Abstract
We present an applied study in cancer genomics for integrating data and inferences from laboratory experiments on cancer cell lines with observational data obtained from human breast cancer studies. The biological focus is on improving understanding of transcriptional responses of tumors to changes in the pH level of the cellular microenvironment. The statistical focus is on connecting experimentally defined biomarkers of such responses to clinical outcome in observational studies of breast cancer patients. Our analysis exemplifies a general strategy for accomplishing this kind of integration across contexts. The statistical methodologies employed here draw heavily on Bayesian sparse factor models for identifying, modularizing and correlating with clinical outcome these signatures of aggregate changes in gene expression. By projecting patterns of biological response linked to specific experimental interventions into observational studies where such responses may be evidenced via variation in gene expression across samples, we are able to define biomarkers of clinically relevant physiological states and outcomes that are rooted in the biology of the original experiment. Through this approach we identify microenvironment-related prognostic factors capable of predicting long term survival in two independent breast cancer datasets. These results suggest possible directions for future laboratory studies, as well as indicate the potential for therapeutic advances though targeted disruption of specific pathway components.
Keywords: Acidosis and neutralization pathways in cancer, Bayesian latent factor models, breast cancer genomics, gene expression signatures, integrative cancer genomics, micro-environmental parameters in cancer, Weibull survival models
1. Introduction
Cancer progression involves a complex interaction of genetic and genomic factors that jointly subvert normal cell development. The genomic component, which encompasses gene expression and regulation, is substantially impacted by the biochemical composition of the local environment in which a cell grows. So-called micro-environmental parameters, including levels of oxidation, lactate, acidity, nutrients of various kinds and other factors affecting physical interactions between cells, are increasingly studied for their potential to improve our understanding of cancer biology, and for their promise to lead to new therapeutic strategies. Changes in such parameters can impact gene transcription, which in turn impacts protein production. Variation in these fundamental parameters can therefore induce a cascade of effects, producing disruptions of normal cellular processes in downstream biological pathways [Hanahan and Weinberg (2000)]. For example, changes to the pH level in the cellular environment may effect glycolysis, thus impacting on numerous genes involved in the glycolysis pathway. Some of these genes may also play roles in the regulation of cell growth, and their suppression may engender tumorigenesis and promote the aggressive advance of existing cancerous states. Microarray gene expression assays can be used to generate data on the transcriptional response of cancer cells to controlled manipulations of environmental factors such as pH. This data is useful for characterizing these micro-environmental response pathways.
Our study concerns changes to cellular pH levels, and the resulting neutralization and lactic acidosis response pathways. Section 2 describes the application of sparse Bayesian regression models [Lucas et al. (2006); Seo, Goldschmidt-Clermont and West (2007)] to microarray data generated through a series of laboratory experiments on cultured breast tumor cells in which cellular pH levels were manipulated in a controlled manner. These analyses yield statistical expression signatures of the cellular responses to various interventions on the pH level. The main challenge lies in relating these signatures, and the biological pathways they characterize, to variation in gene expression across large samples of human breast tumors. This integration of in vitro and in vivo data sets is the driving focus of this and related studies. In addition to comprising a detailed study of new data and experimental results, through which are generated several directions for biomedical research, this work exemplifies an overall strategy for cross-study, integrative analysis of gene expression data for exploring and relating pathway-related experimental findings to clinical contexts and patient outcomes.
When considering variability in expression patterns of genes in observational tumor data, we face questions of differences due to the differing contexts. It is to be expected that a tumor in vivo evidences far more complex and heterogeneous biological variation than in the controlled in vitro setting, and this will be manifest in measures of gene expression. Normal cell processes held in quiescence in cell cultures may when active co-regulate the expression of relevant signature genes in in vivo, confounding the pattern of expression that was evident in vitro. Hence, when aiming to translate experimental findings to tumor populations, thereby providing a mapping of an in vitro signature to its in vivo counterpart, we require statistical models capable of discovering and representing the additional complexity surrounding and interacting with the original response signature. Section 3 describes our analysis of a large and heterogeneous breast cancer data set using sparse latent factor models [West (2003); Carvalho et al. (2008)] that satisfy these desiderata. This analysis includes a targeted factor search that facilitates estimation of statistical factors associated with an initial set of genes underlying the in vitro experimental signatures. The factors discovered in this way represent a modular decomposition of the biological patterns evident in the in vivo breast cancer data, while retaining connections to the experimental signatures.
Section 4 discusses aspects of the biological and clinical interpretations of these estimated factors, which can be viewed as a refined in vivo set of summary bio-markers of variation in the neutralization and lactic acidosis response pathways of these breast cancers. In survival analyses, we find that these factor model derived biomarkers have substantial prognostic value in connection with long-term survival and, hence, the sets of genes comprising these factors warrant further study. We present predictive validation of this key finding in analyses of two separate breast cancer data sets. We then provide biological interpretation of one key factor that emerged from the evolutionary factor search, which plays a key role as a predictive variable in the survival analyses. It turns out that this factor is a single component of a specific biological pathway that has previously been noted as a risk biomarker in cancer, but not, to date, connected at all into response pathways linked with variation in cellular pH. This finding has generated follow-on biological research and initiated a new line of experimentation on the role of this pathway in connection with cancer cell micro-environmental influences.
2. Neutralization experiments and analysis
2.1 Biological and experimental context
Investigating the effects of changes in the micro-environment in which cells grow is of increasing interest in cancer research. The tumor micro-environment is typically characterized by oxygen depletion, high lactate and extracellular acidosis coupled with vascular leakage, glucose and energy deprivation. These and other micro-environmental features vary widely across tumors and generally exhibit substantial temporal and spatial differences in a tumor. Micro-environmental stresses trigger biochemical changes in cancer cells that directly modulate physiological, metabolic and ultimately clinical phenotypes. Improved understanding of the molecular mechanisms of such tumor responses holds promise for immediate translational impact and clinical care, as relevant therapies can be brought to bear to modify the micro-environment.
Currently, with the exception of hypoxia, very little is understood about how each individual stress affects cellular phenotypes and tumor progression. To examine how cancer cells respond to increased acidity or pH neutralization at different time points, MCF7 cell cultures (a commonly-used breast tumor cell line) were grown in neutral media and then exposed to varying interventions in several assays in parallel. For some cells, lactic acid was added to the medium (25 mM lactic acid at pH 6.7) for 1 and 4 hours; others cells experienced strong lactic acidosis conditions (25 nM lactic acid at pH 5.5) for 4 hours. Similarly, the effects of neutralization were assayed by shifting the MCF7 cultures from overnight lactic acidosis conditions at pH 6.7 to neutral regular media at pH 7.4 for 1 and 4 hours. Control cells were grown in each starting condition (neutral conditions and lactic acidosis conditions). The complete set of experiments is summarized in Table 1. The mRNA extracted from each of the resulting n = 27 batches of MCF7 cultures was purified using Ambion miRVana RNA purification kits and standard microarray assays were performed using Affymetrix U133 Plus 2 Genechip platforms. All raw microarray data were preprocessed using RMA [Irizarry et al. (2003)], the log (base 2) scale output of which were used in all ensuing statistical analyses.
Table 1.
Summary of neutralization/acidosis experiments. Cell entries indicate the number of replicates per experimental group
| Exposure condition |
|||||
|---|---|---|---|---|---|
| pH 7.4 |
pH 6.7 |
pH 5.5 |
|||
| Growth condition | 1hr | 4hr | 1hr | 4hr | 4hr |
| pH 7.4 | 3× | 3× | 3× | 3× | 3× |
| pH 6.7 | 3× | 3× | 3× | 3× | |
2.2 Cellular response signatures
Quantitative summaries of the cellular responses to lactic acidosis and neutralization treatments were obtained using a standard sparse multivariate regression model [Lucas et al. (2006); Seo, Goldschmidt-Clermont and West (2007)]. We analyzed 19,375 genes (technically, probe-sets from the Affymetrix array; we will use “gene” and “probe” interchangeably) whose median expression level is at least 5.5 and whose expression ranges more than 0.5-fold across the n = 27 experimental samples. Let Xexp denote the 19,375 × 27 matrix of expression values. Rows represent genes and columns correspond to three replicate samples for each of the following experimental groups: (i) control (pH 7.4 → 7.4) at 1 hour; (ii) control at 4 hours; (iii) lactic acidosis (pH 7.4 → 6.7) at 1 hour; (iv) lactic acidosis at 4 hours; (v) neutralization (pH 6.7 → 7.4) at 1 hour; (vi) neutralization at 4 hours; (vii) acidic growth (constant pH of 6.7) at 1 hour; (viii) acidic growth at 4 hours; (ix) strong lactic acidosis (pH 7.4 → 5.5) at 4 hours. Let Hexp denote the 11 × 27 design matrix where the first 8 rows contain binary indicators for effects associated with differential expression relative to the 1hr control group: 1hr lactic acidosis effect, 1hr neutralization effect, 1hr acidic growth effect, 4hr control effect, 4hr lactic acidosis effect (relative to 4hr control), 4hr neutralization (relative to 4hr control), 4hr acidic growth effect (relative to 4hr control) and 4hr strong lactic acidosis effect (relative to 4hr control). The last three rows contain artefact control factors derived from the first three principle components of the expression levels associated with the AFFX series control genes included on the Affymetrix microarrays. These control genes are not variably expressed in humans, and so patterns of variation across samples manifest in control genes represents systematic errors arising from different experimental conditions. Use of these artefact control factors provides opportunity for sample-specific correction of artefactual effects on genes that may otherwise result in false-discovery or obscure meaningful biological variation [following Lucas et al. (2006) and Carvalho et al. (2008)]. After deriving the artefact control factors, rows corresponding to Affymetrix control genes are removed from subsequent analyses.
The model for the expression of gene g in sample i is
or in matrix from
where μg denotes the mean expression of gene g in the 1hr control samples, each βg,k is the change in expression of gene g due to design factor k, and the νg,i are independent, normally distributed idiosyncratic noise terms representing residual biological variation, experimental and measurement errors with individual variances ψg. Sparsity is induced via prior distributions that place positive probability on βg,k = 0 for each g, k pair, and resulting posterior analysis allows investigation of posterior sparsity patterns via probabilities . Full details follow Lucas et al. (2006) and prior specifications, including priors for the μk, variance parameters and all hyper-parameters, are given in Supplement B. Posterior inference via MCMC is achieved using the BFRM software [Wang et al. (2007)].
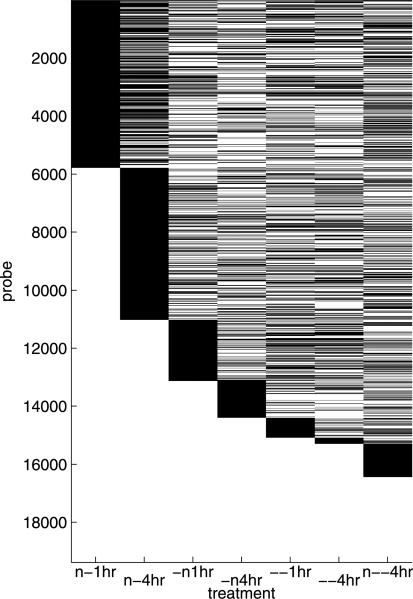
Figure 1 broadly illustrates genes uniquely associated with individual treatment effects as well as those involved in multiple responses. This gives some indication of the degree of intersection of the cellular pathways being queried by the different treatments. Across the 8 treatments, the sparsity, as measured by the percent of genes for which , ranges from 29% (4 hour neutralization) to 46% (4 hour lactic acidosis). The fold-change associated with the involved genes (2|βg,k| for g such that ) ranges from 1.06× to 13×, with a mean of 1.4 ×.
Fig. 1.
Neutralization signature skeleton: black indicates genes g (rows) with posterior probability for each experimental group k (columns). Genes are ordered to emphasize which genes are unique to each successive experiment relative to the previous.
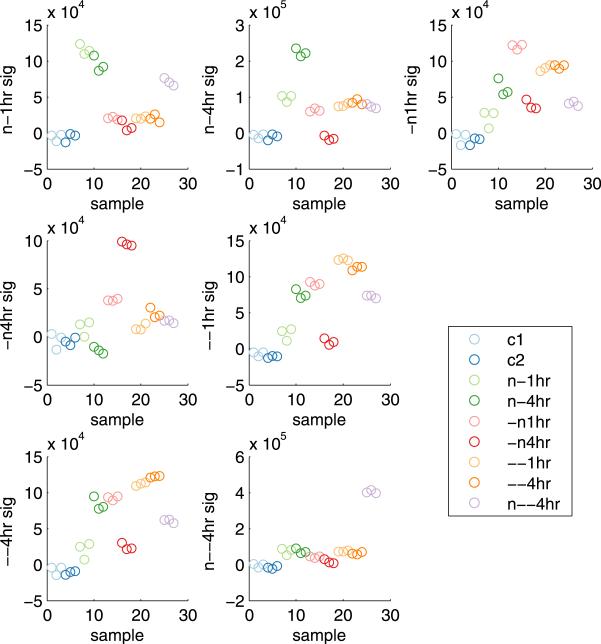
The cellular response to each treatment, also called the signature of the treatment, is characterized by estimated effects together with the . The ability of each signature to uniquely identify the treatment it reflects can be further explored using summary signature scores as defined in Lucas, Carvalho and West (2009). Based on posterior means and , let
define the score for treatment signature k on sample i. This expression is derived from the data-driven component of the Bayes factor that weighs the evidence in favor of the given signature describing the variation in a sample (p(xi|hk,i = 1)//p(xi|hk,i = 0)). Figure 2 shows the values of the scores associated with 7 treatment signatures plotted across samples. As expected, the highest scoring samples for each signature are those upon which that signature is based, but important connections between signatures can be identified on the basis of other high- or low-scoring treatment groups. For example, there is a inverse relationship between the 4hr acidosis score and the 4hr neutralization score. Also evident is the similarity between the 1hr and 4hr acidic growth signatures, which can also be inferred through the large intersection of the genes defining the two signatures (Figure 1).
Fig. 2.
Neutralization treatment signature scores (sk,i ) for each sample in the original study. Separate treatment groups are color coded.
3. Latent factor analysis of breast tumor gene expression
3.1 In vivo breast cancer data
The primary goals of this study are to uncover shared structures in the cell response signatures defined above, and to quantify the extent to which these structures can be used to predict clinical phenotypes in real human cancers. Here we make use of the gene expression data for a collection of 251 surgically removed breast tumors as reported in Miller et al. (2005). Affymetrix 133A and 133B GeneChip microarrays were generated for each tumor sample, and relevant clinico-pathological variables were collected for each patient. This included age at diagnosis, tumor size, lymph node status (an indicator of metastatic cancer) and Elston histological grade (a categorical rating of malignancy as deemed by pathologists). Molecular assays to identify the presence of absence of mutations in the estrogen receptor (ER), progesterone receptor (PgR) and P53 genes were also performed. These data are representative of a variety of different presentations of human breast cancer on these clinical measures.
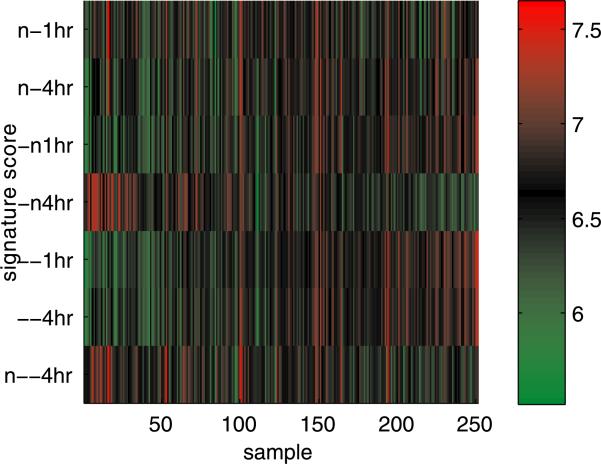
We first evaluate the signature score as defined above for each tumor. These scores are then standardized across samples so that each vector of 27 scores for a particular signature has mean and variance equal to the mean gene-specific expression and mean gene-specific variance. This transformation places the signature scores on the same scale as gene expression in the tumor data set, thus enabling a “metagene” interpretation of a vector of scores [West et al. (2001); Pittman et al. (2004)]; see Figure 3. The relationships between the tumor signature scores bear some similarities to those observed in the cell line study. There is once again evidence of correlation between the two acidic growth signatures and the 1hr neutralization signature. These three signatures, in turn, display patterns opposite that of the 4hr neutralization signature. The patterns are less prominent, however, than was evident in the cell culture data. Although the variation in these scores presumably relates, in part, to underlying biological variation in the activity of the lactic acidosis and neutralization response pathways within these tumors, as mentioned above, the set of genes characterizing the in vivo effects of lactic acidosis and neutralization may differ substantially from those characterizing the in vitro responses as a result of the more complex interactions with other cellular processes.
Fig. 3.
Initial evaluation of neutralization signature levels across tumor samples. Samples are ordered by first principle component to emphasize dominant signature gradients.
We thus aim to refine our evaluation of the response pathway activity levels in the tumor data by using the signature scores as initial “anchors” in an analysis using sparse latent factor models. The main idea is to define statistical factors on sets of genes related to these initial scores, and to link in other genes that may connect with the different response pathways active in vivo. This is accomplished as follows.
3.2 Sparse factor model specification
Sparse latent factor models represent common patterns in gene expression via latent factors in which the factor-gene relationships are sparse; this notion of statistical sparsity is key for representing the intersecting subsets of genes potentially related to underlying networks of biological pathways [West (2003); Seo, Goldschmidt-Clermont and West (2007); Lucas et al. (2009); Carvalho et al. (2008)]. The form of the statistical model is an extension of the sparse regression model. A key part of our analysis strategy stems from augmenting the 44,592 × 251 matrix of gene expression data for the tumor data with the 7 values of the projected treatment signature scores. Let p = 44,592 + 7 = 44,599 and n = 251, and let Xobs denote the p × n matrix in which the first 7 rows are the projected scores across tumor samples, and rows 8−p are the gene expression values. Here we will make use of K = 4 artefact control factors derived from the first four principal components of the control genes of the breast tumor microarrays. A latent factor model consisting of L latent factors is therefore
or,in matrix form,
where: (i) the first K rows of the (K + L) × n matrix Λ are the known artefact controls; (ii) the remaining L rows contain latent factor scores; (iii) the first K columns of the p × (K+L) matrix A are regression parameters on the artefact controls (changing notation from the earlier β to α for notational convenience here); (iv) the remaining L columns of A are factor loadings parameters relating factors to genes and to the projected scores; and (v) A is sparse, with sparsity pattern to be inferred along with estimation of nonzero values. The model is completed by assigning sparsity priors over columns of A, precisely as was done for B in the sparse regression model; prior specification for A, variance components and other hyper-parameters follows default recommendations for the BFRM framework (see Supplement B).
Flexibility in representing potentially complicated patterns underlying expression is achieved using nonparametric Bayesian Dirichlet process models for the factor scores. The L-vectors (λK+1,i,...,λK+L,i)′, representing the latent factor values on tumor sample i, are modeled as draws from an unknown latent factor distribution subject to a Dirichlet process prior with a multivariate normal base measure. This standard nonparametric mixture model allows great flexibility in adapting to nonnormal structures commonly manifest in factor scores [Carvalho et al. (2008); Wang et al. (2007)].
Ensuring the identifiability of latent factors requires the use of a modified prior on A such that the leading L rows have an upper triangle of zeros and positive upper diagonal elements; that is, for g = 1: L, we have αg,g = K > 0 and αg,l = 0 for l > g + K. The first L variables in Xobs then represent “founders” of the L latent factors, with variable g associated with a αg,g-fold change in expression due to factor g, (g = 1,...,L). It also defines an hierarchical dependence on the factors, namely,
and so on. This structure aids the interpretation of the latent factor loadings as representing interconnected components of a complex biological process. The latent factor scores λi,l quantify variation across tumors for these expanding levels of complexity, with each additional factor accounting for variation in observed gene expression unaccounted for by the previous set of factors. With our use of projected in vitro signature scores here as the first 7 variables, the first 7 factors will now represent patterns underlying co-variation in expression of sets of genes that link indirectly to these treatment signatures. Additional factors then reflect other dimensions of common variation in the set of genes analyzed.
3.3 Targeted factor search
Decomposition of the patterns of variation evident in the tumor gene expression data into latent factors proceeds through evolutionary model search, full details of which appear in Carvalho et al. (2008) and Wang et al. (2007). The evolutionary model search provides a computationally efficient approximation to the computationally prohibitive full factor analysis on the entire set of genes, and produces full posterior results for the final set of factors and genes. A key novelty of this approach is that we exploit the sensitivity of the model search procedure to its initial configuration in order to explore the space of factor models surrounding an initial model containing 7 latent factors and representing only the 7 response metagenes. By construction, these initial factors are each defined, or “founded,” by the neutralization/lactic acidosis treatment scores, thereby ensuring that the model search is primarily concerned with patterns of variation related to these particular response pathways.
Evolution of this initial model proceeds as follows. Samples from the joint posterior distribution of model parameters are obtained through MCMC. Based on these fitted values, we impute inferences for all genes g > 7 that are not currently included in the model, as described in Carvalho et al. (2008). Thus, after fitting the initial factor model which considers only the signature scores, we examine expression levels of the full set of 44,000+ genes for evidence of association with the current factors. The imputation process generates approximate probabilities for all such genes g. Genes are ranked on the basis of these probabilities, and the model is then expanded to include a small number of the genes with largest values of the projected . The model is then refitted to this expanded sample, and if appropriate, the number of factors is increased in order to adapt to additional common patterns of expression variation now evident in the increased set of variables being modeled. This process is repeated until no new genes or factors can be added, or until the model reaches a designated maximum size. More details on the search strategy, including control parameters governing model expansion, are given in Supplement B.
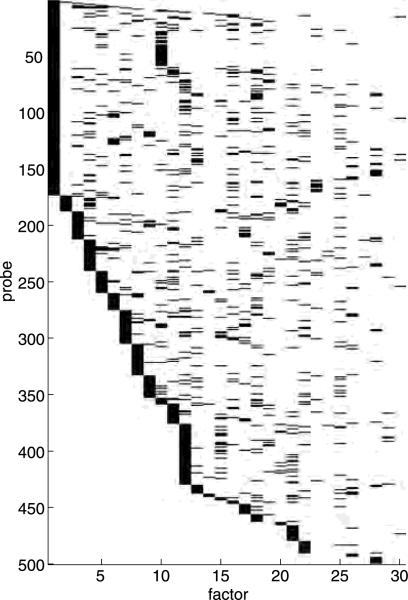
The initial 7-gene, 7-factor model evolved under this process to reach a terminal size of 500 variables (the designated maximum) incorporating 30 latent factors. Figure 4 shows the skeleton of the factor structure, in terms of major patterns of gene-factor relationships. The ordering of the factors is determined by the model search procedure, and represents the incremental improvement to model fit provided by each subsequent factor. In this sense, each subsequent factor builds upon the complexity modeled by the previous factors. The leading 7 factors correspond to the following signatures, respectively: 4hr lactic acidosis, 1hr lactic acidosis, 1hr neutralization, 4hr strong lactic acidosis, 4hr acidic growth, 1hr acidic growth, and 4hr neutralization. Like their in vitro signature counterparts, the in vivo fac- tors loadings contain a great deal of sparsity. Of the 493 genes included in the final model, only 333 are among those identified in the in vitro signature analysis. Factor 1, founded by the the 4hr lactic acidosis signature score, has 173 genes with nonzero loadings at the 0.99 probability threshold, compared to 8909 in the in vitro signature.
Fig. 4.
Skeleton of fitted factor loadings for tumor data. Black indicates variable-factor loadings with . The first 7 variables are the projected neutralization scores, followed by 493 genes reordered for a clear visual presentation of the sparsity structure of, and cross-talk in, gene-factor loadings.
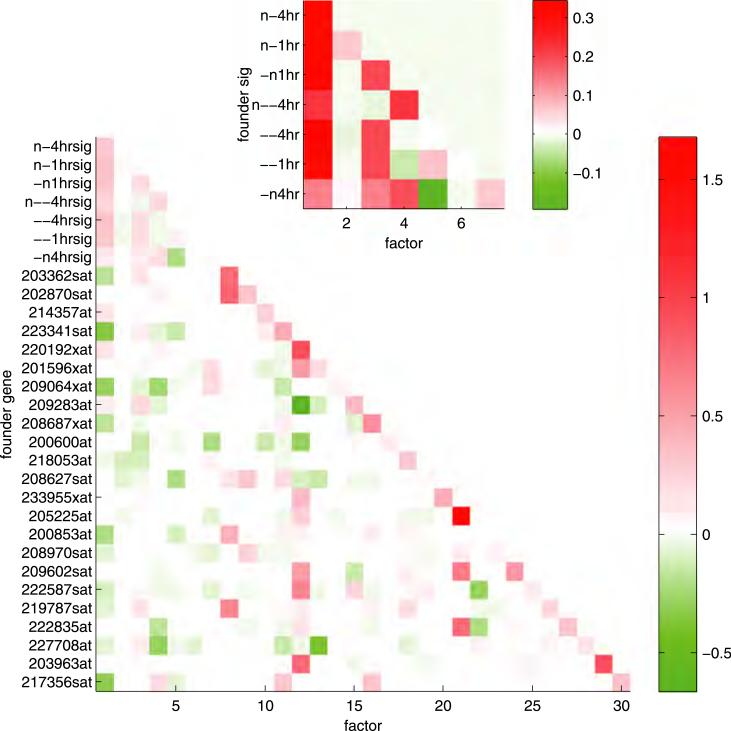
Posterior estimates of the factor loadings () aid in generating further insights. In particular, the upper portion of the estimated loadings matrix sheds light on the structure of connections between latent factors; see Figure 5. As described in Section 3.2, one interpretation of a row A is as a set of coefficients determining a linear combination of factor scores that predict the gene expression vector for the corresponding variable. The inset of Figure 5 shows that the fitted values of all 7 of signature scores involve positive contributions from factor 1, the factor version of the 4hr lactic acidosis signature. Thus, the pattern of 4hr lactic acidosis signature activity across samples describes a fundamental pattern of pathway activation that underlies the activity patterns of the other 6 signatures. The seventh factor (i.e., the factor representation of the 4hr neutralization signature) sits atop this hierarchy of pathway complexity, represented as a linear combination of factors 1 (4hr neutralization), 3 (1hr neutralization), 4 (4hr strong lactic acidosis) and 5 (4hr acidic growth), plus the additional pattern of expression unique to this pathway.
Fig. 5.
Heat map showing the magnitudes of the fitted factor loadings for the first L = 30 rows of A. The founder gene for each factor is designated by its U133 + probe ID. The terms in each row determine a linear combination of latent factors that predict the observed expression levels of the founder gene.
4. Biomedical connections of factor profiles
4.1 Factor-based prediction of long term survival
The in vivo latent factors linked to neutralization pathways represent complexity in the patterns of expres- sion, and therefore in the levels of underlying biological pathway activation evident across the tumor samples. For this reason, latent factors can be regarded as candidate biomarkers of physiological states that link to these pathways. Our study explores this using the posterior mean factor scores as candidate predictors in a survival analysis of the breast cancer patient data.
We use Weibull regression models of patient survival that draw on the 30 estimated neutralization/lactic acidosis pathway factors, the 7 original projected signature scores and the clinical covariates available for this data set [Miller et al. (2005)]. The latter include histologic grade, ER mutation status, node status, P53 mutation status, PgR mutation status, tumor size and age at diagnosis. This analysis allows both integration and comparison of the prognostic value of these traditional markers with specific pathway-related signature scores, and their latent factor representations—an integrative clinico-genomic analysis. Let ti denote the survival time of patient i. The Weibull density function is , where a > 0 is the index parameter and ηi = γ′yi the linear predictor based on a covariate vector yi. We explore subsets of covariates and regression model uncertainty using Bayesian shotgun stochastic search [Hans, Dobra and West (2007); Hans et al. (2007b)]. This generates a list of regression covariate subsets and the corresponding posterior regression model probabilities for use in Bayesian model averaging for survival prediction and in exploring relevance of variables. Details of model and prior specification follow defaults in the SSS software [Hans et al. (2007b)] as noted in Supplement B.
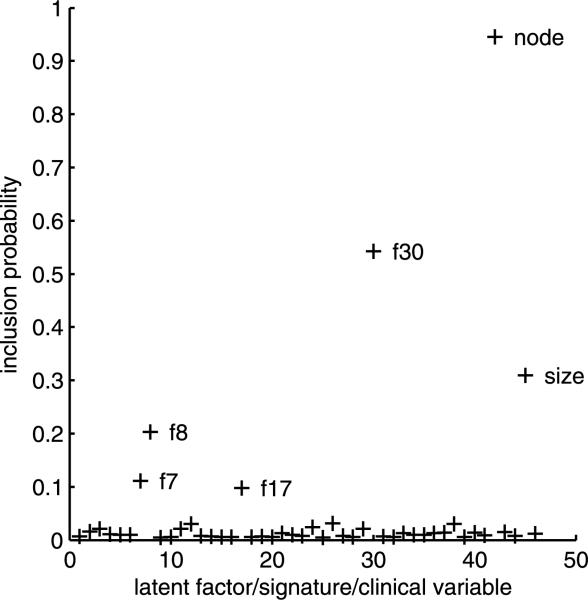
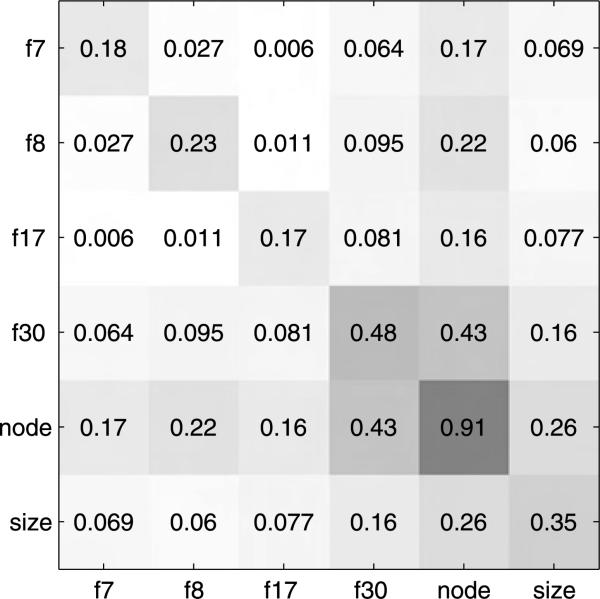
Figure 6 shows posterior probabilities for each of the 46 candidate covariates. Nodal status emerges as the leading predictor of long term survival, followed by latent factor 30 and then tumor size. Note that none of the original signature scores, and no other clinical variables, receive appreciable probability. That nodal status provides the best predictor of survival is to be expected. Previous studies [West et al. (2001); Pittman et al. (2004); Dressman et al. (2006)] have shown that nodal status is not well predicted by gene expression and that combined use of nodal status with gene expression predictors can improve survival prognosis. Hence, it seems that the information content of the nodal status predictor is unlikely to overlap with that of any factor score. This can also be clearly seen through the pairwise inclusion probabilities in Figure 7.
Fig. 6.
Posterior inclusion probabilities for the 46 candidate covariates in the Weibull models. The candidate covariates include the 30 estimated latent factors, followed by the 7 original signature scores, followed by 10 traditional clinical covariates (Elston grades 1, 2 and 3, ER, node status, P53, PgR status, tumor size, age at diagnosis).
Fig. 7.
Pairwise inclusion probabilities for the top 6 predictor variables in breast cancer survival analysis. Darker colored tiles indicate higher probabilities.
The pairwise inclusion probability of factor 30 and nodal status is close to the marginal probability of factor 30; however, the pairwise inclusion probability of factor 30 with any of the other factors is far less than any of the marginal inclusions probabilities of those factors; thus, factor 30 is clearly a dominant and preferred expression-based biomarker of survival risk.
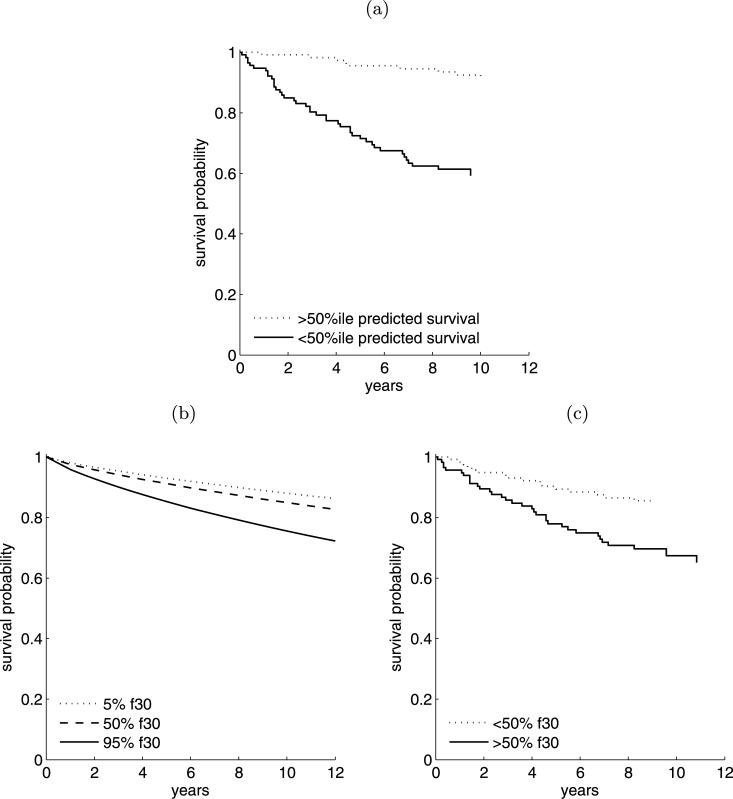
Posterior and predictive inferences are formally based on a mixture of 1000 Weibull survival models, mixed with respect to their posterior probabilities. For each patient i, we can compute the implied predicted survival function at her covariate vector yi and identify the predicted median survival mi; this is the predicted median survival time for a future patient who has the same covariates as patient i. Figure 8(a) shows a Kaplan–Meier survival plot of the Miller et al. data simply stratified by mi ≤ m or mi > m, where m = median{mi, i = 1,...,n}. There is approximately a 30% difference in the empirical 10-year survival probability between patients cohorts stratified crudely on this basis, as a simple visual of the relevance of the included covariates. By way of focusing on factor 30, we plot the model-averaged survival curve for a hypothetical patient whose covariates are held constant at their median values in the data set, save for variation in the factor 30 score; see Figure 8(b), where factor 30 is set at its 5th, 50th and 95th percentiles in the data set, all other covariates remaining fixed. The estimated effect of variation in factor 30 alone accounts for approximately 20% of the difference in 10-year patient survival between the high-risk and low-risk subgroups. This prediction is confirmed by considering the Kaplan–Meier curves formed by stratifying the patients on the basis of the patient-specific factor 30 value compared to the median across samples; see Figure 8(c). The pattern of gene expression comprising the loading associated with factor 30 warrants further investigation, to which we will return in Section 4.3.
Fig. 8.
(a) Kaplan–Meier curves demonstrating stratification of Miller data into high- and low-risk groups, based on the fitted Weibull mixture. (b) Estimated survival curve associated with varying levels of factor 30, holding all other predictors at their median values. (c) Kaplan–Meier curves demonstrating stratification of Miller data into high- and low- risk groups, based solely on the value of factor 30.
4.2 Out-of-sample factor projection
It is critical to evaluate whether or not the above results can be confirmed through out-of-sample prediction. We do this with two additional breast cancer data sets: that of Pawitan et al. (2005), consisting of 159 primary breast tumors assayed on Affymetrix U133A and U133B chips, and that of Sotiriou et al. (2006), consisting of 189 primary breast tumors assayed on U133A chips.
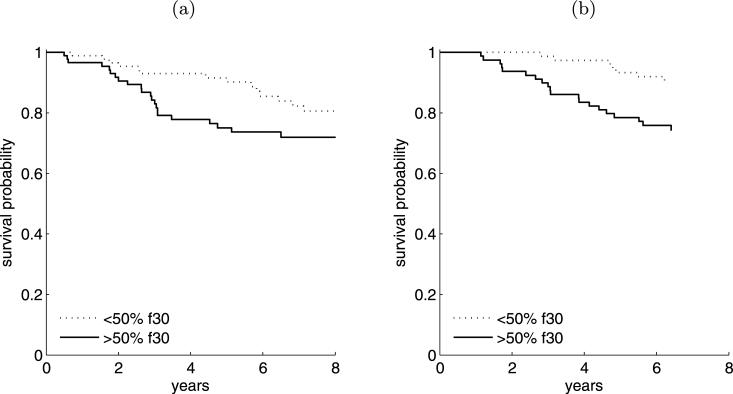
Fixing all factor model parameters at their posterior means, we can directly predict values of the latent factors for each new patient; see Supplement B and [Lucas et al. (2009)]. Note that this calculation is purely predictive; no model fitting nor additional analysis of the two validation data sets was performed. Using the predicted latent factor vectors, we can produce the same survival plots for these data, stratifying each of the two new patient cohorts on the basis of their factor 30 scores as above; see Figure 9, as compared to Figure 8(c). The association between low factor 30 scores and good prognosis remains evident in these out-of-sample predictions that draw on different patient populations. Further, the differences between high and low risk groups is comparable across all three samples.
Fig. 9.
Kaplan–Meier curves demonstrating stratification of (a) the Pawitan et al. (2005) patient samples, and (b) the Sotiriou et al. (2006) patient samples (right) into high- and low- risk groups based on imputed values of factor 30, as identified in the latent factor analysis of the Miller data.
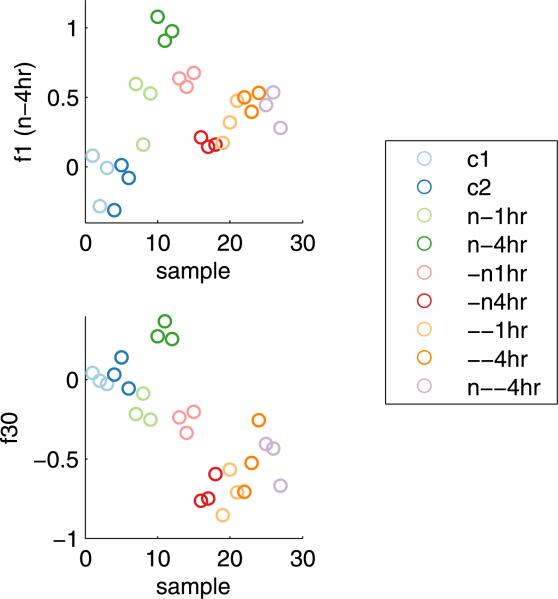
The robustness of factor 30 as a prognostic biomarker provides strong support for the view that it reflects a biologically meaningful module of gene expression. By evaluating the predicted factor scores in the original experimental data, we are able to establish that factor 30, despite its relatively late incorporation to the model, is linked to the 4-hour lactic acidosis pathway. Figure 10 compares the predicted factor scores for factors 1 and 30 as evaluated in the experimental data. Factor 1, which is founded by the 4-hour lactic acidosis signature, is in fact comparable to the original signature score as depicted in Figure 2. Factor 30 has its highest values in the original 4-hour lactic acidosis samples, but shows a different pattern of activity across the other sample cohorts. In particular, factor 30 appears to be repressed in the samples associated with the 4-hour neutralization and acidic growth treatments. This implies that factor 30 may characterize some critical intersection between these pathways that is itself related to tumor aggressiveness.
Fig. 10.
Predicted values of factors 1 and 30 for each sample in the original experimental study. Separate treatment groups are color coded.
4.3 Biological evaluation of prognostic factor 30
Having established the clinical relevance of factor 30, the task remains to ascribe to it biological meaning. The loading of factor 30 is comprised of only 6 gene probe sets for which . Four of these, including the founder gene, are related to the phosphoglycerate kinase 1 (PGK1) gene, while the other two are related to a neuronal cell death-related protein and the CEGP1 protein. The factor is characterized by overexpression (βg,k > 0) of the PGK1 and neuronal cell death proteins and suppression (βg,k < 0) of CEGP1.
A literature search generates detailed biological information on PGK1, and its role in the glycolysis pathway where it is fundamental to cell growth and metabolism. PGK1 catalyzes the reversible conversion of 1,3-diphosphoglycerate to 3-phosphoglycerate with the generation of one molecule of ATP and this represents an important step in glycolysis pathways. In addition, PGK1 has been reported to induce other processes related to cancer progression, such as conferring a multi-drug resistant (MDR) phenotype [Duan et al. (2002)] and affecting tumor angiogenesis through affecting secreted plasmin [Lay et al. (2000)]. Previous studies have also shown that elevated levels of PGK1 predict poor survival outcomes in lung cancers [Chen et al. (2003)], and that PGK1 can often be expressed at high levels in pancreatic [Hwang et al. (2006)] and renal [Unwin et al. (2003)] cancers. The association between high factor 30 levels and poor prognosis indicates a similar relationship between PGK1 and survival may exist for breast cancers.
Since PGK1 is an important component of glycolysis pathways, our findings here may implicate glycolysis activities in poor patient survival. This is supported by previous findings that expression of glycolysis pathways and PGK1 are repressed by lactic acidosis [Chen et al. (2008)]. Factor 30 links the neutralization pathway response signatures to a clear PGK1 factor that may now serve as a bio-marker of one key aspect of tumor responses to changes in pH with the potential to aid in predicting follow-on changes in tumor metabolism via glycolysis pathway activation. Further evaluation of this chain of relationships is now initiated and will be explored using independent methods such as tumor tissue microarrays [Chen et al. (2003)]. Since PGK1 and glycolysis pathways are also controlled by hypoxia [Chi et al. (2006)], these results also highlight their potential roles as integral mediators of multiple micro-environmental factors affecting tumor progression and clinical outcomes.
Supplementary Material
SUPPLEMENTARY MATERIAL
Supplement A: Software and Data (DOI: 10.1214/09-AOAS261SUPPA; url). This site contains all materials needed to reproduce the reported analyses. This includes all data files, control files for the BFRM and SSS software, and MATLAB functions for producing graphical summaries.
Supplement B: Appendix (DOI: 10.1214/09-AOAS261SUPPB; .pdf). The appendix Merl et al. (2009) provides further details on prior specifications in the sparse regression and sparse latent factor models. The appendix also contains details on the control parameters for the evolutionary factor search and shotgun stochastic search, and describes the procedure for imputing factor scores in new samples.
Acknowledgments
The authors are grateful to an editor of AOAS and an anonymous reviewer for constructive comments on this manuscript.
Footnotes
Supported in part by NSF Grant DMS-03-42172 and National Institutes of Health Grant NCI U54-CA-112952. Any opinions, findings and conclusions or recommendations expressed in this work are those of the authors and do not necessarily reflect the views of the NSF or NIH.
Contributor Information
Daniel Merl, Department of Statistical Science Duke University Durham, North Carolina 27708-0251 USA dan@stat.duke.edu.
Julia Ling-Yu Chen, Department of Molecular Genetics & Microbiology Institute for Genome Sciences and Policy Duke University Medical Center Box 3382 Durham, North Carolina 27710-3382 USA julia.chen@duke.edu.
Jen-Tsan Chi, Department of Molecular Genetics & Microbiology Institute for Genome Sciences and Policy Duke University Medical Center Box 3382 Durham, North Carolina 27710-3382 USA jentsan.chi@duke.edu.
Mike West, Department of Statistical Science Institute for Genome Sciences and Policy Duke University Durham, North Carolina 27708-0251 USA mw@stat.duke.edu.
REFERENCES
- Carvalho C, Chang J, Lucas J, Nevins J, Wang Q, West M. High-dimensional sparse factor modelling: Applications in gene expression genomics. J. Amer. Statist. Assoc. 2008;103:1438–1456. doi: 10.1198/016214508000000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Gharib T, Wang H, Huang C, Kuick R, Thomas D, Shedden K, Misek D, Taylor J, Giordano T, Kardia S, Iannettoni M, Yee J, Hogg P, Orringer M, Hanash S, Beer D. Protein profiles associated with survival in lung adenocarcinoma. Proc. Natl. Acad. Sci. 2003;100:13537–13542. doi: 10.1073/pnas.2233850100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL-Y, Lucas J, Schroeder T, Mori S, Nevins J, Dewhirst M, West M, Chi J. Genomic analysis of response to lactic acidosis and acidosis in human cancers. PLoS Genetics. 2008;4:e1000293. doi: 10.1371/journal.pgen.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi J, Wang Z, Nuyten D, Rodriguez E, Schaner M, Salim A, Wang Y, Kristensen G, Helland A, Borresen-Dale A, Giaccia A, Longaker M, Hastie T, Yang G, van de Vijer M, Brown P. Gene expression programs in response to hypoxia: Cell type specificity and prognostic significance in human cancers. PLoS Medicine. 2006;3:e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressman H, Hans C, Bild A, Olson J, Rosen E, Marcom P, Liocheva V, Jones E, Vujaskovic Z, Marks J, Dewhirst M, West M, Nevins J, Black-Well K. Gene expression profiles of multiple breast cancer phenotypes and response to neoadjuvant chemotherapy. Clinical Cancer Research. 2006;12:819–826. doi: 10.1158/1078-0432.CCR-05-1447. [DOI] [PubMed] [Google Scholar]
- Duan Z, Lamendola D, Yusuf R, Penson R, Preffer F, Seiden M. Overexpression of human phosphoglycerate kinase 1 (pgk1) induces a multidrug resistance phenotype. Anticancer Research. 2002;22:1933–1941. [PubMed] [Google Scholar]
- Hanahan D, Weinberg R. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hans C, Dobra A, West M. Shotgun stochastic search in regression with many predictors. J. Amer. Statist. Assoc. 2007;102:507–516. MR2370849. [Google Scholar]
- Hans C, Wang Q, Dobra A, West M. SSS: High-dimensional Bayesian regression model search. Bulletin of the International Society for Bayesian Analysis. 2007b;14:8–9. [Google Scholar]
- Hwang T, Liang Y, Chien K, Yu J. Overexpression and elevated serum levels of phosphoglycerate kinase 1 in pancreatic ductal adenocarcinoma. Proteomics. 2006;6:2259–2272. doi: 10.1002/pmic.200500345. [DOI] [PubMed] [Google Scholar]
- Irizarry R, Bolstad B, Collin F, Cope L, Hobbs B, Speed T. Summaries of affymetrix genechip probe level data. Nucleic Acids Research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay A, Jiang X, Kisker O, Flynn E, Underwood A, Condron R, Hogg P. Phosphoglycerate kinase acts in tumour angiogenesis as a disulphide reductase. Nature. 2000;408:869–873. doi: 10.1038/35048596. [DOI] [PubMed] [Google Scholar]
- Lucas J, Carvalho C, Wang Q, Bild A, Nevins J, West M. Sparse statistical modelling in gene expression genomics. In: Müller P, Do K, Vannucci M, editors. Bayesian Inference for Gene Expression and Proteomics. Cambridge Univ. Press; 2006. pp. 155–176. MR2269095. [Google Scholar]
- Lucas J, Carvalho C, West M. A Bayesian analysis strategy for cross-study translation of gene expression biomarkers. Statist. Appl. Genet. Mol. Biol. 2009;8 doi: 10.2202/1544-6115.1436. art11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas J, Carvalho C, Merl D, West M. In-vitro to in-vivo factor profiling in expression genomics. In: Dey D, Ghosh S, Mallick B, editors. Bayesian Modeling in Bioinformatics. Chapman & Hall/CRC; 2009. To appear. [Google Scholar]
- Merl D, Chen JL-Y, Chi J, West M. Supplement to “An integrative analysis of cancer gene expression studies using Bayesian latent factor modeling.”. 2009. DOI: 10.1214/09-AOAS261SUPPA, 10.1214/09-AOAS261SUPPB. [DOI] [PMC free article] [PubMed]
- Miller L, Smeds J, George J, Vega V, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu E, Bergh J. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawitan Y, Bjohle J, Amler L, Borg A, Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Slaar S, Liu E, Miller M, Nordgren H, Ploner A, Sandelin K, Shaw P, Smeds J, Skoog L, Wedren S, Bergh J. Gene expression profiling spares early breast cancer patients from adjuvant therapy: Derived and validated in two population-based cohorts. Breast Cancer Research. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman J, Huang E, Dressman H, Horng C, Cheng S, Tsou M, Chen C, Bild A, Iversen E, Huang A, Nevins J, West M. Integrated modeling of clinical and gene expression information for personalized prediction of disease outcomes. Proc. Natl. Acad. Sci. 2004;101:8431–8436. doi: 10.1073/pnas.0401736101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo D, Goldschmidt-Clermont P, West M. Of mice and men: Sparse statistical modelling in cardiovascular genomics. Ann. Appl. Statist. 2007;1:152–178. MR2393845. [Google Scholar]
- Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, Desmedt C, Larsimont D, Cardoso F, Peterse H, Nuyten D, Buyse M, Van de Vijver M, Bergh J, Piccart M, Delorenzi M. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. Journal of the National Cancer Institute. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- Unwin R, Craven R, Harnden P, Hanrahan S, Totty N, Knowles M, Eardley I, Selby P, Banks R. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the warburg effect. Proteomics. 2003;3:1620–1632. doi: 10.1002/pmic.200300464. [DOI] [PubMed] [Google Scholar]
- Wang Q, Carvalho C, Lucas J, West M. BFRM: Bayesian factor regression modelling. Bulletin of the International Society for Bayesian Analysis. 2007;14:4–5. [Google Scholar]
- West M. Bayesian factor regression models in the “large p, small n” paradigm. In: Bernardo J, Bayarri M, Berger J, Dawid A, Heckerman D, Smith A, West M, editors. Bayesian Statistics 7. Oxford Univ. Press; 2003. pp. 723–732. MR2003537. [Google Scholar]
- West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, Zuzan H, Marks J, Nevins J. Predicting the clinical status of human breast cancer utilizing gene expression profiles. Proc. Natl. Acad. Sci. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY MATERIAL
Supplement A: Software and Data (DOI: 10.1214/09-AOAS261SUPPA; url). This site contains all materials needed to reproduce the reported analyses. This includes all data files, control files for the BFRM and SSS software, and MATLAB functions for producing graphical summaries.
Supplement B: Appendix (DOI: 10.1214/09-AOAS261SUPPB; .pdf). The appendix Merl et al. (2009) provides further details on prior specifications in the sparse regression and sparse latent factor models. The appendix also contains details on the control parameters for the evolutionary factor search and shotgun stochastic search, and describes the procedure for imputing factor scores in new samples.