Abstract
Helicos™ Single Molecule Sequencing (SMS) provides a unique view of genome biology through direct sequencing of cellular nucleic acids in an unbiased manner, providing both accurate quantitation and sequence information. Sample preparation does not require ligation or PCR amplification, avoiding the GC-content and size biases observed in other technologies. DNA is simply sheared, tailed with poly A, and hybridized to a flow cell surface containing oligo-dT for sequencing-by-synthesis of billions of molecules in parallel. This process also requires far less material than other technologies. Gene expression measurements can be done using 1st-strand cDNA-based methods (RNA- Seq) or using a novel approach that allows direct hybridization and sequencing of cellular RNA for the most direct quantitation possible. A diverse array of applications have been successfully performed including genome sequencing for accurate variant detection, ChIP-Seq using picogram quantities of DNA, copy number variation studies from both fresh tumor tissue and FFPE tissue samples, sequencing of ancient and degraded DNAs, small RNA studies leading to the identification of new classes of RNAs and the direct capture and sequencing of RNA from cell quantities as few as 250 cells. Because most next generation sequencing technologies require amplification and a specific size range of target molecules, DNAs not meeting those criteria cannot be sequenced in a reliable manner. Single-molecule sequencing does not suffer from those limitations as no amplification is necessary and degraded or modified molecules can be used directly as templates. Principles and methods for using the Helicos® Genetic Analysis System will be discussed.
Keywords: RNA Seq, Direct RNA Sequencing, Next Gen Sequencing
Unit Introduction
System Overview
The Helicos™ Genetic Analysis System consists of multiple components that work together as an integrated system. DNA molecules targeted for sequencing are hybridized in place on disposable glass flow cells. Samples are loaded onto the flow cells using the Helicos Sample Loader in which the temperature can be adjusted for optimal hybridization. Each of the 25 channels on one standard flow cell can be addressed individually for addition of sample and any other needed sample preparation steps. Once the flow cells have been appropriately loaded with sample, they are inserted in the HeliScope™ Sequencing System along with all the reagents necessary for sequencing by synthesis and imaging. The Sequencing System is then allowed to sequence as long as necessary with images being processed in real time by the Helicsope™ Analysis Engine. The Analysis Engine processes the images from each physical location and builds sequence reads from those images. Once the run is complete, the images processed, and strand formation complete; the data are downloaded to a compute cluster for reference alignment or assembly as needed.
Two protocols will be described. Basic protocol 1 is for shearing genomic DNA so that it is ready for tailing. This step may not be required for all samples. Basic Protocol 2 is for tailing and blocking samples so they can hybridize to the sequencing flow cell and sequence properly. When supplying samples to a core sequencing facility, samples are generally provided at this stage or after an optional sample concentration determination, depending on the facility.
BASIC PROTOCOL 1: DNA shearing and clean up
The Helicos Genetic Analysis System (http://www.helicosbio.com/) is capable of sequencing nucleic acids over a very broad range of template lengths, from several nucleotides to several thousand nucleotides without the need for size selection in most situations. However, the yield of sequences per unit mass is dependent on the number of 3′ end hydroxyl groups and thus having relatively short templates for sequencing is more efficient than having long templates. If starting with nucleic acids longer than 1000 nt, it is generally advisable to shear the nucleic acids to an average length of 100-200 nt so that more sequence information can be generated from the same mass of nucleic acids. For double stranded DNA, the standard Helicos protocol for shearing employs a Covaris Adaptive Focused Acoustic instrument that allows good control of fragment size and, if used at the recommended power settings, 3′ ends compatible with terminal transferase tailing (see Basic Protocol 2). Not all sonicators provide equivalent results so shearing with an alternative instrument should only be done after testing to ensure that the resulting DNA is not overly damaged. Additionally, there are commercially-available enzymatic shearing approaches that are also compatible with standard sample preparation techniques such as the Nextera system (Epicentre) and NEBNext™ dsDNA Fragmentase™ enzyme (New England Biolabs). For some applications, rather than shearing, it is desirable to cleave with restriction endonucleases or other specific cutters. In other cases, as with DNA from most ChIP, FFPE, and ancient or degraded samples, shearing is completely unnecessary as the starting material is already sufficiently short that further cleavage is not beneficial. In most cases, end repair is not needed as whatever gains that are generated by the process of increasing the usable 3′ ends is counteracted by the loss of material after sample cleanup. For cleavage methodologies that leave a blocked or damaged 3′ end such as micrococcal nuclease, end repair is required but these methods can usually be avoided.
Basic Protocol 1 Materials list:
S2 instrument (Covaris, Inc., Woburn, MA). For higher throughput applications, the E210 instrument may be used.
Preparation Station (Covaris, Inc., Woburn, MA).
MicroTube holder (single tube). (Covaris, Inc., Woburn, MA).
Snap-Cap microTube with AFA fiber and Pre-split Teflon/silicone/Teflon septa (Covaris, Inc., Woburn, MA).
Distilled Water (Invitrogen, Carlsbad, CA).
10 X TE, pH 8.0 (Invitrogen, Carlsbad, CA).
1.5 mL MAXYMum Recovery tubes (Axygen Scientific, Union City, CA) Alternative low-loss tubes may be used but the small quantities of DNA used in these protocols can be easily lost on tubes or tips if care is not taken.
Agencourt ®AMPure® XP Kit (Agencourt Bioscience Corp., Beverly, MA).
100 % Ethanol (Sigma, St Louis, MO).
Dynal® Magnet: DynaMag® −2 Magnet (Invitrogen, Carlsbad, CA) or similar.
Heatblock equipped with block milled for 1.5 mL tubes (VWR, Batavia, IL).
Basic Protocol 1 Steps:
Fill the tank on the Covaris S2 instrument with deionized water to level 12 on the fill line label. The water should cover the visible parts of the microTube when it is in the microTube Holder (i.e., to the bottom of the snap cap).
Set the chiller to 4°C and turn it on.
Turn on the S2 unit by depressing the red switch located at the upper right corner of the instrument.
After the instrument is on, open the software. Click the ON button on the control panel under the word DEGAS to begin the degassing procedure. The instrument is ready to use when the water has been degassed for 30 min and the temperature software display is between 6° and 8°C.
Prepare 500 ng to 3 μg of DNA in 120 μl of TE, pH 8.0. If the DNA is not in 120 μL of TE, add the appropriate amount of 10X TE, pH 8.0 to make the overall concentration of TE in the solution 1X. Smaller quantities of DNA can be sheared if the sample is limiting. As little as 100 ng of high quality DNA can be used at this step as long as the tailing reaction and the SPRI cleanup and elution steps are also scaled down.
Place an unfilled Covaris microTube into the preparation station holder.
Keeping the cap on the tube, use a p200 pipette and 200 μL aerosol-free tip to transfer the 120 μL of DNA sample by inserting the tip through the pre—split septa. Place the tip along the interior wall of the tube. Slowly discharge the fluid into the tube, moving the pipette tip up along the interior wall as the tube fills. Be careful not to introduce a bubble into the bottom of the tube. Should a bubble appear, remove the bubble by briefly (1-2 sec) centrifuging the tube in a low—speed tabletop centrifuge equipped with appropriate adaptors.
Slide the tube into the microTube holder while keeping the tube vertical. Make sure the tube is centered in the holder. Carefully insert the holder into the machine. Take care not to introduce bubbles into the bottom of the tube during this process.
Click on Configure. On the Method Configuration Screen, set the Mode to Frequency Sweeping and the Bath Temperature Limit to 20°C. In the Treatment 1 box, set the Duty Cycle to 10%, the Intensity to 5 and the Cycles/Burst to 200. Set the time to 60 (s) and the Number of Cycles to 3. Click on Return to Main Panel. Click Start and Start again when the second screen appears.
After shearing is complete, remove the tube from the S2 holder and place it into the preparation station. Remove the snap cap with the tool supplied with the preparation station. Use a p200 pipette to transfer the sheared DNA to a new, clean 1.5 mL tube. A brief centrifugation may be used to collect any DNA remaining in the microTube. Samples may be stored at −20°C after this step.
When the shearing is completed, click the OFF button under DEGAS, empty the water tank, turn off the chiller, close the software and power down the instrument.
After shearing, you must do a size selection to remove very small fragments or your sequencing yield will be reduced. This is done with SPRI beads. All alternative clean-up procedures tested have been found to yield inferior results.
Warm the AMPure® XP bead solution to room temperature and vortex thoroughly to resuspend all beads.
Prepare 70% Ethanol. Prepare fresh by diluting 7 mL of absolute ethanol into 3 mL of distilled water. Do not use a stock 70% Ethanol because the ethanol concentration changes over time.
Vortex the AMPure® XP beads and add 360 μL of the AMPure® XP bead slurry to each tube of sheared DNA. Pipette up and down 10 times to mix.
Incubate the sample slurry for 5 to 10 minutes at room temperature.
Capture the AMPure® XP beads by placing the tube(s) on the Dynal™ magnet until the beads are separated from the solution (approximately 5 minutes).
Carefully aspirate the supernatant keeping the tube(s) on the magnet. Do not disturb the beads adhering to the side of the tube. Take care not to remove any AMPure® XP beads. If you notice you are removing beads during aspiration, do not attempt to remove all the beads with a p1000 pipette. Rather, remove the last 20 to 50 μL with a p200 pipette.
Add 700 ml of 70% EtOH to each tube on the Dynal™ magnet. Wait 30 seconds.
Keeping the tubes on the magnet, carefully aspirate the supernatant.
Repeat steps 19 and 20.
Briefly centrifuge the tubes to collect any remaining 70% EtOH to the bottom of the tube. Place the tubes back on the magnet and remove the last drops of 70% EtOH with a p10 pipette.
Dry the pellet at 37°C in a heat block milled for 1.5 mL tubes. Pellets should be dried until cracks appear in them (approximately 1-5 min). Take care not to over dry the pellets as they will be difficult to resuspend. This step can be performed at room temperature with the drying time being extended to a minimum of 10 minutes before cracks appear.
Elute the sheared DNA sample from the AMPure beads by adding 20 μL of distilled water to each tube. A brief (1-2 sec) centrifugation may be necessary to collect all the beads at the bottom of the tube.
Pipette the entire volume of each tube up and down 20 times so that the beads are completely resuspended.
Place the tube back on the magnet. After the beads are separated from the solution, collect the 20 μL of solution and place it into a new 1.5 mL tube. This supernatant contains the sheared, size-selected DNA. Care should be taken to avoid getting beads in the supernatant. This can be achieved more easily by using a p10 pipette to aspirate the supernatant and by leaving the last μL behind.
Add another 20 μL of water to the tube. Repeat steps 25 and 26, this time adding the supernatant to the first elute. The final sheared, size—selected DNA volume should be 40 μL. If low DNA quantities are used (100ng), the DNA should be eluted in 15 μL rather than 20. The DNA can be stored at −20°C after this step.
BASIC PROTOCOL 2: dA-tailing of DNA molecules by terminal transferase
DNA and RNA samples are hybridized to a primer immobilized on a flow cell for sequencing so it is usually necessary to generate a nucleic acid with an end compatible for hybridization to those surfaces. The target sequence attached to the flow cell surface could, in theory, be any sequence which can be synthesized, but, in practice, the standard commercially-available flow cell is oligo-dT50. Other sequences have been used successfully when there is a specific tag on the nucleic acid molecules of interest. Since most work being carried out at the present time is with the oligo-dT surfaces, the discussion here will be restricted to that type of flow cell for simplicity.
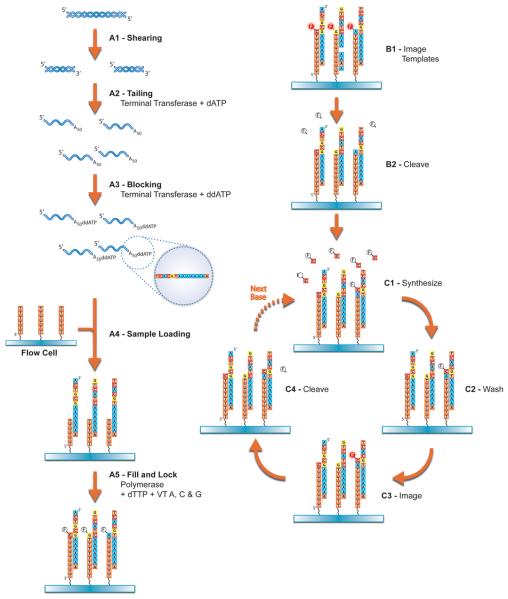
To be compatible with the oligo-dT50 primer on the flow cell surface, it is necessary to generate a poly-dA (or poly-rA) tail of at least 50 nt at the 3′ end of the molecule to be sequenced. Because the fill and lock step (see Figure 1) will fill in excess As but not excess Ts, it is desirable for the A tail to be at least as long as oligo-dT on the surface. It is not clear how long an A-tail can be before it is an issue so it is generally advisable to keep tails less than 200 nt. It is possible for a molecule with an internal poly-dA stretch to hybridize to the surface-bound oligo-dT50 but this process is much slower and energetically less favored due to interference between the unbound 3′ end of the incoming molecule and the glass surface. Few natural sequences contain an internal A-stretch long enough to be stably hybridized.
Figure 1.
Overview of steps required for sequencing. Step A1 is described in Basic Protocol 1 and steps A2 and A3 are described in Basic Protocol 2. Later steps are generally carried out in a sequencing facility and require training for successful completion.
Generation of a 3′ poly-dA or poly-rA tail can be accomplished with a variety of different ligases or polymerases. If the DNA or RNA has a suitable 3′ end, the desired tail (potentially with other sequences of interest such as barcodes) can be ligated to the population of molecules to be sequenced. This sometimes requires pre-treatment of the DNA or RNA to generate a 3′ end compatible with ligation. Furthermore, most ligases have substantial sequence specificities that can cause ligation to occur much more efficiently at some sequences than at others. There can be a length dependence or a base composition dependence; but, in any case, differential ligation can lead to biases in the sequences observed. Thus, while ligation of a poly-dA tail is sometimes readily achievable, we recommend not using ligases for applications that require very quantitative results.
For DNA molecules, we have found terminal transferase to yield the most unbiased tailing of molecules with a free 3′ hydroxyl group. Terminal transferase prefers single stranded ends relative to blunt ends and it tails recessed 3′ ends only very poorly, so, if recessed ends are present, it may be necessary to fill them in with DNA polymerase prior to tailing. The DNA is denatured and snap cooled prior to tailing but, depending on the complexity of the sample and intramolecular folding, some tailing biases could arise. If there is sufficient DNA to measure both mass and average length, it is possible to determine the proper amount of dATP to be added to generate poly dA tails 90-200 nucleotides long. To generate tails of this length, you must first estimate how many 3′ ends you have in your sample and then use the right ratio of DNA, dATP, and terminal transferase to get the optimal size range of tails. If there is insufficient sample to determine mass and length, there is an alternative, low sample mass technique that can be used to generate tails of the proper length.
If the tailed DNA targeted for sequencing is hybridized to the flow cell directly after tailing, it would have a free 3′ hydroxyl that could be extended in the sequencing reaction just like the surface-bound primer and potentially confuse the sequence determination. Thus, prior to sequencing, it is also necessary to block the 3′ ends of the molecules to be sequenced. Any 3′ end treatment that makes the molecule unsuitable for extension can be used. Typically, tailed molecules are blocked using terminal transferase and a dideoxynucleotide but any treatment that leaves a 3′ phosphate or other modification that prevents extension can be similarly effective.
Basic Protocol 2 Materials:
4-20% TBE gel, 1.0 mM, 12 well or similar (Invitrogen, Carlsbad, CA).
Ultrapure 10X TBE buffer (Invitrogen, Carlsbad, CA).
Parafilm
10X BlueJuice™ gel loading buffer (Invitrogen, Carlsbad, CA).
25 bp DNA Ladder (Invitrogen, Carlsbad, CA).
1 kB DNA Ladder (Invitrogen, Carlsbad, CA).
SYBR® Gold Nucleic Acid Gel Stain (Invitrogen, Carlsbad, CA).
Photodocumentation System compatible with a SYBR® Gold photographic filter.
SYBR® Gold Nucleic photographic filter (Invitrogen, Carlsbad, CA).
XCell Surelock™ Mini-Cell (Invitrogen, Carlsbad, CA).
Nanodrop™ 1000, 2000, 2000c or 8000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
Terminal Transferase Kit: includes Terminal Transferase Enzyme (20000 U/mL), CoCl2 at 2.5 mM, 10X Terminal Transferase Buffer (New England BioLabs, Ipswich, MA).
Helicos™DNA Sample Preparation Reagents Kit: Includes Helicos™ PolyA Tailing Control Oligonucleotide TR and Helicos™ PolyA Tailing dATP. Store at −80°C (Helicos BioSciences Corporation, Cambridge, MA). Not all commercial preparations have a functional concentration of dATP as stated. It is important to use a functionally characterized source of dATP like that provided by Helicos BioSciences.
Distilled Water (Invitrogen, Carlsbad, CA).
0.2 mL MAXYMum Recovery Thin Wall PCR tubes and 1.5 mL MAXYMum Recovery tubes (Axygen Scientific, Union City, CA).
Aluminum Block milled for 0.2 mL tubes (VWR, Batavia, IL).
DNA Engine Thermal Cycler (BioRad Laboratories, Hercules, CA).
100 bp DNA Ladder (Invitrogen, Carlsbad, CA).
1 mM Biotin-11-ddATP (PerkinElmer, Waltham, MA)
Basic Protocol 2 Steps:
You should use an XCell SureLock™ Mini-Cell and Invitrogen 4—20% gradient gels. An equivalent electrophoresis apparatus compatible with 10 × 10 cm gel cassettes can also be used. Non-gradient gels should not be used because the DNA will be too dispersed to easily visualize.
Load 2 μl aliquots of the samples in 1X BlueJuice™ buffer in a total volume of 10 μl. Make a 1:10 dilution of the 1 kilobase (kB) and 25 base pair (bp) ladders in distilled water. Load 1 μl of the diluted markers in 1X BlueJuice™ buffer in a total volume of 10 μl.
Run the gel at 180 volts for 45 minutes.
After removing the gel from the cassette using the tool provided with the XCell SureLock™ Mini-Cell, stain the gel for 10 minutes in freshly prepared SYBR® Gold Nucleic Acid Gel Stain diluted 1:10,000 in water. Use clean gloves when handling the gel.
Destain the gel in water for 10 to 15 minutes, changing the water every 2 minutes.
Image with a photodocumentation system compatible with a SYBR® Gold photographic filter.
Determine the average size of your sample by comparing the size of the middle of the sample smear to the size standards.
Determine the double-stranded DNA concentration in ng DNA/μL at this step using a Nanodrop™ 1000, 2000, 2000c or 8000 spectrophotometer.
Calculate the pmoles of ends in the sample using the following formula: pmoles ends/μL= (X ng DNA/μL) × (1000 pg/ng) × (pmole/660 pg) × (1/ average # bp as determined from the gel photo) × 2 ends/molecule
Based on the calculations in step 9, prepare a Sample DNA tube for each DNA to be tailed by determining the volume of DNA that would give 3 pmoles of ends. Put that volume of DNA into a 0.2 mL PCR tube along with distilled water to bring the final volume to 26 μL.
Prepare a Control DNA tube for each DNA to be tailed by determining the volume of DNA that would give 0.8 pmoles of ends. Put that volume of DNA into a 0.2 mL PCR tube along with 1 μL of Helicos™ PolyA Tailing Control Oligo TR and distilled water to bring the final volume to 26 μL.
Prepare a separate Oligo TR Control DNA tube (without DNA sample) by putting 4 μL of Helicos™ PolyA Tailing Control Oligo TR in a tube of 22 μL of distilled water.
Prepare a Sample Master Mix by adding 4.4 μL of 10X Terminal Transferase Buffer, 4.4 μL of CoCl2 (2.5 mM), 4.2 μL of Helicos™ PolyA Tailing dATP, and 2.2 μL Terminal Transferase Enzyme (20U/μL) per sample. The Master Mix volume includes a 10% scale-up. Mix thoroughly by pipetting the entire mix up and down several times. Keep on ice.
Prepare a Control Master Mix by adding 4.4 μL of 10X Terminal Transferase Buffer, 4.4 μL of CoCl2 (2.5 mM), 3 μL of distilled water, 1.4 μL of Helicos™ PolyA Tailing dATP, and 2.2 μL Terminal Transferase Enzyme (20U/μL) per control reaction. The Master Mix includes a 10% scale-up. Mix thoroughly by pipetting the entire mix up and down several times. Keep on ice.
Heat the Sample and Control Tubes DNA tubes to 95°C for 5 minutes in a thermocycler. Immediately remove the DNA tubes from the thermocycler and snap cool for a minimum of 2 minutes by placing the tubes in an aluminum block milled for 0.2 μL tubes that has been prechilled in ice water. It is essential to chill the block to 0°C in an ice and water slurry and cool the DNA as quickly as possible to 0°C to prevent re—annealing of the denatured, single—stranded DNA products.
Add 14 μL of Sample Master Mix to the Sample DNA tubes and 14 μL of Control Master Mix to the Control DNA tubes. Mix thoroughly by pipetting up and down 10 times.
Collect the contents of the tubes into the bottom by briefly centrifuging.
Place the tubes in the thermocycler and incubate at 37°C for 60 minutes, 70°C for 10 minutes followed by a 4°C hold. The tailed DNA can be stored at −20°C after this step.
To determine the success of the tailing reaction, you should run a gradient gel as described above.
Load 20 μl aliquots of the Control reactions in 1X BlueJuice™ buffer (18 μL of the Control reaction and 2 μL of 10X BlueJuice™).
Make a 1:10 dilution of the 100 bp ladder in distilled water. Load 1 μl of the diluted markers in 1X BlueJuice™ buffer in a total volume of 20 μl.
The sample itself is difficult to visualize. The band corresponding to the TR oligo spike is visible in the control lanes and monitors the tail length of the sample. All control reactions should migrate at the size of the Oligo TR Control Sample. A longer polyA tail may be indicative of a sample with a reduced number of strands ending in a 3′OH. Only strands having a 3′OH can be tailed. Tailed oligos with 90 to 200 dA are expected to migrate below the 600 bp band to midway between the 200 and 300 bp bands on the 100 bp ladder. The size of the single stranded polyA tailed samples cannot be determined by direct comparison to the double stranded DNA ladders. The migration patterns of TR oligos containing dA90 and dA200 were determined experimentally. If the TR oligo band in the Control reaction lane migrates anywhere between 250 bp and 600 bp, you may proceed to the 3′ Blocking Reaction.
- In the rare instances where the band in the Control reaction lane migrates below 250 bp, the sample has a polyA tail shorter than 90 nucleotides. Proceed to step 23a: Short Tail Correction. If the band in the Control reaction lane migrates above 600 bp, the polyA tail contains more than 200 dA. In this case the sample could be run on the Helicos™ Genetic Analysis System. However, if sample is not limiting, you should repeat the PolyA Tailing Reaction on another sheared DNA aliquot using twice the amount of input DNA. If the tail length is within the expected range, proceed to step 24.
- 23a. To ensure comparability, both the control and sample reactions undergo the correction. The denaturation step and incubation conditions are as for the PolyA Tailing Reactions in Steps 15-18.
- 23b. After snap cooling the tubes, the following reagents are added. For the Sample reactions, add 3.9 μL of dATP and 2 μL of terminal transferase. For the Control reactions, prepare a 1:2 dilution of the dATP stock in water. Add 1.3 μL of diluted dATP and 1 μL of terminal transferase. Mix by pipetting up and down thoroughly 10 times.
The 3′ Blocking reaction is performed only on the Sample reactions. The denaturation step and incubation conditions are as for the PolyA Tailing Reactions in Steps 15-18. The biotin is used for an optional determination of sample concentration.
Dilute the 1 mM Biotin-11-ddATP 1:6 in water. After snap cooling the Sample tubes, add 1 μL of the diluted Biotin-11-ddATP and 2 μL of terminal transferase to each tube.
After the blocking reaction is completed, add 1 μL of 500 mM EDTA to the samples. Samples should be stored at −20°C until ready to use.
DNA sequencing
Helicos Single Molecule Sequencing is carried out on a glass flow cell with 25 channels for the same or different samples. The system can be run with either one or two flow cells at a time. In the standard configuration, each channel is equivalent and holds approximately 8 μl. Samples are generally loaded with higher volume (usually 20 μl or more) to ensure even hybridization along the length of the flow cell. Samples are inserted into the flow cell via the sample loader included with the overall system. Each channel is individually addressable and sample is applied using a vacuum. Hybridization to the flow cell is typically carried out at 55°C for 1 hr. Generally, samples for sequencing are prepared in such a way that the polyA tail is longer than the oligo-dT50 on the surface of the flow cell. To avoid sequencing the unpaired A residues, a fill and lock protocol is carried out on either the Sample Loader or the HeliScope Sequencing System. After hybridization, the temperature is lowered to 37° C and then dTTP and Virtual Terminator™ nucleotides (Bowers et al. 2009) corresponding to dATP, dCTP, and dGTP are added along with DNA polymerase. Virtual Terminator nucleotides incorporate opposite the complementary base and prevent further incorporation because of the chemical structure appended to the nucleotide. Thus, all of the unpaired dAs present in the polyA tail are filled in with dTTP. The hybridized molecule is locked in place when the polymerase encounters the first non-A residue and inserts the appropriate Virtual Terminator nucleotide. If flow cells with a specific sequence are used instead of oligo-dT50, the fill step is omitted and all four Virtual Terminator nucleotides are used so that each hybridized DNA is locked in place and becomes labeled. If not already loaded on the HeliScope Sequencing System, the flow cells are inserted and the first template picture is taken. Because every DNA molecule should now have a dye attached, an image will include all molecules capable of nucleotide incorporation. Also, because the label could correspond to any base, no sequence information is obtained at this stage. This process is generally 80-90% efficient though a small fraction of DNA molecules will under- or overfill by a base, especially if the first non-A base is a T or there are As immediately after the first base. Thus, for most molecules, sequencing commences with the second base of the original molecule.
Sequencing
In order to sequence the hybridized DNAs, it is first necessary to cleave off the fluorescent dye and terminator moieties present on the Virtual Terminator nucleotides. The current generation of nucleotides is synthesized with a disulfide linkage that can be rapidly and completely cleaved. Following cleavage, the now separated fluorescent dyes are washed away and then new polymerase and a single fluorescent nucleotide are added. After excitation of the fluorescent moiety by the system laser, another image is taken and, on a standard sequencing run, this cyclic process is repeated 120 times. The number of sequencing cycles is user-adjustable and can be modified depending on user needs for run time and length of read.
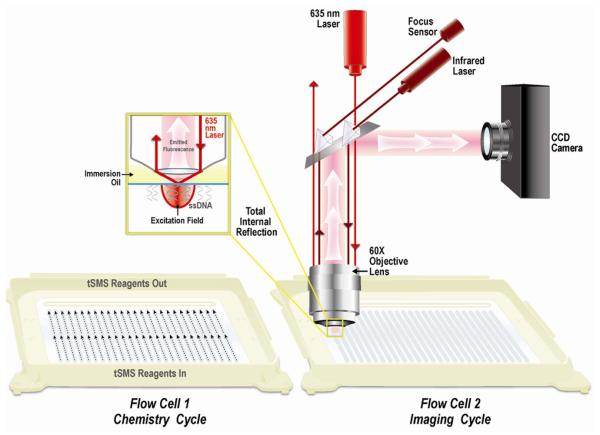
During a standard run, two 25 channel flow cells are used with each flow cell alternating between the chemistry cycle (cleavage of dye/terminator from previous cycle, rinsing, incorporation of next base, rinsing) and the imaging cycle. During the imaging process, four lasers illuminate 1100 Fields of View (FOV) per channel with pictures taken by four CCD cameras via a confocal microscope (see Figure 2 ). Though single molecules are visualized, multiple photon emissions are registered for each molecule with the time spent at each FOV dependent on the brightness of the dye in the particular nucleotide as well as camera speed and detection efficiency. At the present time, the imaging process is the rate determining step and run time could be reduced at the expense of throughput by reducing the number of FOV per channel. Similarly, improvements in camera technology or improved dyes could reduce the run time by lowering the amount of time spent with each image. At the other end of the spectrum, up to 2200 FOVs are possible per channel so it is possible to get increased output but this comes at the expense of increased run time.
Figure 2.
Schematic view of the optical path of the HeliScope Genetic Analysis System and its two flow cells (published with permission).
COMMENTARY
Background Information
Massively parallel DNA sequencing has revolutionized many fields of biology by allowing the generation of sequence information on an unprecedented scale (Kahvejian et al. 2008). The incredible sequence output has been used for many purposes, but primarily for whole genome sequencing of a myriad of species and individuals. The genome information has been complemented by a host of ancillary applications making use of sequencing technologies to shed light on epigenetics, transcription, protein binding, and diagnosis of various medical and other conditions. Most of the sequence data generated thus far has been achieved with amplification-based sequencing systems (reviewed in (Metzker 2010)) but single-molecule, non-amplification based sequencing is now possible including at the scale of resequencing whole human genomes (Pushkarev et al. 2009).
Throughput and read lengths from amplification-based systems has grown at a prodigious rate, straining the capacity of informatics resources, storage capacity, and the ability of biologists to connect function with much of the sequence. Because the technology has evolved at such a rapid rate, it has been sometimes difficult to fully validate all protocols and assess the limitations of the data generated. While some of these limitations can be overcome by sheer mass of data, there are some sequencing applications for which the amplification of the target sequence is not possible or occurs in such a biased or unpredictable manner that the data quality suffers as a result. Single molecule sequencing can be used for virtually all sequencing applications, but, in some situations, it is absolutely required to sequence in an amplification-free manner.
To circumvent amplification issues, various methods for single molecule sequencing have been envisioned for many years because of the inherent advantages of examining single molecules rather than ensembles of molecules (Efcavitch and Thompson, 2010). An early report of single molecule sequencing by synthesis employed FRET to detect incorporation (Braslavsky et al. 2003). As the technology advanced, the mode of detection was changed to measure the fluorescence directly from labeled nucleotides (Harris et al. 2008). While numerous single-molecule approaches are being actively developed, the first commercially-available system was the Helicos Genetic Analysis System. With each sequencing run, this system can generate more than 1,000,000,000 usable reads with a median read length of about 35 nt. The lack of amplification and ligation in sample preparation and sequencing leads to exquisite quantitative abilities and a virtual lack of GC bias in sequencing. It is likely that other single molecule systems will be available soon; each with their own set of read yields, error rates, and read lengths. A brief overview of the theoretical background for the technology and selected applications benefitting from single molecule attributes will be described.
Imaging
For images to be useful, the signal from the molecules of interest must be significantly higher than the background noise level. Any accidental or random source of light emission will be read as a base incorporation and hence appear as an insertion in the sequence. Thus, background signal must be kept to an absolute minimum. This is accomplished by attention to both reagent purity and through the use of Total Internal Reflectance Fluorescence (TIRF) to minimize the ability of molecules far from the surface to fluoresce. This technology (Axelrod et al. 1984) takes advantage of light's differential reflection properties upon passing through media with differing refractive indices. By proper choice of light angle, light absorption and thus fluorescence can be restricted to a very narrow layer near the surface of the flow cell where the desired nucleic acid molecules are located. This minimizes the contribution of the solution to emitted light and enhances the signal to noise so that photons from a single molecule can be visualized. The thickness of the TIRF layer could, in theory, limit the DNA length that can be sequenced because the DNA could become long enough to escape the TIRF limit. However, the DNA length is also limited laterally by neighboring molecules that could potentially overlap. The current lateral resolution limit of the camera is about 300 nM and the thickness of the evanescent wave allowed by TIRF is about 150 nM. The length of DNA required to approach these limits is dependent upon the DNA persistence length which is affected by many factors. The DNA length limit at which signal is lost has not been determined but lengths of several kilobases have been observed to provide a signal.
After incorporation of the fluorescent nucleotide and rinsing to remove any unincorporated molecules, the flow cells are irradiated with a solid state 635 nm red laser to excite molecules on the surface. The flow cell is mounted on a movable platform so that each FOV can be localized under the laser beam and one of four CCD cameras can take pictures via a confocal microscope. Optical focus on the flow cell, critical for maintaining resolution between molecules, is maintained via a separate laser. Prior to each run, this laser goes through a process of focus-finding on one channel so the selected focus channel needs to contain a reliable sample. When flow cells generated by random deposition of oligonucleotides are used, the optimal number of molecules is about 1 per μM2. This limit is caused by the diffraction limit of light and thus the inability to distinguish molecules that are physically located too close to each other. Ordered surfaces will be capable of sequencing about five times as many molecules without danger of having two molecules within an unresolvable distance because it eliminates the random overlap of molecules that can be created in disordered deposition strategies (Schwartz and Quake, 2007).
Image Analysis
Because each cycle of base addition during a standard run involves saving 1100 images in each of 50 channels, a tremendous amount of storage space would be required for both saving and processing the images. These images are processed in real time by the HeliScope Analysis Engine which is dedicated to such processing so that sequence data can be ready within an hour or so after the run finishes. Typically, most images are discarded as soon as they are processed. About 1% of the images are saved from specific camera positions for potential future use in evaluating machine and run performance. Two full runs of data can be stored on the machine before a run needs to be deleted. A new run will not start if it is judged to have insufficient data storage for the next run. Spot finding is performed in real time which greatly reduces the amount of data that needs to be saved. At the end of the run, the images are registered to each other and strands are formed via a basecalling algorithm. This algorithm looks at the overlap of the spot centers in the sequences of images and makes a determination as to whether or not a spot belongs to a strand. If the location of the spot agrees with other images, it is called a base addition while spots not agreeing with a sufficient number of other images are discarded as noise or “rinse objects”.
Sequence Data
Transformation of the base addition calls into sequence data occurs immediately after the run completes. The analysis engine initiates strand formation in which the results from each physical location are converted to a set of sequences. The presence or absence of a base addition at each cycle is retained in the Short Read Format (SRF) file which contains all DNA sequences from all channels as well as other significant run information in the file header such as process parameters and reagent identifiers. This large file can be reduced to files containing simply the sequence data. At present, there have been no parameters identified that predict sequence quality so all bases are given the same sequence quality score. Data in this format has been submitted to the Short Read Archive for public access.
Specific Sequencing Applications - DNA
Because of the power of single-molecule sequencing, all standard DNA sequencing applications as well as many additional single-molecule specific methods can be carried out. Many different applications can be used with variations in the up-front sample preparation to generate samples that can be run on different channels of the same sequencing run. Starting with large amounts (≥100 ng) of DNA, whole genome or targeted sequencing can be accomplished by sonicating the target DNA, tailing, and sequencing. Enzymatic approaches such as Nextera (Epicentre) or NEBNext dsDNA Fragmentase (NEB) can also be used. For DNAs that are already short, as with ancient DNA, FFPE DNA, or ChIP DNA (Goren et al. 2010), shearing is not necessary and only tailing is needed prior to sequencing. Indeed, Helicos single-molecule sequencing can yield usable sequence data for DNA fragments that are shorter and more highly modified than can be achieved with other technologies. It is not necessary to copy the short/modified DNA prior to sequencing and this single-molecule methodology allows sequencing right up to the point of a fatal modification or cleavage. ChIP methods can be used to characterize a wide variety of regulatory and epigenetic phenomena. Methylation studies can be carried out in multiple ways including using 5meC specific binding protein to pull down DNA regions rich in methylated residues and following ChIP-like protocols; cutting DNA with methylation sensitive restriction enzymes and either tailing or ligating an enzyme-specific tail; and sequencing bisulfite-treated DNA and determining which residues have changed from C to U. The standard method for barcoding any of these samples involves ligating a bar code and a tail onto the ends of the DNA though other approaches are possible. Bar coding is another application that is greatly enhanced by single molecule sequencing where even coverage of different samples is important, making avoidance of amplification advantageous. Other quantitative applications such as copy number variation are particularly well suited to amplification-free sequencing because biases introduced by differential amplification are not present. Similarly, sequencing of DNA or genomes with extremes of GC content is much more efficient with a single-molecule approach because it is much less sensitive to GC content (Goren et al. 2010).
Specific Sequencing Applications – Unique to Single Molecule Methods
Because most experimenters have not considered how the unique properties of single-molecule systems can be used to their advantage, there is still a tremendous potential for new approaches that could simplify or expand areas of biological interest. A few unique applications will be discussed here, but it is clear that many more will be developed as the possibilities are considered by a broader scientific population. Among the applications unique to single-molecule sequencing that have been demonstrated are paired reads and multiple reads. With paired reads (or zebra reads), two or more reads can be made from the same molecule, overcoming the limitations of short read length. Multiple strategies are possible. For example, following one sequencing read, one can carry out a controlled dark fill in which alternating sets of three nucleotides are added for a desired number of cycles followed by another sequencing read. If necessary, additional dark fills and sequencing reads can follow the first ones. Instead of controlling insert size by cycling nucleotides, it is also possible to carry out a timed dark fill in which the length of the insert is controlled by time and nucleotide concentration. Paired or zebra reads can be carried out on the same runs as other DNA sequencing but require additional time for subsequent sequencing reactions so are frequently carried out on independent runs.
In addition to multiple reads along different segments of the same DNA molecule, it is also possible to sequence the same segment of DNA multiple times for highly accurate sequences (melt and resequence, (Harris et al. 2008)). Because sequencing errors have been observed to be random with Helicos single-molecule sequencing, repeated reads of the same DNA yield an error rate that is multiplicative. For substitution SNPs, the current error rate is typically 0.2% so two reads of the same molecule that are in agreement will yield a consensus error rate of 0.0004% for SNPs. Insertion and deletion errors, which typically occur at 1-3%, would yield consensus error rates of 0.01-0.09% and become correspondingly lower with additional reads. Thus, if the highest accuracy possible is desired and throughput is less of a concern, as may be the case for tumor samples with low tumor cell content or with miRNAs, two or more reads can yield substantially higher accuracy than possible with any amplification-based sequencing system.
An additional application of single-molecule sequencing is the arena of very small amounts of starting material. The eventual combination of microfluidics with single-molecule sequencing will allow tremendous advances to be made in single cell analysis but, even now, as few as hundreds of cells can be manipulated and their nucleic acids sequenced (Ozsolak et al. 2010). Many manipulations can be carried out on the flow cell surface, minimizing loss and allowing efficient transformation of molecules into sequence data.
Summary
Single-molecule sequencing has already been shown to have substantial power for a variety of applications and provides the potential for many new advances to be made as its full power is unleashed. Already, projects ranging from sequencing RNA in a few hundred cells to sequencing an entire human genome have been described. Similarly, the unparalleled quantitative nature of the technology and ability to sequence very short or modified DNA make it ideal for many projects that are poorly addressed by amplification-based sequencing.
Acknowledgements
We would like to thank Drs. Parris Wellman, Patrice Milos, and J. William Efcavitch for comments and critical reading of the manuscript. Also, this overview summarizes the accomplishments of many engineers, chemists, biochemists, informaticists, molecular biologists and others without whom this system would not have been possible. This publication was made possible, in part, by grant numbers R01 HG004144 and 1RC2HG005598 from the National Human Genetics Research Institute (NHGRI) at the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NHGRI.
Footnotes
Internet Resources with Annotations
Helicos BioSciences website for applications and updates http://www.helicosbio.com/Applications/tabid/66/Default.aspx
Sites for information on DNA shearing http://www.covarisinc.com/ http://www.neb.com/nebecomm/products/productM0348.asp http://www.epibio.com/nextera/nextera_tech_overview.asp
Literature Cited
- Axelrod D, Burghardt TP, Thompson NL. Total internal reflection fluorescence. Annu Rev Biophys Bioeng. 1984;13:247–268. doi: 10.1146/annurev.bb.13.060184.001335. [DOI] [PubMed] [Google Scholar]
- Virtual terminator nucleotides for next-generation DNA sequencing. Nat Methods. 2009;6(8):593–595. doi: 10.1038/nmeth.1354. *Detailed discussion of the chemistry of the nucleotides. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braslavsky I, Hebert B, Kartalov E, Quake SR. Sequence information can be obtained from single DNA molecules. Proc Natl Acad Sci U S A. 2003;100(7):3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efcavitch JW, Thompson JF. Single Molecule DNA Analysis. Ann. Rev. Anal. Chem. 2010;3:000–000. doi: 10.1146/annurev.anchem.111808.073558. [DOI] [PubMed] [Google Scholar]
- Goren A, Ozsolak F, Shoresh N, Ku M, Adli M, Hart C, Gymrek M, Zuk O, Regev A, Milos PM, et al. Chromatin profiling by directly sequencing small quantities of immunoprecipitated DNA. Nat Methods. 2010;7(1):47–49. doi: 10.1038/nmeth.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320(5872):106–109. doi: 10.1126/science.1150427. *The first single molecule sequencing of a genome though with an older version of the chemistry. [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nat Biotechnol. 2008;26(10):1125–1133. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson D, Raz T, Kieu A, Jones DR, Giladi E, Thayer E, Thompson JF, Letovsky S, Milos P, Causey M. Quantification of the yeast transcriptome by single-molecule sequencing. Nat Biotechnol. 2009;27(7):652–658. doi: 10.1038/nbt.1551. [DOI] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Goren A, Gymrek M, Guttman M, Regev A, Bernstein BE, Milos PM. Digital transcriptome profiling from attomole-level RNA samples. Genome Res. 2010;20(4):519–525. doi: 10.1101/gr.102129.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM. Direct RNA sequencing. Nature. 2009;461(7265):814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- Pushkarev D, Neff NF, Quake SR. Single-molecule sequencing of an individual human genome. Nat Biotechnol. 2009;27(9):847–852. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JJ, Quake SR. High density single molecule surface patterning with colloidal epitaxy. App. Phys. Letters. 2007;91:083902. [Google Scholar]