNA polymerase II pausing, which magnifies AID activity. Likewise, B cell lines or memory IgM cells may have mutations in S regions but not switch because some proteins involved in NHEJ are not functioning. It would be interesting to compare the time course of mutations in V and S regions following immunization in vivo, to see if they occur simultaneously or differentially.
3.3. Fine targeting to hotspots
As mentioned above, SHM occurs at a greater frequency in a defined sequence motif, WRC (W = A/T, R = A/G) (Rogozin and Kolchanov, 1992). Prior to the discovery of AID, sequence analysis of V genes indicated that the complementarity-determining regions are heavily biased in using the serine codons AGC or AGT, while framework regions utilized the TCN serine codons (Wagner et al., 1995). Additionally, SHM occurred in AGY (Y = C/T) codons at a higher rate than in TCN, indicating a bias to focus mutation within the complementarity-determining regions for AM. Further analyses using in vivo mouse models deficient for different DNA repair enzymes, have defined WGCW in V and S regions as the mutational hotspot for SHM (Delbos et al., 2007; Ehrenstein and Neuberger, 1999; Martomo et al., 2004; Rada et al., 1998). Characterization of AID activity in vitro indicates that deamination events occur with high frequency within the WRC context (Bransteitter et al., 2004; Larijani et al., 2005; Yu et al., 2004). As with AM in V genes, CSR has evolved to utilize these hotspots to focus AID activity to the WGCW motif within the repetitive core repeat (Davis et al., 1980; Dunnick et al., 1980; Kataoka et al., 1981; Sakano et al., 1980). Additionally, the frequency and palindromic nature of the motif allows for deamination events on both strands to occur in close proximity to promote double strand break formation versus mutagenic repair.
In addition to the catalytic residues in the AID protein, a loop at residues 113–123 has recently been identified as the main determinant in directing activity to the WRC hotspot (Fig. 1). Altering this loop to resemble the homologous loop from APOBEC family members switches the hotspot motif to that of the APOBEC enzyme (Kohli et al., 2009; Wang et al., 2010). Taken together, this indicates a co-evolution of both the Ig loci sequence and the AID enzyme.
3.4. Access to both DNA strands
An additional phenomenon of AID activity within the Ig loci is the ability of AID to access and mutate both strands. Mutational analysis in Ung−/−Msh2−/− and Ung−/−Msh6−/− mice indicates that both the transcribed and non-transcribed strands are mutated at an equal frequency (Rada et al., 2004; Shen et al., 2006; Xue et al., 2006). Most models for AID deamination suggest that AID can access single strand DNA within transcription bubbles and/or R-loops. However, these models would only allow for deamination of the non-template strand as the template strand would be associated with either the RNA polymerase or contained within an RNA-DNA complex. To achieve access to both strands, it has been proposed that antisense transcription occurs throughout the Ig loci. In support of this model, RT-PCR has been utilized to identify low levels of antisense transcripts in V and S regions (Chowdhury et al., 2008; Perlot et al., 2008; Ronai et al., 2007). Although these findings have been called into question (Zhao et al., 2009), the identification of ~11 nt single strand DNA patches on both strands of DNA in Ramos cells supports the presence of transient transcription bubbles moving in opposite directions (Ronai et al., 2007). However, the finding that cytosines are also mutated on both strands in the S regions ((Xue et al., 2006) is particularly perplexing, since the transcribed strand contains an RNA-DNA hybrid in the R-loop structures. Two other theories have been proposed to make this strand available for attack by AID. (1) The DNA upstream of an elongating RNA polymerase II may be supercoiled and unwound, which allows AID access to both strands (Shen and Storb, 2004). (2) The DNA in R-loops may be collapsed by endogenous RNase H digestion, which would expose single strand regions on the transcribed strand (Huang et al., 2007). Taken together, this data suggests that the models for AID activity within the Ig loci are still in a state of flux and require further experimentation to fully define AID targeting.
3.5. Protein co-factors
In addition to the mechanisms discussed above, extensive work has been performed in an attempt to identify AID protein partners. It has been hypothesized that targeting will be tightly regulated by protein co-factors which coordinate the recruitment and activity of AID. With recent advances in AID protein biochemistry, the intricate network of AID interactions is just beginning to emerge.
As discussed in section 2.4, AID is phosphorylated by PKA and PKC (Basu et al., 2005; McBride et al., 2008; Pasqualucci et al., 2006). The phosphorylation by PKA is required for interaction with RPA and disruption of this interaction inhibits AID activity (Basu et al., 2005; Cheng et al., 2009; McBride et al., 2006; Vuong et al., 2009). While these interactions are well characterized, the precise mechanism by which RPA assists AID is unclear. Recently Chaudhuri and colleagues found that neither PKA or RPA are required to physically recruit AID to DNA (Vuong et al., 2009). In vitro analysis suggests that RPA stabilizes the transcription bubble to allow AID activity on single strand DNA (Chaudhuri et al., 2004). Yet it remains unclear whether the interaction between the proteins promotes a coordinated hand-off of the DNA between the two proteins, and if the RPA-AID interaction exists during the deamination step.
While a genetic interaction between transcription and SHM has been well documented, very little is know about the role of the transcription complex in physically recruiting AID to the DNA. AID has been shown to physically interact with RNA polymerase II; however no further analysis was done to examine which subunit is responsible for this interaction (Nambu et al., 2003). Recently, Neuberger and colleagues identified an interaction between AID and CTNNBL1. Deletion of CTNNBL1 or mutation of AID residues 39–42 abolished AID activity (Conticello et al., 2008). Interestingly, CTNNBL1 physically interacts with proteins associated with the RNA polymerase II spliceosome, suggesting that AID may travel with the transcription complex. This interaction also highlights the potential role for splice sites in initiating AID activity on DNA. In a critical experiment, Radbruch and colleagues (Hein et al., 1998) reported that switching was abrogated when the splice donor site for Iγ1 was deleted, even though Iγ1 transcripts were made. Another study showed that mice lacking the splice donor site for the Iμ exon had switching, but splicing still occurred in transcripts using pseudo-splice donor sites (Kuzin et al., 2000). Thus, AID could be brought to the Ig loci through interaction with cis-acting elements (potentially using E2A family members (Michael et al., 2003; Schoetz et al., 2006; Tanaka et al., 2010)), bind to the RNA polymerase II spliceosome complex through CTNNBL1, and load onto DNA at donor splice sites to interact with RPA and deaminate cytosine residues. This may explain why mutation is highest in V genes after donor splice sites in the leader and V exons, and in S regions after donor splice sites in intronic exons preceding Sμ, Sγ, and Sα.
4. ROGUE URACILS
4.1. Deoxyuracil in DNA
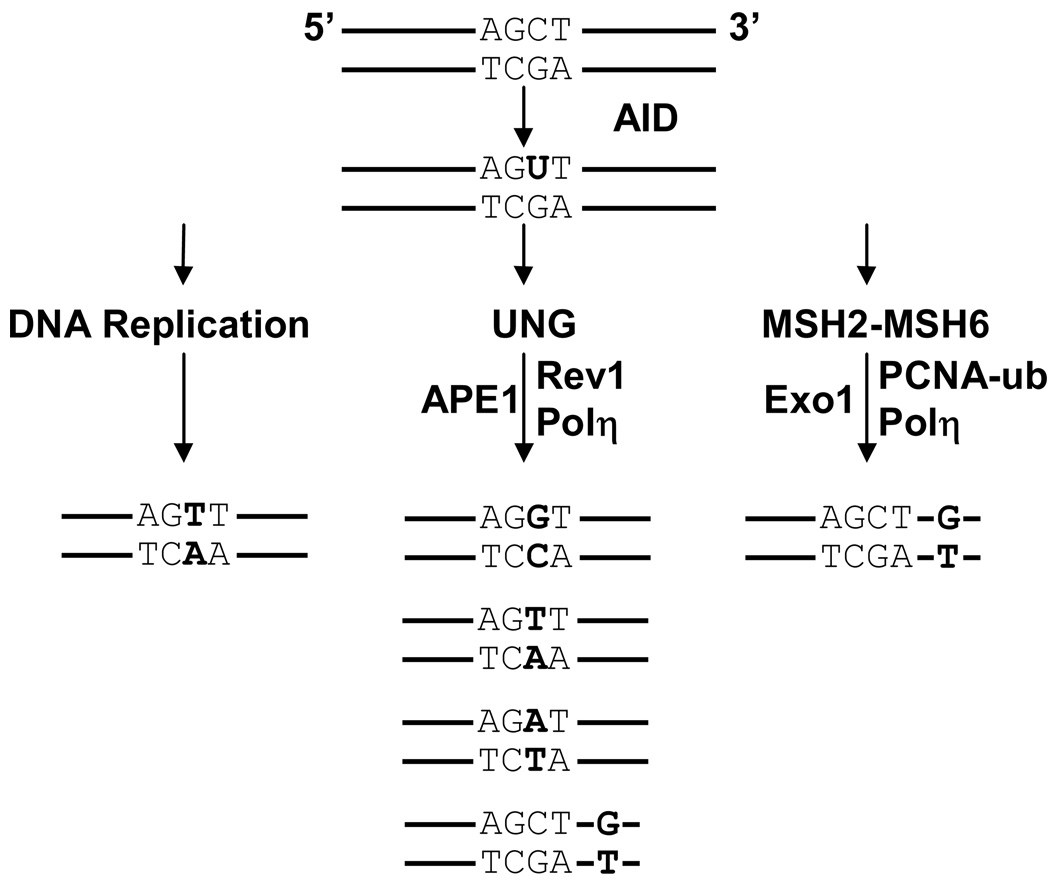
Since the first identification of AID, extensive work has been performed in an attempt to elucidate the mechanism of how it promotes genomic mutation. Initial identification of the sequence similarity between AID and APOBEC-1 suggested that AID may function as a RNA deaminase (Muramatsu et al., 1999). Honjo and colleagues proposed a RNA editing model where AID binds to an unidentified mRNA partner in the cytoplasm and deaminates C to U. The edited mRNA would then produce a protein, perhaps an endonuclease, that cleaves DNA during the immune response (Honjo et al., 2005). Alternatively, identification of a mutational hotspot in SHM (WRC, discussed above) suggested a mechanism where alterations occur directly at dC:dG basepairs (Rada et al., 1998). Neuberger and colleagues proposed a DNA deamination model where AID deaminates dC bases to dU, which initiates error-prone processing by some proteins in the base excision repair (BER) and mismatch repair (MMR) pathways. Support for direct deamination of DNA came from the finding that uracil DNA glycosylase (UNG or UDG) is required for CSR and GC, and alters SHM frequency and mutational spectra (Di Noia and Neuberger, 2002; Di Noia and Neuberger, 2004; Petersen-Mahrt et al., 2002; Rada et al., 2002b; Saribasak et al., 2006). The importance of UNG to the mechanism of CSR is further confirmed by genetic mutations in the human UNG gene that block CSR and cause hyper-IgM syndrome (Imai et al., 2003; Kavli et al., 2005). During classical BER, UNG binds to dU:dG mispairs in DNA, and the uracil base is cleaved to form an abasic site. Abasic sites are then cleaved by apurinic/apryimidinic endonuclease (APE1) to remove the abasic nucleotide, and DNA polymerase (Pol) β re-synthesizes the DNA strand. However, during SHM and CSR, the canonical mechanism of BER is impaired by altering the re-synthesis step with low-fidelity polymerases, which introduce mutations and single strand DNA breaks. Consistent with this model shown in Fig. 3, deletion or inhibition of UNG and APE1 results in decreased CSR (Guikema et al., 2007; Rada et al., 2002b; Schrader et al., 2005). Alternatively, deletion of Polβ supports increased CSR and double strand breaks by inhibiting the faithful re-synthesis step of canonical BER (Wu and Stavnezer, 2007).
FIGURE 3.
Three error-prone pathways for processing U:G mismatches. AID deaminates dC to dU in a hotspot, WGCW. Mutations are shown in bold. (1) DNA replication. U mimics T, and replication from the 5’ strand will incorporate C:G to T:A transitions. (2) UNG recognition. UNG removes the U, and several possibilities could occur. Rev1 may bypass the abasic site with a C:G to G:C transversion. APE1 may nick the abasic site, and the 5’ strand would be extended by a low-fidelity polymerase to insert other transversions. Polη may extend the synthesis to insert G opposite a downstream template T, to cause A:T to G:C transitions. (3) MSH2-MSH6 recognition. The two proteins bind to the U:G mismatch and recruit Exo1 to form a gap. It is not known what triggers a nick for Exo1 to access the DNA. Mono-ubiquitinated PCNA brings in Polη to synthesize predominantly A:T mutations downstream of the initial deamination event.
In opposition to the existence of uracil in DNA, several reports from Honjo and colleagues have suggested that UNG is important for CSR through an alternative mechanism not requiring DNA glycosylase activity (Begum et al., 2004). Mutants of active site residues in UNG had no detectable glycosylase activity in vitro, but they were proficient for CSR when complementing Ung−/− cells (Begum et al., 2004; Begum et al., 2009). Additionally, the identification of normal γH2AX foci formation and strand break junctions in the absence of UNG supports a model by which UNG is involved in resolving the double strand breaks, perhaps as a scaffold for other proteins, but not in the direct formation of the breaks (Begum et al., 2007). However, these results have been called into question due to a possible dissociation between in vitro UNG glycosylase activity and in vivo CSR (Di Noia et al., 2007; Kavli et al., 2005; Stivers, 2004). It has been shown that several UNG active site mutants with severally diminished in vitro activity may still retain enough glycosylase activity in vivo to promote CSR. Honjo and colleagues also report that deletion of either APE1 or APE2 had no effect on CSR (Sabouri et al., 2009), in contrast to an earlier finding by Stavnezer and colleagues (Guikema et al., 2007). Furthermore, the results looking at γH2AX and strand breaks did not take into account the interplay between BER and MMR in causing double strand breaks and CSR in the S region, since it is possible that breaks were still being formed due to cleavage during MMR (Di Noia et al., 2007; Rada et al., 2004; Shen et al., 2006).
Additional evidence for AID acting as a DNA deaminase comes from direct analysis of the purified protein in vitro. Using specific single strand DNA substrates, recombinant AID is able to convert a single dC residue to dU, creating a UNG/APE1 sensitive substrate (Bransteitter et al., 2003; Dickerson et al., 2003). Additionally, looking at whole cellular extracts from either splenic B cells or HEK293T cells expressing recombinant AID, single strand DNA oligomers containing multiple hotspot motifs become susceptible to treatment with UNG and APE1 (Chaudhuri et al., 2003). Futhermore, double strand DNA substrates were protected from AID deamination except in the presence of transcription, which explains the requirement for a single strand DNA substrate (Chaudhuri et al., 2003; Ramiro et al., 2003). However, in these experiments, AID was able to bind to RNA molecules, albeit with lower affinity than the single strand DNA template, allowing for the slight possibility that AID may also function on RNA (Dickerson et al., 2003). Additionally, the known RNA editing enzyme APOBEC1 has residual activity on single strand DNA in similar assays, suggesting these enzymes might act promiscuously (Harris et al., 2002). Taken together with the genetic data, most evidence suggests that AID functions as a DNA deaminase. However, the true test of AID activity would be to directly detect the accumulation of dU residues in genomic DNA during an immune response, which has yet to be characterized at this time.
4.2. Mismatch repair and DNA polymerases
In addition to being processed by UNG, deoxyuracil can be recognized by some proteins in the MMR pathway (Fig. 3). The canonical role for MMR is to remove DNA mismatches and repair DNA in an accurate manner. The MSH2-MSH6 or MSH2-MSH3 heterodimer binds to a mismatch, and recruits MLH1-PMS2 to the site. This nicks the DNA downstream of the mismatch, and attracts exonuclease 1 (Exo1) to remove the strand containing the mismatch. The gap is then filled in by high fidelity Polδto restore the original sequence. However, during the immune response, deficiency in some MMR proteins resulted in decreased, not increased, mutation frequencies, suggesting that these proteins actually generate mutations. Extensive analyses have examined the roles of the above proteins in processing mismatches in V and S regions (Bardwell et al., 2004; Ehrenstein and Neuberger, 1999; Ehrenstein et al., 2001; Frey et al., 1998; Jacobs et al., 1998; Kim et al., 1999; Kong and Maizels, 1999; Li et al., 2004b; Li et al., 2006; Martin et al., 2003; Martomo et al., 2004; Phung et al., 1999; Phung et al., 1998; Rada et al., 2004; Rada et al., 1998; Schrader et al., 2003; Shen et al., 2006; Wiesendanger et al., 2000; Winter et al., 1998). The prevailing model is that MSH2-MSH6 binds to a U:G mismatch and recruits DNA Polη (Wilson et al., 2005), a low-fidelity polymerase that preferentially synthesizes mispairs when copying T nucleotides (Matsuda et al., 2001). This explains why the frequency of mutations at A:T bp drops dramatically in the absence of MSH2, MSH6, and Exo1, which interact with Polη. In contrast, mice deficient for the other MMR proteins had no alteration in the SHM spectra.
Throughout evolution, specialized DNA polymerases have evolved to copy DNA with low fidelity to bypass DNA damaging lesions. To see if these polymerases are recruited to the Ig loci to increase sequence diversity, SHM was examined in mice deficient for 8 polymerases. Pols β,μ, λ, and ι are not involved in SHM (Bertocci et al., 2002; Esposito et al., 2000; Martomo et al., 2006; McDonald et al., 2003), while the role of Polθ is currently unclear due to conflicting results (Martomo et al., 2008; Masuda et al., 2007; Masuda et al., 2006; Masuda et al., 2005; Zan et al., 2005). However, there is well defined evidence that Polζ, Rev1, and Polη, have distinct roles during SHM. Conditional inactivation of Polζ in mice resulted in ~2–3 fold decrease in mutation frequency without altering mutation spectra, consistent with a role for Polζ in extending DNA mismatches (Diaz et al., 2001; Schenten et al., 2009; Zan et al., 2001). However, due to the embryonic lethality and genomic instability seen in Polζ deficient mice and B cells, the full extent for a role of Polζ has yet to be defined. Rev1 is a cytidyl transferase which causes G:C to C:G transversions in SHM (Arakawa et al., 2006; Jansen et al., 2006; Masuda et al., 2009; Ross and Sale, 2006). More recently, a catalytically inactive Rev1 mutant has been examined, and suggests a minor role for mouse Rev1 contributing to transition mutations as well (Masuda et al., 2009), however this was not seen in DT40 cells (Ross and Sale, 2006).
As compared to the modest roles for Polζ and Rev1, Polη has been shown to contribute significantly to diversity during SHM. Genetic mutation of Polη in humans with xeroderma pigmentosum variant disease or deletion of the gene in mice, resulted in a dramatic decrease in mutations at A:T residues with a modest decrease in overall mutation frequency (Delbos et al., 2005; Faili et al., 2004; Zeng et al., 2004; Zeng et al., 2001). Significantly, the decrease in A:T mutations is similar to the effects seen in mice with deficiencies in MSH2, MSH6, and Exo1, suggesting that they all act in the same pathway. Additionally, the MSH2-MSH6 heterodimer physically and functionally interacts with Polη, suggesting a shared role in producing A:T mutations during SHM (Wilson et al., 2005). However, close examination of the spectra from Msh6−/− mice or double deletion of both MSH2 and Polη indicates that the effects are not completely overlapping (Delbos et al., 2007; Martomo et al., 2005). Individually Polh−/− (which encodes Polη) or Msh2−/− mice display ~15% residual A:T mutations, while the double knockout shows only ~1% A:T mutations. This suggests that while Polη and MSH2 function together, they are also able to contribute to SHM independently of each another. Interestingly, a recent paper by Reynaud and colleagues analyzed a Polk−/− Polh−/− mouse strain which shows ~7% mutations at A:T, suggesting that Polκ may function during SHM and contribute a modest amount of mutations to the overall spectra (Faili et al., 2009). The identification of Polκ dependent mutations is significant as previous reports failed to identify such a role (Schenten et al., 2002; Shimizu et al., 2005; Shimizu et al., 2003). However, it is difficult to differentiate between the possibility that Polκ function is obscured by the high mutation rate of Polη, or if the normal presence of Polη blocks Polκ access in wildtype mice. The residual A:T mutations seen in the absence of both Polη and Polκ also suggests a third polymerase can function in SHM. The total lack of A:T mutations in the Polh−/−Msh2−/− mice indicates that MSH2 is responsible for recruiting Polη, Polκ and perhaps other polymerases.
One key aspect which promotes error-prone replication is the role of proliferating cell nuclear antigen (PCNA) monoubiquitination. PCNA is a replication accessory factor which functions in recruiting, tethering, and switching DNA polymerases at the primer-template junction. To coordinate these events, PCNA is post-translationally modified at residue K164 to either initiate error-free or error-prone repair (Ulrich, 2009). Mutation of the K164 residue, or deletion of PCNA ubiquitin ligase Rad18, resulted in a dramatic decrease in A:T mutations during SHM (Arakawa et al., 2006; Bachl et al., 2006; Langerak et al., 2007; Roa et al., 2008). This suggests that PCNA modification is regulated to cause DNA synthesis by Polη in activated B cells. Additionally, loss of PCNA ubiquitination in DT40 cell lines, but not mice, showed a decrease in overall mutation frequency, indicating an increased utilization of PCNA-Ub in DT40 (Arakawa et al., 2006; Bachl et al., 2006). Interestingly, in DT40 cells, the combination of a PCNA mutant and deletion of Rev1 showed almost complete loss on SHM, suggesting a potential inability of the canonical DNA replication machinery to bypass dU (Arakawa et al., 2006). Therefore it would be interesting to further understand the role of PCNA modification in SHM, as the K164 mutation also effects other modifications such as sumoylation and polyubiquitination.
Finally, do the UNG and MSH2-MSH6 pathways shown in Fig. 3 operate at the same time and compete for the same dU? Some models suggest they do (Rada et al., 2004; Schanz et al., 2009), whereas others propose the pathways are temporally separated during the G1 and S phases of the cell cycle (Krijger et al., 2009; Weill and Reynaud, 2008). A non-competitive model is appealing in that MSH2-MSH6 could recognize U:G in double strand DNA during G1, and UNG would be most active on single strand DNA in replication during S phase. This latter model is based on the recent finding that UNG is weakly expressed during G1 and is upregulated during early S phase (Hagen et al., 2008). However, Stavnezer and colleagues show that UNG is fully active during the G1 phase and that double strand DNA breaks are produced during G1 (Schrader et al., 2007). This process of breaks is dependent upon both UNG and MSH2-MSH6, suggesting that both pathways function in G1. Taken together, it is currently unclear when and by what nature UNG and MSH2-MSH6 function in relation to each other when processing AID-generated uracils.
5. CONCLUSION
Even with the shear girth of information on AID biology, it is still unclear how the cell fully coordinates deamination events with mutagenic repair. As mentioned above, the mechanisms of AM, GC, and CSR start with a single protein, yet require extensive cellular coordination to produce the initiating deamination. It has been established that AID is tightly regulated at the levels of transcription, translation, phosphorylation, ubiquitination, cellular localization, protein stability, and protein-protein interaction. While much has been discovered, many components are still unknown. It is clear that cis-regulatory elements and transcription are involved, yet no true recruiting factor has been identified for AID. AID specifically interacts with RPA, yet it is unclear how or if this interaction assists in AID localizing to single strand DNA. Does AID travel with the transcription machinery in association with CTNNBL1 alone or is it more complicated? These questions and many more require further studies to understand how AID is targeted to the Ig loci to cause SHM.
In addition to regulation of AID deamination, the processing of dU also plays a significant role in achieving efficient antibody diversity. Of specific interest is how a B cell can manipulate DNA repair to function either less efficiently or less faithfully at the Ig loci, while still maintaining overall genomic integrity. Does the frequency and/or proximity of deamination events overwhelm the faithful capacities of BER and MMR? Is DNA repair specifically inhibited at the Ig loci during an immune response, or does repair become error-prone throughout the cell? It has been reported that DNA repair can be more or less efficient in different regions of the genome (Alrefai et al., 2007; Liu et al., 2008), however, it is not known what mechanisms coordinate this phenotype. Therefore, it remains to be fully understood what role DNA repair plays in transforming AID-dependent uracils into mutations.
ACKNOWLEDGMENTS
This work was supported entirely by the Intramural Research program of the NIH, National Institute on Aging. We gratefully thank Sebastian Fugmann and Huseyin Saribasak for insightful comments.
REFERENCES
- Alrefai RH, Winter DB, Bohr VA, Gearhart PJ. Nucleotide excision repair in an immunoglobulin variable gene is less efficient than in a housekeeping gene. Mol Immunol. 2007;44:2800–2805. doi: 10.1016/j.molimm.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoufouchi S, Faili A, Zober C, D'Orlando O, Weller S, Weill JC, Reynaud CA. Proteasomal degradation restricts the nuclear lifespan of AID. J Exp Med. 2008;205:1357–1368. doi: 10.1084/jem.20070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H, Hauschild J, Buerstedde JM. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- Arakawa H, Moldovan GL, Saribasak H, Saribasak NN, Jentsch S, Buerstedde JM. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 2006;4:e366. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachl J, Carlson C, Gray-Schopfer V, Dessing M, Olsson C. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J Immunol. 2001;166:5051–5057. doi: 10.4049/jimmunol.166.8.5051. [DOI] [PubMed] [Google Scholar]
- Bachl J, Ertongur I, Jungnickel B. Involvement of Rad18 in somatic hypermutation. Proc Natl Acad Sci U S A. 2006;103:12081–12086. doi: 10.1073/pnas.0605146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell PD, Woo CJ, Wei K, Li Z, Martin A, Sack SZ, Parris T, Edelmann W, Scharff MD. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat Immunol. 2004;5:224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- Basu U, Chaudhuri J, Alpert C, Dutt S, Ranganath S, Li G, Schrum JP, Manis JP, Alt FW. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–511. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- Begum NA, Izumi N, Nishikori M, Nagaoka H, Shinkura R, Honjo T. Requirement of non-canonical activity of uracil DNA glycosylase for class switch recombination. J Biol Chem. 2007;282:731–742. doi: 10.1074/jbc.M607439200. [DOI] [PubMed] [Google Scholar]
- Begum NA, Kinoshita K, Kakazu N, Muramatsu M, Nagaoka H, Shinkura R, Biniszkiewicz D, Boyer LA, Jaenisch R, Honjo T. Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch. Science. 2004;305:1160–1163. doi: 10.1126/science.1098444. [DOI] [PubMed] [Google Scholar]
- Begum NA, Stanlie A, Doi T, Sasaki Y, Jin HW, Kim YS, Nagaoka H, Honjo T. Further evidence for involvement of a noncanonical function of uracil DNA glycosylase in class switch recombination. Proc Natl Acad Sci U S A. 2009;106:2752–2757. doi: 10.1073/pnas.0813252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertocci B, De Smet A, Flatter E, Dahan A, Bories JC, Landreau C, Weill JC, Reynaud CA. Cutting edge: DNA polymerases mu and lambda are dispensable for Ig gene hypermutation. J Immunol. 2002;168:3702–3706. doi: 10.4049/jimmunol.168.8.3702. [DOI] [PubMed] [Google Scholar]
- Betz AG, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger MS. Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- Blagodatski A, Batrak V, Schmidl S, Schoetz U, Caldwell RB, Arakawa H, Buerstedde JM. A cis-acting diversification activator both necessary and sufficient for AID-mediated hypermutation. PLoS Genet. 2009;5:e1000332. doi: 10.1371/journal.pgen.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Calabrese P, Goodman MF. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J Biol Chem. 2004;279:51612–51621. doi: 10.1074/jbc.M408135200. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar SS, Watson M, Diaz M. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J Biol Chem. 2004;279:26395–26401. doi: 10.1074/jbc.M403503200. [DOI] [PubMed] [Google Scholar]
- Bulfone-Paus S, Reiners-Schramm L, Lauster R. The chicken immunoglobulin lambda light chain gene is transcriptionally controlled by a modularly organized enhancer and an octamer-dependent silencer. Nucleic Acids Res. 1995;23:1997–2005. doi: 10.1093/nar/23.11.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canugovi C, Samaranayake M, Bhagwat AS. Transcriptional pausing and stalling causes multiple clustered mutations by human activation-induced deaminase. FASEB J. 2009;23:34–44. doi: 10.1096/fj.08-115352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji M, Unniraman S, McBride KM, Schatz DG. Role of activation-induced deaminase protein kinase A phosphorylation sites in Ig gene conversion and somatic hypermutation. J Immunol. 2007;179:5274–5280. doi: 10.4049/jimmunol.179.8.5274. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–998. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Vuong BQ, Basu U, Franklin A, Schwer B, Astarita J, Phan RT, Datta A, Manis J, Alt FW, Chaudhuri J. Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc Natl Acad Sci U S A. 2009;106:2717–2722. doi: 10.1073/pnas.0812304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury M, Forouhi O, Dayal S, McCloskey N, Gould HJ, Felsenfeld G, Fear DJ. Analysis of intergenic transcription and histone modification across the human immunoglobulin heavy-chain locus. Proc Natl Acad Sci U S A. 2008;105:15872–15877. doi: 10.1073/pnas.0808462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Ganesh K, Xue K, Lu M, Rada C, Neuberger MS. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol Cell. 2008;31:474–484. doi: 10.1016/j.molcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Crouch EE, Li Z, Takizawa M, Fichtner-Feigl S, Gourzi P, Montano C, Feigenbaum L, Wilson P, Janz S, Papavasiliou FN, Casellas R. Regulation of AID expression in the immune response. J Exp Med. 2007;204:1145–1156. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dariavach P, Williams GT, Campbell K, Pettersson S, Neuberger MS. The mouse IgH 3'-enhancer. Eur J Immunol. 1991;21:1499–1504. doi: 10.1002/eji.1830210625. [DOI] [PubMed] [Google Scholar]
- Davis MM, Kim SK, Hood LE. DNA sequences mediating class switching in alpha-immunoglobulins. Science. 1980;209:1360–1365. doi: 10.1126/science.6774415. [DOI] [PubMed] [Google Scholar]
- de Yebenes VG, Belver L, Pisano DG, Gonzalez S, Villasante A, Croce C, He L, Ramiro AR. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J Exp Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeoglu F, Horwitz B, Chaudhuri J, Alt FW, Geha RS. Induction of activation-induced cytidine deaminase gene expression by IL-4 and CD40 ligation is dependent on STAT6 and NFkappaB. Int Immunol. 2004;16:395–404. doi: 10.1093/intimm/dxh042. [DOI] [PubMed] [Google Scholar]
- Delbos F, Aoufouchi S, Faili A, Weill JC, Reynaud CA. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J Exp Med. 2007;204:17–23. doi: 10.1084/jem.20062131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbos F, De Smet A, Faili A, Aoufouchi S, Weill JC, Reynaud CA. Contribution of DNA polymerase eta to immunoglobulin gene hypermutation in the mouse. J Exp Med. 2005;201:1191–1196. doi: 10.1084/jem.20050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Immunoglobulin gene conversion in chicken DT40 cells largely proceeds through an abasic site intermediate generated by excision of the uracil produced by AID-mediated deoxycytidine deamination. Eur J Immunol. 2004;34:504–508. doi: 10.1002/eji.200324631. [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Williams GT, Chan DT, Buerstedde JM, Baldwin GS, Neuberger MS. Dependence of antibody gene diversification on uracil excision. J Exp Med. 2007;204:3209–3219. doi: 10.1084/jem.20071768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M, Verkoczy LK, Flajnik MF, Klinman NR. Decreased frequency of somatic hypermutation and impaired affinity maturation but intact germinal center formation in mice expressing antisense RNA to DNA polymerase zeta. J Immunol. 2001;167:327–335. doi: 10.4049/jimmunol.167.1.327. [DOI] [PubMed] [Google Scholar]
- Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan I, Bertocci B, Vilmont V, Delbos F, Megret J, Storck S, Reynaud CA, Weill JC. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- Doi T, Obayashi K, Kadowaki T, Fujii H, Koyasu S. PI3K is a negative regulator of IgE production. Int Immunol. 2008;20:499–508. doi: 10.1093/intimm/dxn009. [DOI] [PubMed] [Google Scholar]
- Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, San-Martin BR, Heidkamp G, Schwickert TA, Eisenreich T, Rajewsky K, Nussenzweig MC. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick W, Rabbitts TH, Milstein C. An immunoglobulin deletion mutant with implications for the heavy-chain switch and RNA splicing. Nature. 1980;286:669–675. doi: 10.1038/286669a0. [DOI] [PubMed] [Google Scholar]
- Dunnick WA, Collins JT, Shi J, Westfield G, Fontaine C, Hakimpour P, Papavasiliou FN. Switch recombination and somatic hypermutation are controlled by the heavy chain 3' enhancer region. J Exp Med. 2009;206:2613–2623. doi: 10.1084/jem.20091280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenstein MR, Neuberger MS. Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation. EMBO J. 1999;18:3484–3490. doi: 10.1093/emboj/18.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenstein MR, Rada C, Jones AM, Milstein C, Neuberger MS. Switch junction sequences in PMS2-deficient mice reveal a microhomology-mediated mechanism of Ig class switch recombination. Proc Natl Acad Sci U S A. 2001;98:14553–14558. doi: 10.1073/pnas.241525998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Texido G, Betz UA, Gu H, Muller W, Klein U, Rajewsky K. Mice reconstituted with DNA polymerase beta-deficient fetal liver cells are able to mount a T cell-dependent immune response and mutate their Ig genes normally. Proc Natl Acad Sci U S A. 2000;97:1166–1171. doi: 10.1073/pnas.97.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faili A, Aoufouchi S, Weller S, Vuillier F, Stary A, Sarasin A, Reynaud CA, Weill JC. DNA polymerase eta is involved in hypermutation occurring during immunoglobulin class switch recombination. J Exp Med. 2004;199:265–270. doi: 10.1084/jem.20031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faili A, Stary A, Delbos F, Weller S, Aoufouchi S, Sarasin A, Weill JC, Reynaud CA. A backup role of DNA polymerase kappa in Ig gene hypermutation only takes place in the complete absence of DNA polymerase eta. J Immunol. 2009;182:6353–6359. doi: 10.4049/jimmunol.0900177. [DOI] [PubMed] [Google Scholar]
- Frasca D, Landin AM, Alvarez JP, Blackshear PJ, Riley RL, Blomberg BB. Tristetraprolin, a negative regulator of mRNA stability, is increased in old B cells and is involved in the degradation of E47 mRNA. J Immunol. 2007;179:918–927. doi: 10.4049/jimmunol.179.2.918. [DOI] [PubMed] [Google Scholar]
- Frasca D, Landin AM, Lechner SC, Ryan JG, Schwartz R, Riley RL, Blomberg BB. Aging down-regulates the transcription factor E2A, activation-induced cytidine deaminase, and Ig class switch in human B cells. J Immunol. 2008;180:5283–5290. doi: 10.4049/jimmunol.180.8.5283. [DOI] [PubMed] [Google Scholar]
- Frey S, Bertocci B, Delbos F, Quint L, Weill JC, Reynaud CA. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity. 1998;9:127–134. doi: 10.1016/s1074-7613(00)80594-4. [DOI] [PubMed] [Google Scholar]
- Fukita Y, Jacobs H, Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9:105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- Gauld SB, Dal Porto JM, Cambier JC. B cell antigen receptor signaling: roles in cell development and disease. Science. 2002;296:1641–1642. doi: 10.1126/science.1071546. [DOI] [PubMed] [Google Scholar]
- Goldfarb AN, Flores JP, Lewandowska K. Involvement of the E2A basic helix-loop-helix protein in immunoglobulin heavy chain class switching. Mol Immunol. 1996;33:947–956. doi: 10.1016/s0161-5890(96)00047-8. [DOI] [PubMed] [Google Scholar]
- Gonda H, Sugai M, Nambu Y, Katakai T, Agata Y, Mori KJ, Yokota Y, Shimizu A. The balance between Pax5 and Id2 activities is the key to AID gene expression. J Exp Med. 2003;198:1427–1437. doi: 10.1084/jem.20030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guikema JE, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, Stavnezer J, Schrader CE. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med. 2007;204:3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen L, Kavli B, Sousa MM, Torseth K, Liabakk NB, Sundheim O, Pena-Diaz J, Otterlei M, Horning O, Jensen ON, Krokan HE, Slupphaug G. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 2008;27:51–61. doi: 10.1038/sj.emboj.7601958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- Hauser J, Sveshnikova N, Wallenius A, Baradaran S, Saarikettu J, Grundstrom T. B-cell receptor activation inhibits AID expression through calmodulin inhibition of E-proteins. Proc Natl Acad Sci U S A. 2008;105:1267–1272. doi: 10.1073/pnas.0708220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein K, Lorenz MG, Siebenkotten G, Petry K, Christine R, Radbruch A. Processing of switch transcripts is required for targeting of antibody class switch recombination. J Exp Med. 1998;188:2369–2374. doi: 10.1084/jem.188.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltemes-Harris LM, Gearhart PJ, Ghosh P, Longo DL. Activation-induced deaminase-mediated class switch recombination is blocked by anti-IgM signaling in a phosphatidylinositol 3-kinase-dependent fashion. Mol Immunol. 2008;45:1799–1806. doi: 10.1016/j.molimm.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T, Nagaoka H, Shinkura R, Muramatsu M. AID to overcome the limitations of genomic information. Nat Immunol. 2005;6:655–661. doi: 10.1038/ni1218. [DOI] [PubMed] [Google Scholar]
- Huang FT, Yu K, Balter BB, Selsing E, Oruc Z, Khamlichi AA, Hsieh CL, Lieber MR. Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Mol Cell Biol. 2007;27:5921–5932. doi: 10.1128/MCB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FT, Yu K, Hsieh CL, Lieber MR. Downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc Natl Acad Sci U S A. 2006;103:5030–5035. doi: 10.1073/pnas.0506548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, Ochs HD, Fischer A, Durandy A. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- Inlay MA, Gao HH, Odegard VH, Lin T, Schatz DG, Xu Y. Roles of the Ig kappa light chain intronic and 3' enhancers in Igk somatic hypermutation. J Immunol. 2006;177:1146–1151. doi: 10.4049/jimmunol.177.2.1146. [DOI] [PubMed] [Google Scholar]
- Ito S, Nagaoka H, Shinkura R, Begum N, Muramatsu M, Nakata M, Honjo T. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci U S A. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabara HH, Chaudhuri J, Dutt S, Dedeoglu F, Weng Y, Murphy MM, Franco S, Alt FW, Manis J, Geha RS. B-cell receptor cross-linking delays activation-induced cytidine deaminase induction and inhibits class-switch recombination to IgE. J Allergy Clin Immunol. 2008;121:191–196. doi: 10.1016/j.jaci.2007.08.008. e192. [DOI] [PubMed] [Google Scholar]
- Jacobs H, Fukita Y, van der Horst GT, de Boer J, Weeda G, Essers J, de Wind N, Engelward BP, Samson L, Verbeek S, de Murcia JM, de Murcia G, te Riele H, Rajewsky K. Hypermutation of immunoglobulin genes in memory B cells of DNA repair-deficient mice. J Exp Med. 1998;187:1735–1743. doi: 10.1084/jem.187.11.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Miyata T, Honjo T. Repetitive sequences in class-switch recombination regions of immunoglobulin heavy chain genes. Cell. 1981;23:357–368. doi: 10.1016/0092-8674(81)90131-8. [DOI] [PubMed] [Google Scholar]
- Kavli B, Andersen S, Otterlei M, Liabakk NB, Imai K, Fischer A, Durandy A, Krokan HE, Slupphaug G. B cells from hyper-IgM patients carrying UNG mutations lack ability to remove uracil from ssDNA and have elevated genomic uracil. J Exp Med. 2005;201:2011–2021. doi: 10.1084/jem.20050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamlichi AA, Glaudet F, Oruc Z, Denis V, Le Bert M, Cogne M. Immunoglobulin class-switch recombination in mice devoid of any S mu tandem repeat. Blood. 2004;103:3828–3836. doi: 10.1182/blood-2003-10-3470. [DOI] [PubMed] [Google Scholar]
- Kim N, Bozek G, Lo JC, Storb U. Different mismatch repair deficiencies all have the same effects on somatic hypermutation: intact primary mechanism accompanied by secondary modifications. J Exp Med. 1999;190:21–30. doi: 10.1084/jem.190.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998;188:1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–2582. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- Kohli RM, Abrams SR, Gajula KS, Maul RW, Gearhart PJ, Stivers JT. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J Biol Chem. 2009;284:22898–22904. doi: 10.1074/jbc.M109.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Maizels N. PMS2-deficiency diminishes hypermutation of a lambda1 transgene in young but not older mice. Mol Immunol. 1999;36:83–91. doi: 10.1016/s0161-5890(99)00027-9. [DOI] [PubMed] [Google Scholar]
- Kothapalli N, Norton DD, Fugmann SD. Cutting edge: a cis-acting DNA element targets AID-mediated sequence diversification to the chicken Ig light chain gene locus. J Immunol. 2008;180:2019–2023. doi: 10.4049/jimmunol.180.4.2019. [DOI] [PubMed] [Google Scholar]
- Krijger PH, Langerak P, van den Berk PC, Jacobs H. Dependence of nucleotide substitutions on Ung2, Msh2, and PCNA-Ub during somatic hypermutation. J Exp Med. 2009;206:2603–2611. doi: 10.1084/jem.20091707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T. Regulation of B-cell signal transduction by adaptor proteins. Nat Rev Immunol. 2002;2:354–363. doi: 10.1038/nri801. [DOI] [PubMed] [Google Scholar]
- Kuzin II, Ugine GD, Wu D, Young F, Chen J, Bottaro A. Normal isotype switching in B cells lacking the I mu exon splice donor site: evidence for multiple I mu-like germline transcripts. J Immunol. 2000;164:1451–1457. doi: 10.4049/jimmunol.164.3.1451. [DOI] [PubMed] [Google Scholar]
- Langerak P, Nygren AO, Krijger PH, van den Berk PC, Jacobs H. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J Exp Med. 2007;204:1989–1998. doi: 10.1084/jem.20070902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larijani M, Frieder D, Basit W, Martin A. The mutation spectrum of purified AID is similar to the mutability index in Ramos cells and in ung(−/−)msh2(−/−) mice. Immunogenetics. 2005;56:840–845. doi: 10.1007/s00251-004-0748-0. [DOI] [PubMed] [Google Scholar]
- Le Morvan CL, Pinaud E, Decourt C, Cuvillier A, Cogne M. The immunoglobulin heavy-chain locus hs3b and hs4 3' enhancers are dispensable for VDJ assembly and somatic hypermutation. Blood. 2003;102:1421–1427. doi: 10.1182/blood-2002-12-3827. [DOI] [PubMed] [Google Scholar]
- Lebecque SG, Gearhart PJ. Boundaries of somatic mutation in rearranged immunoglobulin genes: 5' boundary is near the promoter, and 3' boundary is approximately 1 kb from V(D)J gene. J Exp Med. 1990;172:1717–1727. doi: 10.1084/jem.172.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Luo Z, Scharff MD. Differential regulation of histone acetylation and generation of mutations in switch regions is associated with Ig class switching. Proc Natl Acad Sci U S A. 2004a;101:15428–15433. doi: 10.1073/pnas.0406827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Scherer SJ, Ronai D, Iglesias-Ussel MD, Peled JU, Bardwell PD, Zhuang M, Lee K, Martin A, Edelmann W, Scharff MD. Examination of Msh6- and Msh3-deficient mice in class switching reveals overlapping and distinct roles of MutS homologues in antibody diversification. J Exp Med. 2004b;200:47–59. doi: 10.1084/jem.20040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhao C, Iglesias-Ussel MD, Polonskaya Z, Zhuang M, Yang G, Luo Z, Edelmann W, Scharff MD. The mismatch repair protein Msh6 influences the in vivo AID targeting to the Ig locus. Immunity. 2006;24:393–403. doi: 10.1016/j.immuni.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Lieberson R, Giannini SL, Birshtein BK, Eckhardt LA. An enhancer at the 3' end of the mouse immunoglobulin heavy chain locus. Nucleic Acids Res. 1991;19:933–937. doi: 10.1093/nar/19.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Liu ZM, George-Raizen JB, Li S, Meyers KC, Chang MY, Garrard WT. Chromatin structural analyses of the mouse Igkappa gene locus reveal new hypersensitive sites specifying a transcriptional silencer and enhancer. J Biol Chem. 2002;277:32640–32649. doi: 10.1074/jbc.M204065200. [DOI] [PubMed] [Google Scholar]
- Luby TM, Schrader CE, Stavnezer J, Selsing E. The mu switch region tandem repeats are important, but not required, for antibody class switch recombination. J Exp Med. 2001;193:159–168. doi: 10.1084/jem.193.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Groudine M. Identification of a locus control region in the immunoglobulin heavy-chain locus that deregulates c-myc expression in plasmacytoma and Burkitt's lymphoma cells. Genes Dev. 1994;8:2212–2226. doi: 10.1101/gad.8.18.2212. [DOI] [PubMed] [Google Scholar]
- Martin A, Li Z, Lin DP, Bardwell PD, Iglesias-Ussel MD, Edelmann W, Scharff MD. Msh2 ATPase activity is essential for somatic hypermutation at a-T basepairs and for efficient class switch recombination. J Exp Med. 2003;198:1171–1178. doi: 10.1084/jem.20030880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair (Amst) 2008 doi: 10.1016/j.dnarep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo SA, Yang WW, Gearhart PJ. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J Exp Med. 2004;200:61–68. doi: 10.1084/jem.20040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo SA, Yang WW, Vaisman A, Maas A, Yokoi M, Hoeijmakers JH, Hanaoka F, Woodgate R, Gearhart PJ. Normal hypermutation in antibody genes from congenic mice defective for DNA polymerase iota. DNA Repair (Amst) 2006;5:392–398. doi: 10.1016/j.dnarep.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Martomo SA, Yang WW, Wersto RP, Ohkumo T, Kondo Y, Yokoi M, Masutani C, Hanaoka F, Gearhart PJ. Different mutation signatures in DNA polymerase eta- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc Natl Acad Sci U S A. 2005;102:8656–8661. doi: 10.1073/pnas.0501852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Ouchida R, Hikida M, Kurosaki T, Yokoi M, Masutani C, Seki M, Wood RD, Hanaoka F, J OW. DNA polymerases eta and theta function in the same genetic pathway to generate mutations at A/T during somatic hypermutation of Ig genes. J Biol Chem. 2007;282:17387–17394. doi: 10.1074/jbc.M611849200. [DOI] [PubMed] [Google Scholar]
- Masuda K, Ouchida R, Hikida M, Nakayama M, Ohara O, Kurosaki T, J OW. Absence of DNA polymerase theta results in decreased somatic hypermutation frequency and altered mutation patterns in Ig genes. DNA Repair (Amst) 2006;5:1384–1391. doi: 10.1016/j.dnarep.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Masuda K, Ouchida R, Li Y, Gao X, Mori H, Wang JY. A critical role for REV1 in regulating the induction of C:G transitions and A:T mutations during Ig gene hypermutation. J Immunol. 2009;183:1846–1850. doi: 10.4049/jimmunol.0901240. [DOI] [PubMed] [Google Scholar]
- Masuda K, Ouchida R, Takeuchi A, Saito T, Koseki H, Kawamura K, Tagawa M, Tokuhisa T, Azuma T, J OW. DNA polymerase theta contributes to the generation of C/G mutations during somatic hypermutation of Ig genes. Proc Natl Acad Sci U S A. 2005;102:13986–13991. doi: 10.1073/pnas.0505636102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase eta. J Mol Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- Matthias P, Baltimore D. The immunoglobulin heavy chain locus contains another B-cell-specific 3' enhancer close to the alpha constant region. Mol Cell Biol. 1993;13:1547–1553. doi: 10.1128/mcb.13.3.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul RW, Gearhart PJ. Women, autoimmunity, and cancer: a dangerous liaison between estrogen and activation-induced deaminase? J Exp Med. 2009;206:11–13. doi: 10.1084/jem.20080086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J Exp Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KM, Gazumyan A, Woo EM, Barreto VM, Robbiani DF, Chait BT, Nussenzweig MC. Regulation of hypermutation by activation-induced cytidine deaminase phosphorylation. Proc Natl Acad Sci U S A. 2006;103:8798–8803. doi: 10.1073/pnas.0603272103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KM, Gazumyan A, Woo EM, Schwickert TA, Chait BT, Nussenzweig MC. Regulation of class switch recombination and somatic mutation by AID phosphorylation. J Exp Med. 2008;205:2585–2594. doi: 10.1084/jem.20081319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Frank EG, Plosky BS, Rogozin IB, Masutani C, Hanaoka F, Woodgate R, Gearhart PJ. 129-derived strains of mice are deficient in DNA polymerase iota and have normal immunoglobulin hypermutation. J Exp Med. 2003;198:635–643. doi: 10.1084/jem.20030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KB, Neuberger MS. The immunoglobulin kappa locus contains a second, stronger B-cell-specific enhancer which is located downstream of the constant region. EMBO J. 1989;8:1959–1964. doi: 10.1002/j.1460-2075.1989.tb03601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael N, Shen HM, Longerich S, Kim N, Longacre A, Storb U. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity. 2003;19:235–242. doi: 10.1016/s1074-7613(03)00204-8. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Muto T, Okazaki IM, Yamada S, Tanaka Y, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. Negative regulation of activation-induced cytidine deaminase in B cells. Proc Natl Acad Sci U S A. 2006;103:2752–2757. doi: 10.1073/pnas.0510970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, Agata Y, Yokota Y, Shimizu A. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- Niiro H, Clark EA. Regulation of B-cell fate by antigen-receptor signals. Nat Rev Immunol. 2002;2:945–956. doi: 10.1038/nri955. [DOI] [PubMed] [Google Scholar]
- Odegard VH, Kim ST, Anderson SM, Shlomchik MJ, Schatz DG. Histone modifications associated with somatic hypermutation. Immunity. 2005;23:101–110. doi: 10.1016/j.immuni.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- Omori SA, Cato MH, Anzelon-Mills A, Puri KD, Shapiro-Shelef M, Calame K, Rickert RC. Regulation of class-switch recombination and plasma cell differentiation by phosphatidylinositol 3-kinase signaling. Immunity. 2006;25:545–557. doi: 10.1016/j.immuni.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Park SR, Zan H, Pal Z, Zhang J, Al-Qahtani A, Pone EJ, Xu Z, Mai T, Casali P. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat Immunol. 2009;10:540–550. doi: 10.1038/ni.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Kitaura Y, Gu H, Dalla-Favera R. PKA-mediated phosphorylation regulates the function of activation-induced deaminase (AID) in B cells. Proc Natl Acad Sci U S A. 2006;103:395–400. doi: 10.1073/pnas.0509969103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude AM, Orthwein A, Hu Y, Campo VA, Kavli B, Buschiazzo A, Di Noia JM. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat Struct Mol Biol. 2009;16:517–527. doi: 10.1038/nsmb.1598. [DOI] [PubMed] [Google Scholar]
- Pauklin S, Petersen-Mahrt SK. Progesterone inhibits activation-induced deaminase by binding to the promoter. J Immunol. 2009;183:1238–1244. doi: 10.4049/jimmunol.0803915. [DOI] [PubMed] [Google Scholar]
- Pauklin S, Sernandez IV, Bachmann G, Ramiro AR, Petersen-Mahrt SK. Estrogen directly activates AID transcription and function. J Exp Med. 2009;206:99–111. doi: 10.1084/jem.20080521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc Natl Acad Sci U S A. 2005;102:14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlot T, Li G, Alt FW. Antisense transcripts from immunoglobulin heavy-chain locus V(D)J and switch regions. Proc Natl Acad Sci U S A. 2008;105:3843–3848. doi: 10.1073/pnas.0712291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Coker HA, Pauklin S. DNA deaminases: AIDing hormones in immunity and cancer. J Mol Med. 2009;87:893–897. doi: 10.1007/s00109-009-0496-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- Pettersson S, Cook GP, Bruggemann M, Williams GT, Neuberger MS. A second B cell-specific enhancer 3' of the immunoglobulin heavy-chain locus. Nature. 1990;344:165–168. doi: 10.1038/344165a0. [DOI] [PubMed] [Google Scholar]
- Phung QH, Winter DB, Alrefai R, Gearhart PJ. Hypermutation in Ig V genes from mice deficient in the MLH1 mismatch repair protein. J Immunol. 1999;162:3121–3124. [PubMed] [Google Scholar]
- Phung QH, Winter DB, Cranston A, Tarone RE, Bohr VA, Fishel R, Gearhart PJ. Increased hypermutation at G and C nucleotides in immunoglobulin variable genes from mice deficient in the MSH2 mismatch repair protein. J Exp Med. 1998;187:1745–1751. doi: 10.1084/jem.187.11.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quong MW, Harris DP, Swain SL, Murre C. E2A activity is induced during B-cell activation to promote immunoglobulin class switch recombination. EMBO J. 1999;18:6307–6318. doi: 10.1093/emboj/18.22.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Rada C, Ehrenstein MR, Neuberger MS, Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- Rada C, Jarvis JM, Milstein C. AID-GFP chimeric protein increases hypermutation of Ig genes with no evidence of nuclear localization. Proc Natl Acad Sci U S A. 2002a;99:7003–7008. doi: 10.1073/pnas.092160999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002b;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- Rajagopal D, Maul RW, Ghosh A, Chakraborty T, Khamlichi AA, Sen R, Gearhart PJ. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med. 2009;206:1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro A, San-Martin BR, McBride K, Jankovic M, Barreto V, Nussenzweig A, Nussenzweig MC. The role of activation-induced deaminase in antibody diversification and chromosome translocations. Adv Immunol. 2007;94:75–107. doi: 10.1016/S0065-2776(06)94003-6. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, Nussenzweig MC. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Roa S, Avdievich E, Peled JU, Maccarthy T, Werling U, Kuang FL, Kan R, Zhao C, Bergman A, Cohen PE, Edelmann W, Scharff MD. Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proc Natl Acad Sci U S A. 2008;105:16248–16253. doi: 10.1073/pnas.0808182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim Biophys Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- Ronai D, Iglesias-Ussel MD, Fan M, Li Z, Martin A, Scharff MD. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J Exp Med. 2007;204:181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner K, Winter DB, Kasmer C, Skovgaard GL, Tarone RE, Bohr VA, Gearhart PJ. Impact of age on hypermutation of immunoglobulin variable genes in humans. J Clin Immunol. 2001;21:102–115. doi: 10.1023/a:1011003821798. [DOI] [PubMed] [Google Scholar]
- Ross AL, Sale JE. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol Immunol. 2006;43:1587–1594. doi: 10.1016/j.molimm.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Roy D, Lieber MR. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol. 2009;29:3124–3133. doi: 10.1128/MCB.00139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush JS, Hasbold J, Hodgkin PD. Cross-linking surface Ig delays CD40 ligand- and IL-4-induced B cell Ig class switching and reveals evidence for independent regulation of B cell proliferation and differentiation. J Immunol. 2002;168:2676–2682. doi: 10.4049/jimmunol.168.6.2676. [DOI] [PubMed] [Google Scholar]
- Sabouri Z, Okazaki IM, Shinkura R, Begum N, Nagaoka H, Tsuchimoto D, Nakabeppu Y, Honjo T. Apex2 is required for efficient somatic hypermutation but not for class switch recombination of immunoglobulin genes. Int Immunol. 2009;21:947–955. doi: 10.1093/intimm/dxp061. [DOI] [PubMed] [Google Scholar]
- Sakano H, Maki R, Kurosawa Y, Roeder W, Tonegawa S. Two types of somatic recombination are necessary for the generation of complete immunoglobulin heavy-chain genes. Nature. 1980;286:676–683. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- Saribasak H, Saribasak NN, Ipek FM, Ellwart JW, Arakawa H, Buerstedde JM. Uracil DNA glycosylase disruption blocks Ig gene conversion and induces transition mutations. J Immunol. 2006;176:365–371. doi: 10.4049/jimmunol.176.1.365. [DOI] [PubMed] [Google Scholar]
- Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- Schanz S, Castor D, Fischer F, Jiricny J. Interference of mismatch and base excision repair during the processing of adjacent U/G mispairs may play a key role in somatic hypermutation. Proc Natl Acad Sci U S A. 2009;106:5593–5598. doi: 10.1073/pnas.0901726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D, Gerlach VL, Guo C, Velasco-Miguel S, Hladik CL, White CL, Friedberg EC, Rajewsky K, Esposito G. DNA polymerase kappa deficiency does not affect somatic hypermutation in mice. Eur J Immunol. 2002;32:3152–3160. doi: 10.1002/1521-4141(200211)32:11<3152::AID-IMMU3152>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Schenten D, Kracker S, Esposito G, Franco S, Klein U, Murphy M, Alt FW, Rajewsky K. Pol zeta ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J Exp Med. 2009;206:477–490. doi: 10.1084/jem.20080669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoetz U, Cervelli M, Wang YD, Fiedler P, Buerstedde JM. E2A expression stimulates Ig hypermutation. J Immunol. 2006;177:395–400. doi: 10.4049/jimmunol.177.1.395. [DOI] [PubMed] [Google Scholar]
- Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol. 2007;179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- Schrader CE, Linehan EK, Mochegova SN, Woodland RT, Stavnezer J. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J Exp Med. 2005;202:561–568. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader CE, Vardo J, Stavnezer J. Mlh1 can function in antibody class switch recombination independently of Msh2. J Exp Med. 2003;197:1377–1383. doi: 10.1084/jem.20022190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe MJ, Milstein C, Jarvis JM, Neuberger MS. Somatic hypermutation of immunoglobulin kappa may depend on sequences 3' of C kappa and occurs on passenger transgenes. EMBO J. 1991;10:2139–2145. doi: 10.1002/j.1460-2075.1991.tb07748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Peters A, Kao D, Storb U. The 3' Igkappa enhancer contains RNA polymerase II promoters: implications for endogenous and transgenic kappa gene expression. Int Immunol. 2001;13:665–674. doi: 10.1093/intimm/13.5.665. [DOI] [PubMed] [Google Scholar]
- Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci U S A. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Tanaka A, Bozek G, Nicolae D, Storb U. Somatic hypermutation and class switch recombination in Msh6(−/−)Ung(−/−) double-knockout mice. J Immunol. 2006;177:5386–5392. doi: 10.4049/jimmunol.177.8.5386. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Azuma T, Ishiguro M, Kanjo N, Yamada S, Ohmori H. Normal immunoglobulin gene somatic hypermutation in Pol kappa-Pol iota double-deficient mice. Immunol Lett. 2005;98:259–264. doi: 10.1016/j.imlet.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Shinkai Y, Ogi T, Ohmori H, Azuma T. The absence of DNA polymerase kappa does not affect somatic hypermutation of the mouse immunoglobulin heavy chain gene. Immunol Lett. 2003;86:265–270. doi: 10.1016/s0165-2478(03)00046-4. [DOI] [PubMed] [Google Scholar]
- Shinkura R, Ito S, Begum NA, Nagaoka H, Muramatsu M, Kinoshita K, Sakakibara Y, Hijikata H, Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- Stivers JT. Comment on "Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch". Science. 2004;306:2042. doi: 10.1126/science.1105225. author reply 2042. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Shen HM, Ratnam S, Kodgire P, Storb U. Attracting AID to targets of somatic hypermutation. J Exp Med. 2010 doi: 10.1084/jem.20090821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Tornaletti S, Park-Snyder S, Hanawalt PC. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J Biol Chem. 2008;283:12756–12762. doi: 10.1074/jbc.M705003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TH, Nakata M, Suzuki K, Begum NA, Shinkura R, Fagarasan S, Honjo T, Nagaoka H. B cell-specific and stimulation-responsive enhancers derepress Aicda by overcoming the effects of silencers. Nat Immunol. 2009 doi: 10.1038/ni.1829. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Nagai Y, Kariyone A, Shibata T, Kaisho T, Akira S, Miyake K, Takatsu K. Toll-like receptor 7 cooperates with IL-4 in activated B cells through antigen receptor or CD38 and induces class switch recombination and IgG1 production. Mol Immunol. 2009;46:1278–1288. doi: 10.1016/j.molimm.2008.11.022. [DOI] [PubMed] [Google Scholar]
- Tumas-Brundage K, Manser T. The transcriptional promoter regulates hypermutation of the antibody heavy chain locus. J Exp Med. 1997;185:239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst) 2009;8:461–469. doi: 10.1016/j.dnarep.2009.01.006. [DOI] [PubMed] [Google Scholar]
- van der Stoep N, Gorman JR, Alt FW. Reevaluation of 3'Ekappa function in stage- and lineage-specific rearrangement and somatic hypermutation. Immunity. 1998;8:743–750. doi: 10.1016/s1074-7613(00)80579-8. [DOI] [PubMed] [Google Scholar]
- Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KG, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong BQ, Lee M, Kabir S, Irimia C, Macchiarulo S, McKnight GS, Chaudhuri J. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat Immunol. 2009;10:420–426. doi: 10.1038/ni.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner SD, Milstein C, Neuberger MS. Codon bias targets mutation. Nature. 1995;376:732. doi: 10.1038/376732a0. [DOI] [PubMed] [Google Scholar]
- Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, Difilippantonio S, Wesemann DR, Zarrin AA, Rajewsky K, Nussenzweig A, Alt FW. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009a;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Whang N, Wuerffel R, Kenter AL. AID-dependent histone acetylation is detected in immunoglobulin S regions. J Exp Med. 2006;203:215–226. doi: 10.1084/jem.20051774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J Exp Med. 2009b;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Rada C, Neuberger MS. Altering the spectrum of immunoglobulin V gene somatic hypermutation by modifying the active site of AID. J Exp Med. 2010;207:141–153. doi: 10.1084/jem.20092238. S141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill JC, Reynaud CA. DNA polymerases in adaptive immunity. Nat Rev Immunol. 2008;8:302–312. doi: 10.1038/nri2281. [DOI] [PubMed] [Google Scholar]
- Wiesendanger M, Kneitz B, Edelmann W, Scharff MD. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J Exp Med. 2000;191:579–584. doi: 10.1084/jem.191.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TM, Vaisman A, Martomo SA, Sullivan P, Lan L, Hanaoka F, Yasui A, Woodgate R, Gearhart PJ. MSH2-MSH6 stimulates DNA polymerase eta, suggesting a role for A:T mutations in antibody genes. J Exp Med. 2005;201:637–645. doi: 10.1084/jem.20042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter DB, Phung QH, Umar A, Baker SM, Tarone RE, Tanaka K, Liskay RM, Kunkel TA, Bohr VA, Gearhart PJ. Altered spectra of hypermutation in antibodies from mice deficient for the DNA mismatch repair protein PMS2. Proc Natl Acad Sci U S A. 1998;95:6953–6958. doi: 10.1073/pnas.95.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter DB, Sattar N, Mai JJ, Gearhart PJ. Insertion of 2 kb of bacteriophage DNA between an immunoglobulin promoter and leader exon stops somatic hypermutation in a kappa transgene. Mol Immunol. 1997;34:359–366. doi: 10.1016/s0161-5890(97)00073-4. [DOI] [PubMed] [Google Scholar]
- Woo CJ, Martin A, Scharff MD. Induction of somatic hypermutation is associated with modifications in immunoglobulin variable region chromatin. Immunity. 2003;19:479–489. doi: 10.1016/s1074-7613(03)00261-9. [DOI] [PubMed] [Google Scholar]
- Wu X, Stavnezer J. DNA polymerase beta is able to repair breaks in switch regions and plays an inhibitory role during immunoglobulin class switch recombination. J Exp Med. 2007;204:1677–1689. doi: 10.1084/jem.20070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Garrard WT. The Downstream Transcriptional Enhancer, Ed, positively regulates mouse Ig kappa gene expression and somatic hypermutation. J Immunol. 2008;180:6725–6732. doi: 10.4049/jimmunol.180.10.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue K, Rada C, Neuberger MS. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J Exp Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A, Olaru A, Saltis M, Setren A, Cerny J, Livak F. Identification of a ubiquitously active promoter of the murine activation-induced cytidine deaminase (AICDA) gene. Mol Immunol. 2006;43:529–541. doi: 10.1016/j.molimm.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Yang SY, Fugmann SD, Schatz DG. Control of gene conversion and somatic hypermutation by immunoglobulin promoter and enhancer sequences. J Exp Med. 2006;203:2919–2928. doi: 10.1084/jem.20061835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelamos J, Klix N, Goyenechea B, Lozano F, Chui YL, Gonzalez Fernandez A, Pannell R, Neuberger MS, Milstein C. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature. 1995;376:225–229. doi: 10.1038/376225a0. [DOI] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- Yu K, Huang FT, Lieber MR. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J Biol Chem. 2004;279:6496–6500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]
- Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P. The translesion DNA polymerase zeta plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 2001;14:643–653. doi: 10.1016/s1074-7613(01)00142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H, Shima N, Xu Z, Al-Qahtani A, Evinger Iii AJ, Zhong Y, Schimenti JC, Casali P. The translesion DNA polymerase theta plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–3769. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrin AA, Alt FW, Chaudhuri J, Stokes N, Kaushal D, Du Pasquier L, Tian M. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat Immunol. 2004;5:1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- Zeng X, Negrete GA, Kasmer C, Yang WW, Gearhart PJ. Absence of DNA polymerase eta reveals targeting of C mutations on the nontranscribed strand in immunoglobulin switch regions. J Exp Med. 2004;199:917–924. doi: 10.1084/jem.20032022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Dunn-Walters DK, Barone F, Spencer J. Antisense transcripts of V(D)J rearrangements; artifacts caused by false priming? Mol Immunol. 2009;46:2357–2362. doi: 10.1016/j.molimm.2009.03.020. [DOI] [PubMed] [Google Scholar]