Abstract
Base excision repair (BER) is a critical pathway in cellular defense against endogenous or exogenous DNA damage. This elaborate multistep process is initiated by DNA glycosylases that excise the damaged base, and continues through the concerted action of additional proteins that finally restore DNA to the unmodified state. BER has been subject to detailed biochemical analysis in bacteria, yeast and animals, mainly through in vitro reproduction of the entire repair reaction in cell-free extracts. However, an understanding of this repair pathway in plants has consistently lagged behind. We report the extension of BER biochemical analysis to plants, using Arabidopsis cell extracts to monitor repair of DNA base damage in vitro. We have used this system to demonstrate that Arabidopsis cell extracts contain the enzymatic machinery required to completely repair ubiquitous DNA lesions, such as uracil and abasic (AP) sites. Our results reveal that AP sites generated after uracil excision are processed both by AP endonucleases and AP lyases, generating either 5′- or 3′-blocked ends, respectively. We have also found that gap filling and ligation may proceed either through insertion of just one nucleotide (short-patch BER) or several nucleotides (long-patch BER). This experimental system should prove useful in the biochemical and genetic dissection of BER in plants, and contribute to provide a broader picture of the evolution and biological relevance of DNA repair pathways.
Keywords: abasic sites, Arabidopsis, DNA polymerase, DNA repair, uracil
Introduction
Maintaining genome integrity in the face of constant DNA damage induced by spontaneous chemical changes or genotoxic agents is an essential task for all organisms (Friedberg et al., 2006; Lindahl, 1993). A major pathway for protecting DNA from endogenous or exogenous damage is base excision repair (BER). This is a multistep process initiated by DNA glycosylases that excise damaged or incorrect bases and generate an abasic (apurinic/apyrimidinic, AP) site (Stivers and Jiang, 2003; Zharkov and Grollman, 2005). AP sites may also arise from normal bases by spontaneous hydrolysis of the N-glycosylic bond (Lindahl, 1993). The subsequent steps required to complete the repair include incision at the AP site, generation of a gap, repair synthesis and ligation (Fortini and Dogliotti, 2007).
BER has been extensively studied in higher eukaryotes. In mammalian cells, AP sites generated during BER are usually processed by AP endonucleases that create an incision in the 5′ side of the AP site, leaving 3′-OH and 5′-deoxyribose-5-phosphate (5′-dRP) termini (Dianov et al., 1992; Levin and Demple, 1990). The ensuing gap filling may take place either by insertion of a single nucleotide (short-patch repair, SP) or by DNA synthesis involving several nucleotides (long-patch repair, LP) (Fortini and Dogliotti, 2007). In the SP BER subpathway, gap filling is performed by DNA polymerase β (Pol β), which is also endowed with dRPase activity that releases the blocking 5′-dRP end and allows DNA ligation (Srivastava et al., 1998). In the LP BER subpathway, DNA synthesis may be performed by replicative DNA polymerases δ or ε, which carry out displacement of the strand containing the 5′-dRP terminus, and generate a ‘flap’ structure that is excised by the 5′-flap endonuclease FEN1 before ligation (Pascucci et al., 1999). Both SP and LP BER have been reconstituted in vitro using purified proteins (Dianov and Lindahl, 1994; Klungland and Lindahl, 1997; Kubota et al., 1996; Nicholl et al., 1997; Pascucci et al., 1999; Srivastava et al., 1998), and the choice between the subpathways in cells may depend on various factors, such as the nature of the lesion and the type of DNA glycosylase that initiates the BER process (Fortini and Dogliotti, 2007).
As experimental data accumulate for non-mammalian systems, it is becoming clear that there are significant differences in the strategies employed by different species during the post-excision events that take place in BER (Kelley et al., 2003). Therefore, a complete understanding of this repair pathway will require mechanistic studies involving species from all main groups of organisms. However, plants have usually been neglected in the biochemical and genetic analysis of DNA repair pathways, which have generally focused on bacterial, yeast and mammalian systems (Friedberg et al., 2006). Recent research efforts are rapidly changing this, and substantial knowledge has begun to accumulate about plant DNA repair processes (Bray and West, 2005; Britt, 2002; Hays, 2002; Kunz et al., 2005). Numerous studies mostly performed in Arabidopsis have shown that plants are equipped with structural and/or functional homologs of most of the BER proteins identified in other organisms (Britt, 2002; Hays, 2002; Roldan-Arjona and Ariza, 2009). Additionally, recent genetic and biochemical evidence has revealed that plant BER performs a key role in epigenetic regulation, through active DNA demethylation initiated by 5-methylcytosine DNA glycosylases (Agius et al., 2006; Gehring et al., 2006; Morales-Ruiz et al., 2006). Therefore, unraveling the enzymatic mechanisms of BER will be critical to understand its full relevance for plant physiology.
Much of the knowledge gathered during the past 20 years about the enzymology of animal and microbial BER has been obtained through the use of cell-free in vitro assays that allow the steps of the repair process to be monitored (for a review, see Dianov, 2003). The complete BER pathway has been reproduced in vitro using extracts from Escherichia coli (Dianov et al., 1992), Saccharomyces cerevisiae (Wang et al., 1993), Schizosaccharomyces pombe (Alseth et al., 2004), Xenopus laevis (Matsumoto and Bogenhagen, 1989, 1991), Plasmodium falciparum (Haltiwanger et al., 2000), Caenorhabditis elegans (Shatilla and Ramotar, 2002) and human cells (Dianov et al., 1992). However, to date, there are no reports describing assays for specifically detecting BER in plant cells. This has been a persistent obstacle to understanding the relevance of this DNA repair pathway in plants, leaving many significant questions unanswered. In this study, we have extended the biochemical analysis of BER to plants by developing a specific assay to monitor the repair of damaged bases in Arabidopsis whole-cell extracts. Using this assay we have found that repair intermediates arising during plant BER may be processed both by AP lyases and AP endonucleases. We have also established that uracil and AP site repair may be completed both via single-nucleotide insertion and by long-patch DNA synthesis. We have also explored the DNA polymerase requirements for the gap-filling step. We discuss our findings in the light of what is currently known about plant DNA polymerases.
Results
Uracil excision repair in Arabidopsis whole-cell extracts
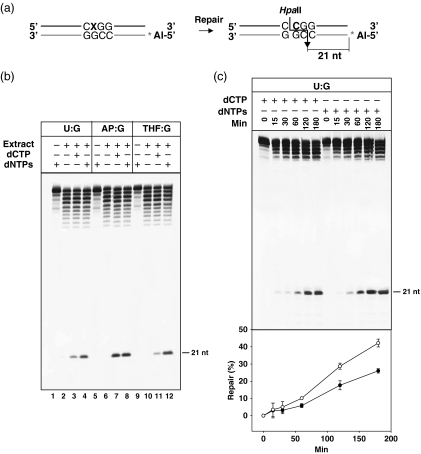
We chose uracil as a convenient model lesion to investigate BER in Arabidopsis cells, as uracil-initiated BER is well documented in different prokaryotic and eukaryotic systems (Friedberg et al., 2006). Uracil in DNA may arise either from spontaneous deamination of cytosine or from erroneous incorporation of dUMP instead of dTMP, leading to mutagenic U:G mispairs or to U:A pairs, respectively (Kavli et al., 2007). To establish an in vitro system for BER, we used a 51-mer duplex oligonucleotide labeled at the 5′ end of the lower strand, with a single U residue within an HpaII recognition site on the upper strand, as the DNA substrate (Figure 1a). The presence of U instead of C at the restriction site makes the DNA substrate refractory to HpaII cleavage. In this system, uracil repair is detected as a conversion of duplex DNA to a form susceptible to HpaII digestion, visualized as a 21-nt-labeled fragment following polyacrylamide gel electrophoresis (Figure 1a).
Figure 1.
Repair of base DNA damage by Arabidopsis cell extracts. (a) Schematic diagram of molecules used as DNA substrates. Double-stranded oligonucleotides contained a lesion (X = uracil or AP site) at an HpaII site on the upper strand. The Alexa Fluor-labeled 5′ end of the lower strand is indicated by a star. The size of the 5′ end-labeled fragment generated after HpaII digestion of the fully repaired product is indicated on the right. (b) DNA duplexes containing U, a natural AP site, or THF opposite G were incubated with Arabidopsis cell extract at 30°C for 3 h in a reaction mixture containing either dCTP or all four dNTPs, as indicated. Reaction products were digested with HpaII, separated in a 12% denaturing polyacrylamide gel and quantified by fluorescence scanning. (c) A DNA duplex containing a U:G mispair was incubated with Arabidopsis cell extracts at 30°C in a reaction mixture containing either dCTP or all four dNTPs. Reactions were stopped at the indicated times and products were analyzed as described above. The percentage of fully repaired DNA product for reactions containing only dCTP (•) or all four dNTPs (○) is shown in the bottom panel. Values are means with standard errors from two independent experiments.
Extracts obtained from snap-frozen 15-day-old seedlings (see Experimental procedures) efficiently and reproducibly converted the uracil-containing DNA to a form susceptible to HpaII digestion (Figure 1b). Importantly, repair was dependent on the presence of deoxyribonucleotide triphosphates (dNTPs) in the reaction mixture, but was also detected when only deoxycytidine triphosphate (dCTP) was present (Figure 1b, lanes 3 and 4). Equivalent results were observed when the whole-cell extract was incubated with DNA substrates containing either a natural AP site or a synthetic AP site analog (tetrahydrofuran, THF), instead of uracil (Figure 1b, lanes 5–12). Fragments shorter than 51 nt detected in the upper part of the gel are probably the result of exonuclease activity in the extracts, as they were not observed when incubations were performed in the absence of Mg2+ (data not shown). We also examined the repair kinetics of a U:G mispair, measuring the level of fully repaired product at different times (Figure 1c). We found that repair was faster when all four deoxynucleotides were present in the reaction mixture, but the rate of repair was also significant when the only deoxynucleotide available was dCTP.
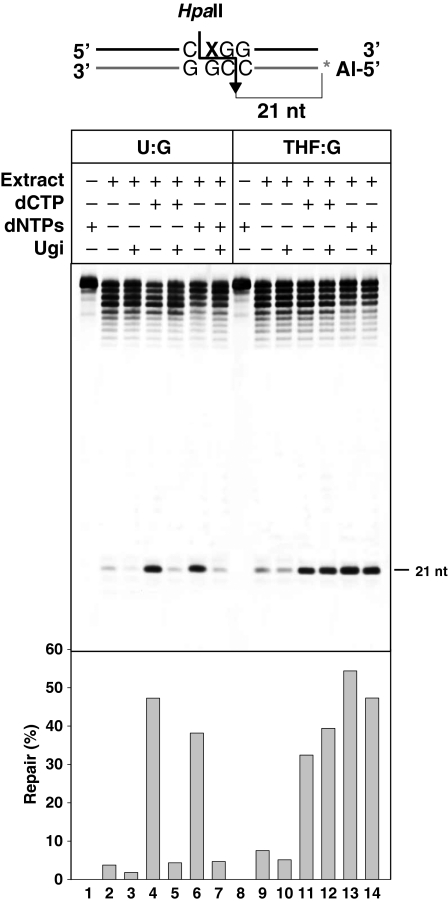
To assess whether the first step of the repair process is dependent on uracil DNA glycosylase (UDG) activity, we examined the effect of uracil-DNA glycosylase inhibitor (Ugi) on the reaction. This small peptide is a specific inhibitor of UDGs from the UNG family, and has been successfully used to detect UDG-dependent uracil excision in cell extracts from different organisms (Bennett et al., 2001; Shatilla and Ramotar, 2002; Zeitlin et al., 2005). We found that repair of uracil-containing DNA was strongly reduced when Ugi was added to the reaction, in the presence of either dCTP or of all four dNTPs (Figure 2, lanes 5 and 7). In contrast, the addition of Ugi had no significant effect on the repair of an analogous DNA substrate containing a synthetic AP site (Figure 2, lanes 12 and 14). We conclude that Arabidopsis cell extracts are able to fully repair a U:G mismatch through a BER pathway initiated by UDG activity.
Figure 2.
Uracil repair by Arabidopsis cell extracts is dependent on uracil DNA glycosylase (UDG) activity. DNA duplexes containing either U or THF opposite G were incubated with Arabidopsis cell extracts at 30°C for 3 h in a reaction mixture containing either dCTP or all four dNTPs, both in the absence or presence of 2 U of uracil-DNA glycosylase inhibitor (Ugi). Reaction products were digested with HpaII, separated in a 12% denaturing polyacrylamide gel and quantified by fluorescence scanning. The percentage of fully repaired DNA product is shown in the bottom panel.
AP sites generated after uracil excision may be processed both by AP endonucleases and AP lyases
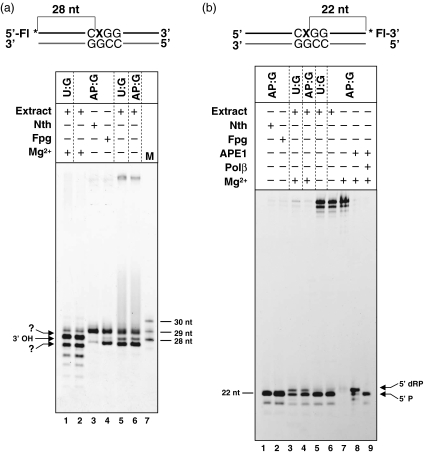
We next turned our attention to the nature of the 3′ and 5′ termini generated on the damaged DNA strand following uracil excision. AP sites generated after base excision may be processed by AP endonucleases, but also by the AP lyase activity associated with bifunctional DNA glycosylases/lyases. These two kinds of activities produce very different types of 5′ and 3′ termini (Figure S1). AP endonucleases are usually Mg2+-dependent, and they perform hydrolysis at the 5′ side of the AP site, yielding a 3′-OH end and a 5′-dRP terminus (Levin and Demple, 1990). By contrast, AP lyases cleave 3′ to the AP site through a Mg2+-independent β-elimination mechanism, leaving a 5′-phosphate (5′-P) end and a 3′-α,β-unsaturated aldehyde (Levin and Demple, 1990); in some cases, they additionally catalyze δ-elimination, generating a 3′-phosphate (3′-P) terminus (Zharkov et al., 2003). Thus, AP endonucleases and AP lyases generate 5′- and 3′-blocked ends, respectively. To determine whether either or both of these two types of termini are generated during repair of uracil or AP sites by Arabidopsis cell-free extracts, we performed repair reactions in the absence or presence of Mg2+, using DNA substrates in which the lesion-containing strand was labeled at the 5′ or 3′ end (Figure 3).
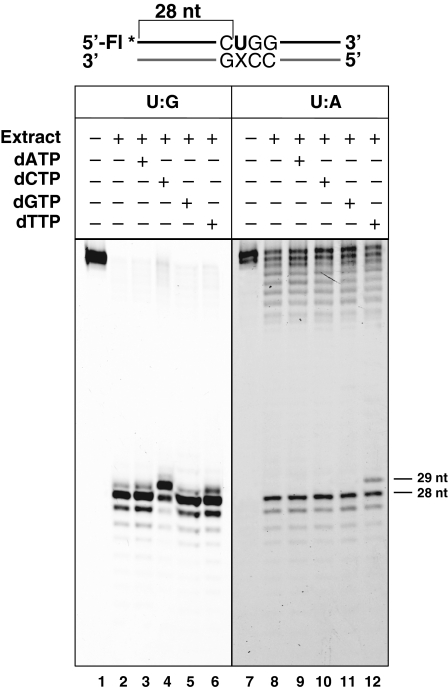
Figure 3.
AP sites generated after uracil excision are processed both by AP endonucleases and AP lyases. (a) Analysis of 3′ termini. DNA duplexes containing either U or a natural AP site at the 5′ end-labeled upper strand were incubated at 30°C for 3 h with different combinations of Arabidopsis cell extract, Escherichia coli Nth (13 ng), E. coli Fpg (20 ng) and MgCl2 (5 mm), as indicated. Reaction products were separated in a 12% denaturing polyacrylamide gel, and detected by fluorescence scanning. Size markers corresponding to 28, 29 and 30 nt were loaded on lane 7. Fragments corresponding to β- and δ-elimination products, and to 3′-OH termini are indicated by arrows. (b) Analysis of 5′ termini. DNA duplexes containing either U or a natural AP site at the 3′ end-labeled upper strand were incubated at 30°C for 3 h with different combinations of Arabidopsis cell extract, E. coli Nth (13 ng), E. coli Fpg (20 ng), human APE1 (10 U), human Pol β (2.4 U) and MgCl2 (5 mm), as indicated. Reaction products were stabilized by reduction with NaBH4, separated in a 12% denaturing polyacrylamide gel and then detected by fluorescence scanning. Fragments corresponding to 5′-deoxyribose-5-phosphate (5′-dRP) and 5′-phosphate (5′-P) products are indicated by arrows.
We first examined the 3′ terminus, using DNA substrates in which the 5′ end-labeled damaged strand contained either a uracil residue or a natural AP site (Figure 3a). When the repair reaction was carried out in the presence of Mg2+, a major 28-nt fragment was detected (Figure 3a, lanes 1 and 2), accompanied by smaller fragments probably arising by exonucleolytic degradation. This fragment corresponds to a 3′-OH end, and is probably generated by AP endonuclease incision at the 5′ side of the AP site. When the DNA substrates were incubated with the cell extract in the absence of Mg2+, two major fragments appeared (Figure 3a, lanes 5 and 6). These two products co-migrated with those generated on an AP site-containing substrate by the β- and β,δ-elimination activities of the E. coli DNA glycosylases/lyases endonuclease III (Nth) and Fpg, respectively (Figure 3a, lanes 3 and 4). Quantification of the relative fluorescence intensity revealed that fragments corresponding to δ-elimination products are threefold more abundant than those representing β-elimination ends (Figure 3a, lanes 5 and 6).
We next analyzed the nature of the 5′ terminus, using DNA substrates in which the damaged strand was labeled at the 3′ end (Figure 3b). As the 5′-blocked ends arising by AP endonuclease activity are known to be alkali labile, the reaction products were stabilized by reduction with sodium borohydride (NaBH4) prior to PAGE analysis (Dianov et al., 1992). We found that in the absence of Mg2+, the extract catalyzed incision at both the U- and AP-containing substrates to generate a single 22-nt fragment (Figure 3b, lanes 5 and 6). This fragment co-migrated with those produced by E. coli Nth or Fpg DNA glycosylases/lyases on an AP site-containing substrate (Figure 3b, lanes 1 and 2), and with the product generated by the dRP-lyase activity of Pol β (Figure 3b, lane 9). Therefore, it probably represents a 5′-P terminus. In contrast, when the reaction was performed in the presence of Mg2+ a slightly longer fragment appeared (Figure 3b, lanes 3 and 4). This co-migrated with the 5′-dRP product generated by incubation of the AP:G pair with human AP endonuclease APE1 (Figure 3b, lane 8), and was undetectable when the reaction products were not stabilized with NaBH4 (data not shown). Therefore, it probably represents a 5′-dRP blocked terminus.
Taken together, these results indicate that AP sites generated after uracil excision by Arabidopsis cell extracts may be processed either by AP endonucleases or by AP lyases, yielding either 5′- or 3′-blocked ends, respectively.
Uracil repair involves single-nucleotide insertion and/or long-patch DNA synthesis
As shown in Figure 1b, excision repair of uracil depends on the presence of dNTPs in the reaction mixture, but can also be completed when only dCTP is present. These results suggest that uracil excision repair in Arabidopsis extracts takes place through short- and long-patch DNA synthesis. To explore this possibility, we analyzed the DNA synthesis intermediates arising during the repair reaction.
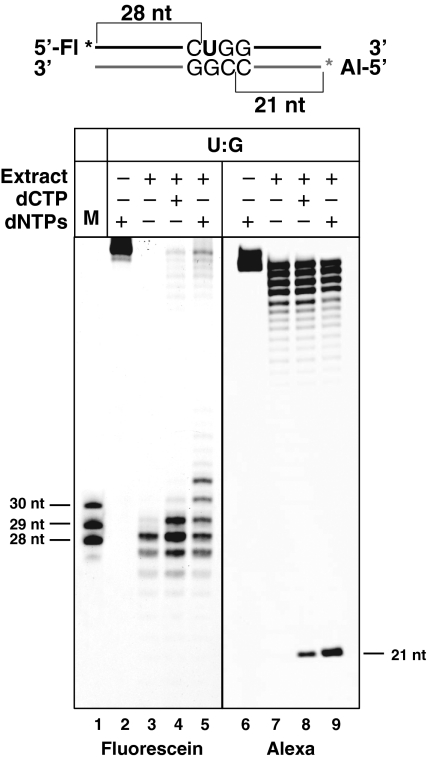
Repair reactions were performed incubating Arabidopsis extracts with a duplex oligonucleotide labeled with fluorescein at the 5′ end of the upper strand, and with Alexa Fluor at the 5′ end of the lower strand (Figure 4). We found that fully repaired products were undetectable in the absence of dNTPs (Figure 4, lane 7), although most of the upper strand was processed to generate a 28-nt fragment corresponding to a free 3′-OH terminus (Figure 4, lane 3). In the presence of dCTP as the only deoxynucleotide, a fully repaired product was detected (Figure 4, lane 8), with the concomitant appearance of a 29-nt fragment on the upper strand, corresponding to the insertion of dCMP in the repair gap (Figure 4, lane 4). Incubation in the presence of all four dNTPs somewhat increased the quantity of fully repaired product (Figure 4, lane 9), and this was accompanied by the accumulation of upper-strand reaction intermediates, which comigrated with 29-, 30- and 31-nt fragments containing free 3′-OH ends (Figure 4, lane 5). These results indicate that up to three nucleotides are inserted when all four dNTPs are present in the repair reaction.
Figure 4.
Uracil excision repair involves single-nucleotide insertion and long-patch DNA synthesis. Arabidopsis cell extract was incubated with duplex DNA that contained a U residue in the upper strand, and which was labeled at the 5′ end of the upper strand with fluorescein and at the 5′ end of the lower strand with Alexa Fluor. Reactions were incubated at 30°C for 3 h in a reaction mixture containing either dCTP or all four dNTPs, as indicated. Reaction products were separated in a 12% denaturing polyacrylamide gel either before (lanes 2–5) or after (lanes 6–9) HpaII digestion. Fluorescein- (lanes 2–5) and Alexa Fluor-labeled fragments (lanes 6–9) were detected by fluorescence scanning.
We then investigated whether the single-nucleotide insertion observed in the presence of dCTP was template-dependent. We performed repair reactions with DNA substrates containing either a U:G or a U:A pair, in the presence of a single deoxynucleotide (dATP, dCTP, dGTP or dGTP) (Figure 5). Only dCMP or dTMP was inserted in the DNA substrates containing either U:G or U:A, respectively (Figure 5, lanes 4 and 12). The faint bands just above the 28-nt position (Figure 5, lanes 4 and 12) might represent some β-elimination product(s) generated by AP lyases. As previously reported for mammalian cells (Visnes et al., 2008), we found that Arabidopsis extracts processed U:G pairs more efficiently than U:A pairs.
Figure 5.
Single-nucleotide insertion after uracil excision is template-dependent. Duplex DNA containing a U residue at the 5′ end-labeled upper strand paired to G (left panel) or A (right panel) was incubated at 30°C for 3 h with Arabidopsis cell extract and a single dNTP, as indicated. Reaction products were separated in a 12% denaturing polyacrylamide gel, and then detected by fluorescence scanning. Fragments corresponding to 28 and 29 nt are indicated by arrows.
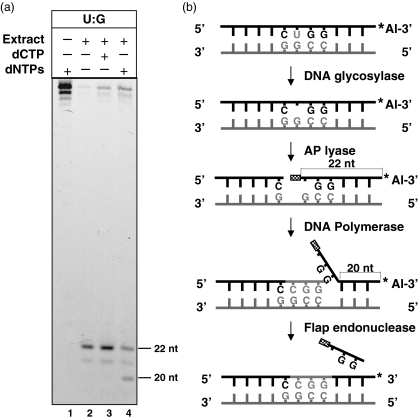
The standard model of LP mammalian BER proposes that, after base excision and AP site incision, the damaged strand undergoes displacement 3′ to the nick, and that the resulting flap structure is finally excised by FEN1 before DNA ligation can occur (Klungland and Lindahl, 1997; Prasad et al., 2000). We therefore looked for evidence of displacement and incision 3′ to the damaged base on the uracil-containing strand when all four dNTPs were present in the reaction mixture. To this end, we performed the repair reaction using a duplex oligonucleotide containing a U:G pair, labeled at the 3′ end of the uracil-containing strand, as the DNA substrate (Figure 6). As expected, irrespective of the presence or absence of dNTPs, a major 22-nt 3′-labeled fragment was generated, which corresponds to the AP incision intermediate (Figure 6a, lane 2). This major intermediate was accompanied by a minor 21-nt fragment that might represent exonucleolytic degradation. However, incubation of the extract and the repair substrate in the presence of all four dNTPs specifically gave rise to a 20-nt 3′-labeled fragment (Figure 6a, lane 4), which is consistent with a 3-nt strand displacement followed by flap excision (Figure 6b).
Figure 6.
Strand displacement and incision during long-patch DNA synthesis. (a) Duplex DNA containing a U residue at the 3′ end-labeled upper strand was incubated at 30°C for 3 h with Arabidopsis cell extract in a reaction mixture containing either dCTP or all four dNTPs, as indicated. Relevant sizes in nucleotides are shown on the right. (b) Schematic diagram of strand displacement and incision during long-patch BER.
Taken together, these results suggest that excision repair of uracil by Arabidopsis whole-cell extracts may arise by single-nucleotide insertion, and by an extended (at least 3-nt-long) DNA synthesis involving strand displacement and flap excision.
DNA repair synthesis is aphidicolin resistant and ddCTP sensitive
In mammalian cells, it has been proposed that SP BER is Pol β-mediated, whereas LP BER is dependent of replicative DNA polymerases δ and/or ε (Fortini and Dogliotti, 2007). This is supported by reconstitution experiments with purified enzymes, and also by evidence obtained using specific inhibitors to estimate the relative contribution of both types of enzymes to each BER subpathway (Parlanti et al., 2002). In an initial examination of the requirement of DNA polymerase(s) for the gap-filling step during plant BER, we measured the repair activity of Arabidopsis extracts in the presence of two DNA polymerase inhibitors: aphidicolin and 2′,3′-dideoxycytidine 5′-triphosphate (ddCTP) (Figure 7). Aphidicolin is a tetracyclic diterpenoid that specifically interferes with eukaryotic replicative DNA polymerases (α, δ and ε) (Weiser et al., 1991), and is commonly used to synchronize plant cells in culture (Menges and Murray, 2002). ddCTP is an inhibitor of Pol β-like polymerases (Garcia-Diaz et al., 2002; Singhal et al., 1995).
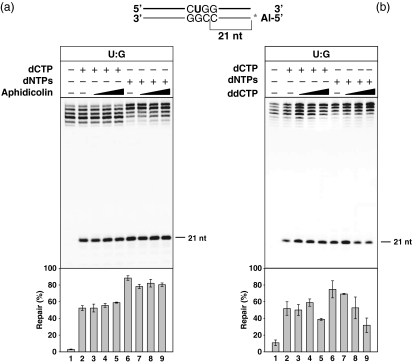
Figure 7.
Effects of DNA polymerase inhibitors on uracil excision repair by Arabidopsis cell extracts. Duplex DNA containing U opposite G at a HpaII site was incubated at 30°C for 3 h with Arabidopsis cell extract in a reaction mixture containing either dCTP or all four dNTPs, as indicated, and increasing levels of aphidicolin (left panel: 0, 300, 600 or 1200 μm) or 2′,3′-dideoxycytidine 5′-triphosphate (ddCTP, right panel: 0, 20, 200 or 2000 μm). Reaction products were digested with HpaII, separated in a 12% denaturing polyacrylamide gel and quantified by fluorescence scanning. The percentage of fully repaired DNA product is shown. Values are means with standard errors from two independent experiments.
Aphidicolin exerted no noticeable effect on the level of fully repaired HpaII-sensitive product; neither in the presence of dCTP nor in the presence of all four dNTPs (Figure 7a, lanes 2–9). In contrast, ddCTP caused a significant decrease in the level of full-repair product when reactions were carried out in the presence of all four dNTPS (Figure 7b, lanes 6–9). In repair reactions performed with dCTP as the only deoxynucleotide source, an inhibitory effect was observed at a 100:1 molar ratio of ddCTP:dCTP (Figure 7b, lanes 2–5). We therefore concluded that DNA synthesis after uracil excision is aphidicolin resistant and ddCTP sensitive.
Partial stimulation of uracil excision repair by addition of exogenous proteins
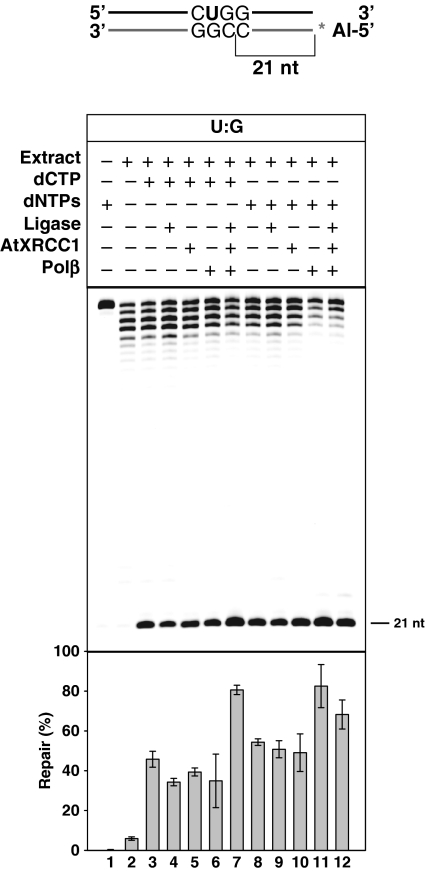
As shown in Figure 4, some DNA repair intermediates accumulate during the reaction catalyzed by cell extracts. This may result from low level and/or partial inhibition of some of the protein factors required to complete repair. To explore this possibility, we tested whether the level of fully repaired product might be increased by adding exogenous proteins to the repair reaction. The repair intermediates detected during the reaction included both AP incision and nucleotide insertion products, implying that the DNA synthesis and ligation steps might be rate-limiting. We therefore supplemented the reaction mixture with T4 DNA ligase, human DNA polymerase β or the Arabidopsis ortholog of the mammalian BER scaffold protein XRCC1, as well as with all three proteins together (Figure 8). The concentrations of DNA polymerase and DNA ligase used were in excess over those required to process all DNA substrate (DCC, TMR, TRA and RRA, unpublished data). We found that the simultaneous addition of all three exogenous proteins significantly increased the quantity of fully repaired product in reactions performed with dCTP as the only deoxynucleotide source (Figure 8, lane 7). In reactions with all four dNTPs, addition of Pol β significantly enhanced full repair (Figure 8, lane 11), and some increase was also observed when all three exogenous proteins were added (Figure 8, lane 12). Notwithstanding the fact that the added ligase and DNA polymerase are heterologous proteins, these results suggest that optimal DNA repair in vitro by Arabidopsis cell extracts may depend on an appropriate balance of different enzymatic activities and accessory factors.
Figure 8.
Partial stimulation of uracil repair by the addition of exogenous proteins. Duplex DNA containing U opposite G at a HpaII site was incubated at 30°C for 3 h with Arabidopsis cell extract in a reaction mixture containing either deoxycytidine triphosphate (dCTP) or all four deoxyribonucleotide triphosphates (dNTPs), as indicated, and different combinations of T4 DNA ligase (1.5 U), Arabidopsis AtXRCC1 (65 nm) and human Pol β (2.4 U). Reaction products were digested with HpaII, separated in a 12% denaturing polyacrylamide gel and quantified by fluorescence scanning. The percentage of fully repaired DNA product is shown. Values are means with standard errors from two independent experiments.
Discussion
An experimental system that specifically detects BER in plants
Previous reports have described DNA repair activities in plant cell extracts, but to our knowledge there is no published description of an experimental in vitro system allowing the specific analysis of the BER pathway in plants. A reliable assay has been developed to detect nucleotide excision repair (NER) in Arabidopsis whole-cell extracts, which measures DNA repair synthesis in plasmid substrates treated with UV-C, methylene blue or cisplatin, but does not allow the unambiguous distinction of the relative contributions of NER and BER, especially for oxidative lesions induced by methylene blue (Li et al., 2002). A recent report has described the incision of osmium tetroxide-treated plasmids in chloroplast extracts by DNA glycosylase/lyase and/or by AP endonuclease activities (Gutman and Niyogi, 2009), but not full BER. A previous investigation of BER using rice extracts (Sarkar et al., 2004) did not allow the discrimination between unrepaired substrate and fully-repaired product, as both were detected as a 51-mer labeled fragment.
A major aim of this work was to extend the biochemical analysis of BER to plants by developing an assay for the specific detection of this important repair pathway in Arabidopsis. We used similar cell extracts to those of previous studies (Li et al., 2002; Qi et al., 2005) in conjunction with defined DNA substrates containing uracil, a lesion known to be repaired by base excision in prokaryotes and eukaryotes. The location of the target lesion at an appropriate restriction site allowed detection of fully repaired products, as previously described in mammalian cell extracts (Dianov et al., 1992; Singhal et al., 1995). Using this in vitro system we have demonstrated that Arabidopsis whole-cell extracts promote uracil excision repair to convert a U:G mismatch to a normal C:G pair. The initiation of uracil repair, but not the AP incision and gap-filling steps, is entirely dependent on enzymatic activity that is blocked by a specific inhibitor of UDGs of the UNG family. The Arabidopsis genome encodes only a putative member of this protein family (At3G18630), but its role in uracil repair is still unknown. Therefore, the assay system described here allows reliable monitoring of uracil-initiated BER in Arabidopsis cells.
Processing of AP sites by AP endonucleases and AP lyases
We have used the in vitro BER assay in Arabidopsis extracts to initiate the functional characterization of this repair pathway in plants. In the classical model of BER, repair of an AP site proceeds through hydrolysis of the phosphodiester bond 5′ to the abasic site by an AP endonuclease, followed by action of a DNA polymerase that simultaneously removes the 5′-dRP blocking residue, and performs gap filling (Dianov et al., 1992; Singhal et al., 1995). However, bifunctional DNA glycosylases with an intrinsic AP lyase can also cleave the phosphodiester bond 3′ to the AP site, and the subsequent 3′-blocked end is processed by an AP endonuclease, thus generating a gap flanked by conventional 3′-OH and 5′-P ends (Levin and Demple, 1990). The biological significance of the AP lyase activity of bifunctional DNA glycosylases during BER is still unclear, as AP endonuclease is still needed to clear the 3′-blocked end, even after AP processing by AP lyase activity. However, there is some evidence that AP lyases may play an important role during BER in some species. In Schizosaccharomyces pombe, for example, genetic and biochemical evidence points to the activity of DNA glycosylase/lyase Nth1 as being mainly responsible for incision at AP sites, which are then processed by the AP endonuclease Apn2 and repaired though an SP subpathway (Alseth et al., 2004).
By performing repair reactions in the presence or absence of Mg2+, we have found that AP sites generated after uracil excision may be processed either by AP endonucleases or by AP lyases, generating either 5′- or 3′-blocked ends, respectively. Interestingly, most of the 3′-termini products generated in the absence of Mg2+ represented 3′-P ends. This suggests that the AP lyase activity (or activities) involved catalyze β,δ-elimination. This avoids the need for an AP endonuclease to clear the 3′-blocked end, but implies that 3′-phosphoesterase activity may be required to restore conventional 3′-OH. A possible candidate for this role in Arabidopsis is a previously described plant 3′-phosphoesterase that specifically binds to single-strand DNA breaks (Petrucco et al., 2002). In any case, our results are compatible with the hypothesis that AP endonuclease-dependent and -independent BER pathways coexist in plant cells, as reported in mammalian cells (Wiederhold et al., 2004).
At this point it is difficult to assign candidate Arabidopsis genes for the AP endonuclease and AP lyase activities detected in cell extracts. The Arabidopsis genome encodes three putative AP endonucleases (AtAPE1, AtAP2 and AtARP), and only the latter has been shown to perform incisions in DNA containing abasic sites (Babiychuk et al., 1994). On the other hand, there are at least nine Arabidopsis genes encoding bifunctional DNA glycosylases, and for seven of these (AtMMH, AtNTH1, AtOGG1, DME, ROS1, DML2 and DML3), an AP lyase activity has been demonstrated in vitro (Agius et al., 2006; Garcia-Ortiz et al., 2001; Gehring et al., 2006; Morales-Ruiz et al., 2006; Ohtsubo et al., 1998; Ortega-Galisteo et al., 2008; Roldan-Arjona et al., 2000).
Single-nucleotide and long-patch base excision repair of uracil and abasic sites in DNA by Arabidopsis cell extracts
In mammalian cells, processing of AP sites generated after excision is carried out either by single-nucleotide replacement or by long-patch DNA synthesis (Fortini and Dogliotti, 2007). Given the absence of plant homologs of DNA ligase III and DNA polymerase β, which are both required for mammalian SP BER (Kubota et al., 1996), it has recently been proposed that plants only use an LP BER pathway (Uchiyama et al., 2008). However, we have found that BER of uracil and abasic sites in Arabidopsis cell extracts occurs by both single-nucleotide insertion and long-patch DNA synthesis. Three main observations indicate that repair of a U:G mispair may take place via single-nucleotide DNA synthesis: (i) dCTP alone is sufficient to support full repair; (ii) only a 29-nt intermediate is detected in single-nucleotide reactions; and (iii) the only nucleotide inserted is complementary to the base opposite the U residue. On the other hand, we also found evidence of an LP BER when the reaction mixtures contained all four dNTPs. In this case, the reaction intermediates detected included not only 29-, but also 30- and 31-nt 5′-labeled fragments. Importantly, a shortened 3′-labeled fragment was detected in these conditions, consistent with strand displacement DNA synthesis followed by the removal of the DNA flap. Homologs of the mammalian flap endonuclease FEN1 are encoded in the genomes of Arabidopsis and Oryza sativa (Kimura and Sakaguchi, 2006), but their role in BER is not known.
In mammalian cells, the factors that determine the selection of either the SP or LP BER pathway are still poorly understood, although it has been proposed that both the nature of the DNA glycosylase and the lesion may influence the choice. Thus, repair of hypoxanthine or ethenoadenine initiated by the ANPG glycosylase is completed via both SP and LP BER (Fortini et al., 1999), whereas excision of 8-oxoG by OGG1 is preferentially followed by an SP BER (Dianov et al., 1998; Fortini et al., 1999). There is evidence that uracil-initiated BER in mammalian cell-free extracts involves SP in about 20% of repair events (Bennett et al., 2001), but the balance between SP and LP may also be modulated by the cell-cycle phase (Fortini and Dogliotti, 2007). Reports on mammalian mitochondrial extracts have variously suggested that uracil excision is followed only by SP BER (Stierum et al., 1999) or by SP and LP BER pathways (Akbari et al., 2008). Findings from non-mammalian systems suggest that the relative relevance of SP and LP pathways in the same lesion may differ, even between related species. Thus, in Saccharomyces cerevisiae most AP sites are repaired through LP BER (Boiteux and Guillet, 2004), whereas in Schizosaccharomyces pombe genetic and biochemical evidence points to the preeminence of the SP pathway (Alseth et al., 2004). We found that the kinetics of BER by Arabidopsis extracts is faster when all dNTPs are present in the repair reaction, but that the rate of SP BER is nonetheless also quite significant. It will be important to determine whether these two pathways occur in competition in plant cells, and to identify the factors determining their relative significance in vivo. Resolving these and other questions will require experimental data on different DNA lesions, and the identification of enzymes specifically involved either in SP or LP BER.
The nature of DNA polymerases involved in plant BER
We have explored the nature of the DNA polymerase(s) involved in the repair synthesis step during plant BER by performing repair assays in the presence of two inhibitors. Our results indicate that DNA synthesis after uracil excision is aphidicolin resistant and ddCTP sensitive. In contrast, the DNA synthesis observed during NER in Arabidopsis cell extracts is both aphidicolin- and ddTTP-resistant (Li et al., 2002). Uracil-initiated BER in mammalian cells is completely inhibited by ddCTP and unaffected by aphidicolin, thus supporting the idea that DNA polymerase β is required for the gap-filling step (Singhal et al., 1995). However, the BER of AP sites in mammalian cells is partially aphidicolin sensitive, and the analysis of Pol β-proficient and -deficient cells has led to the proposal that there is a competition between distributive and processive DNA polymerases for nucleotide addition at the primer terminus (Parlanti et al., 2004). The Arabidopsis genome does not encode any ortholog of DNA polymerase β, but contains a gene for DNA polymerase λ (Garcia-Diaz et al., 2000), another member of the X DNA polymerase family that also contributes to BER in mammalian cells (Braithwaite et al., 2005). Pol λ is the only X family-DNA polymerase identified in plants (Uchiyama et al., 2008), and O. sativa Pol λ is ddTTP sensitive and aphidicolin resistant (Uchiyama et al., 2004), like its mammalian counterpart (Garcia-Diaz et al., 2002; Shimazaki et al., 2002).
Therefore, our results are consistent with a possible role of Arabidopsis Pol λ during uracil-initiated BER. However, it should be noted that inhibition by ddCTP was less intense under conditions of single-nucleotide insertion, and that substantial DNA repair persisted, with both SP and LP, even at a 100:1 molar ratio of ddCTP:dCTP. Therefore, it is possible that, as in mammalian cells, there is competition between different DNA polymerases for access to the repair gap. Nevertheless, the information available about plant DNA polymerases and their role in repair is still too limited to suggest tentative candidates for DNA synthesis during plant BER.
Control of BER rate and accumulation of DNA repair intermediates
BER is a multistep process, and it is likely that the individual enzymatic activities involved in this repair pathway are coordinated in vivo to maximize repair efficiency and minimize the persistence of deleterious repair intermediates. Not unexpectedly, this coordination is to some extent lost when BER is recapitulated in vitro. We have found that the rate of uracil repair by Arabidopsis whole-cell extracts reaches a plateau after about 3 h, and not all DNA substrates are fully repaired even after longer incubation times (data not shown). As this is accompanied by the accumulation of AP incision and nucleotide insertion products, it is likely that the rate-limiting steps are neither uracil excision nor AP processing, but rather some of the factors required during gap filling and/or ligation. In fact, we found that the exogenous addition of heterologous DNA ligase and DNA polymerase β activities, as well as of the Arabidopsis scaffold protein AtXRCC1, to some extent increase the level of fully repaired product. However, it is also conceivable that the repair rate is additionally controlled by some inhibitory factor(s) in the extracts. In mammalian cells, the poly(ADP-ribose) polymerase PARP-1 has a high affinity for the single-strand-break intermediates generated during BER (Lindahl et al., 1995), and has been reported to have a negative effect on BER repair rates in cell extracts (Allinson et al., 2003, 2004). The Arabidopsis genome encodes two PARP proteins that are induced by DNA damage (Babiychuk et al., 1998; Doucet-Chabeaud et al., 2001), but it is not known whether there is any PARP activity in the Arabidopsis extracts used in our study. In any case, we envisage that the experimental system described here may be useful for identifying important regulatory factors that control the rate of BER in plant cells.
Experimental procedures
Plant material
Sterilized seeds of wild-type Arabidopsis thaliana plants (ecotype Columbia) were plated on 10-cm-diameter Petri dishes containing 25 ml of 0.44% (w/v) MS medium (Sigma-Aldrich, http://www.sigmaaldrich.com), supplemented with 3% (w/v) sucrose and 0.8% (w/v) agar, pH 5.8. Plates were transferred to the growth chamber under long-day conditions (16-h light/8-h dark) at 23°C. After 15 days, whole plants were harvested, frozen in liquid nitrogen and stored at −80°C.
Plant cell extract preparation
Seedling extracts were prepared by introducing several modifications to previously published methods (Li et al., 2002; Qi et al., 2005). All steps were performed at 0–4°C. Frozen plant material was ground in a handle mortar with liquid N2, and the resulting powder was resuspended in 2–3 volumes (w/v) of homogenization buffer containing 25 mm HEPES-KOH, pH 7.8, 100 mm KCl, 5 mm MgCl2, 250 mm sucrose, 10% glycerol, 1 mm DTT and 1 μl ml−1 protease inhibitor cocktail (Sigma-Aldrich). The homogenate was incubated for 30 min at 4°C and centrifuged at 13 000 g for 1 h. The supernatant was filtered through a 20-μm nylon mesh and dialyzed overnight against 25 mm HEPES-KOH, pH 7.8, 100 mm KCl, 17% glycerol and 2 mm DTT. Protein concentration was determined by the Bradford assay with BSA standards, and the extract was stored in small aliquots at −80°C.
Reagents and enzymes
Escherichia coli UDG, human AP endonuclease APE1 and Ugi were obtained from New England BioLabs (http://www.neb.com). Human DNA polymerase β was from Trevigen (http://www.trevigen.com). E. coli T4 DNA ligase was purchased from Promega (http://www.promega.com) and HpaII was purchased from Roche (http://www.roche-applied-science.com). E. coli Nth and Fpg DNA glycosylases/lyases were prepared as previously described (Roldan-Arjona et al., 1996, 1997). Partially purified AtXRCC1 from A. thaliana was a kind gift from M.I. Martínez-Macías (University of Córdoba, Spain).
DNA substrates
Oligonucleotides used to prepare DNA substrates (Table S1) were synthesized by Operon (http://www.operon-biotech.com), and were purified by PAGE before use. Double-stranded DNA substrates were prepared by mixing a 5-μm solution of the upper-strand oligonucleotide (labeled at the 5′ or 3′ end with fluorescein, where indicated) with a 10-μm solution of the lower-strand oligonucleotide (labeled at the 5′ end with Alexa Fluor 647, where indicated), heating to 95°C for 5 min and slowly cooling to room temperature (25°C). DNA containing a natural AP site opposite guanine was prepared by incubating a DNA duplex containing a U:G mispair, prepared as above, with E. coli UDG (0.5 U) at 30°C for 5 min.
DNA repair reactions in seedling extracts
Repair reactions (50 μl) contained 45 mm HEPES-KOH, pH 7.8, 70 mm KCl, 5 mm MgCl2, 1 mm DTT, 0.4 mm EDTA, 2 mm ATP, 36 μg BSA, 1 mm NAD, 2% glycerol, 20 μm each deoxynucleotide (dCTP, dGTP, dATP and dTTP, except where indicated), substrate DNA (40 nm) and 70 μg of extract. After incubation at 30°C for 3 h, reactions were stopped by adding 20 mm EDTA, 0.6% SDS and 0.5 mg ml−1 proteinase K, and the mixtures were incubated at 37°C for 30 min. DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and ethanol-precipitated at −20°C in the presence of 0.3 mm NaCl and 16 μg ml−1 glycogen. Samples were resuspended in 10 μl of 90% formamide, and were heated at 95°C for 5 min. Where indicated, DNA was resuspended in 5 μl of SuRE/Cut Buffer L containing 5 U of HpaII (Roche) and incubated at 37°C for 1 h. Reactions were then stopped by adding 5 μl of 90% formamide and then heating at 95°C for 5 min. Reaction products were separated in a 12% denaturing polyacrylamide sequencing gel (40 × 20 cm) containing 7 m urea. Fluorescein- or Alexa-labeled DNA was visualized in an FLA-5100 imager, and data were analyzed using Multigauge (Fujifilm, http://www.fujifilm.com).
Analysis of 5′ and 3′ ends generated during AP site processing
Repair reactions (50 μl) were performed at 30°C for 3 h in the buffer indicated above (with no dNTPs), either in the presence or absence of 5 mm MgCl2. Control AP lyase reactions were performed by incubating a DNA containing a natural AP site opposite guanine with E. coli Nth or Fpg at 37°C, for 30 or 60 min, respectively. Control AP endonuclease reactions were performed by incubating a DNA containing a natural AP site opposite guanine with human APE1 (10 U) at 37°C for 60 min, supplemented with 2.4 U of human DNA polymerase β when generating a control dRP-lyase product. When analyzing 5′ ends, reaction products were stabilized by the addition of freshly prepared sodium borohydride (NaBH4; Sigma-Aldrich) to a final concentration of 300 mm, incubation at 0°C for 30 min and desalting in a microspin G-25 column (GE Healthcare, http://www.gelifesciences.com). All reaction products were purified, separated and analyzed as described above.
Acknowledgments
We thank members of our laboratory for helpful discussions, and M.I. Martínez-Macías for the generous gift of the AtXRCC1 protein. This work was supported by funds from the Ministerio de Educación y Ciencia, Spain (grant number BFU2007-60956/BMC) and from the Junta de Andalucía, Spain (grant number P07-CVI-02770).
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article:
Figure S1. DNA ends generated after AP site cleavage by AP endonucleases and AP lyases.
Table S1. DNA sequences of the oligonucleotides used as substrates.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl Acad. Sci. USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Allinson SL, Dianova II, Dianov GL. Poly(ADP-ribose) polymerase in base excision repair: always engaged, but not essential for DNA damage processing. Acta Biochim. Pol. 2003;50:169–179. [PubMed] [Google Scholar]
- Allinson SL, Sleeth KM, Matthewman GE, Dianov GL. Orchestration of base excision repair by controlling the rates of enzymatic activities. DNA Repair (Amst) 2004;3:23–31. doi: 10.1016/j.dnarep.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Alseth I, Korvald H, Osman F, Seeberg E, Bjoras M. A general role of the DNA glycosylase Nth1 in the abasic sites cleavage step of base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res. 2004;32:5119–5125. doi: 10.1093/nar/gkh851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiychuk E, Kushnir S, Van Montagu M, Inze D. The Arabidopsis thaliana apurinic endonuclease Arp reduces human transcription factors Fos and Jun. Proc. Natl Acad. Sci. USA. 1994;91:3299–3303. doi: 10.1073/pnas.91.8.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiychuk E, Cottrill PB, Storozhenko S, Fuangthong M, Chen Y, O’Farrell MK, Van Montagu M, Inze D, Kushnir S. Higher plants possess two structurally different poly(ADP-ribose) polymerases. Plant J. 1998;15:635–645. doi: 10.1046/j.1365-313x.1998.00240.x. [DOI] [PubMed] [Google Scholar]
- Bennett SE, Sung JS, Mosbaugh DW. Fidelity of uracil-initiated base excision DNA repair in DNA polymerase β-proficient and -deficient mouse embryonic fibroblast cell extracts. J. Biol. Chem. 2001;276:42588–42600. doi: 10.1074/jbc.M106212200. [DOI] [PubMed] [Google Scholar]
- Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA polymerase λ mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J. Biol. Chem. 2005;280:18469–18475. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- Bray CM, West CE. DNA repair mechanisms in plants: crucial sensors and effectors for the maintenance of genome integrity. New Phytol. 2005;168:511–528. doi: 10.1111/j.1469-8137.2005.01548.x. [DOI] [PubMed] [Google Scholar]
- Britt AB. Repair of damaged bases. In: Somerville C, Meyerowitz EM, editors. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. doi 10.1111/tab.0005. [Google Scholar]
- Dianov GL. Monitoring base excision repair by in vitro assays. Toxicology. 2003;193:35–41. doi: 10.1016/s0300-483x(03)00288-9. [DOI] [PubMed] [Google Scholar]
- Dianov G, Lindahl T. Reconstitution of the DNA base excision-repair pathway. Curr. Biol. 1994;4:1069–1076. doi: 10.1016/s0960-9822(00)00245-1. [DOI] [PubMed] [Google Scholar]
- Dianov G, Price A, Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol. Cell. Biol. 1992;12:1605–1612. doi: 10.1128/mcb.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov G, Bischoff C, Piotrowski J, Bohr VA. Repair pathways for processing of 8-oxoguanine in DNA by mammalian cell extracts. J. Biol. Chem. 1998;273:33811–33816. doi: 10.1074/jbc.273.50.33811. [DOI] [PubMed] [Google Scholar]
- Doucet-Chabeaud G, Godon C, Brutesco C, de Murcia G, Kazmaier M. Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis. Mol. Genet. Genomics. 2001;265:954–963. doi: 10.1007/s004380100506. [DOI] [PubMed] [Google Scholar]
- Fortini P, Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 2007;6:398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Fortini P, Parlanti E, Sidorkina OM, Laval J, Dogliotti E. The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J. Biol. Chem. 1999;274:15230–15236. doi: 10.1074/jbc.274.21.15230. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd edn. Washington, D.C.: ASM Press; 2006. [Google Scholar]
- Garcia-Diaz M, Dominguez O, Lopez-Fernandez LA, et al. DNA polymerase λ (Pol λ), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol. 2000;301:851–867. doi: 10.1006/jmbi.2000.4005. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M, Bebenek K, Sabariegos R, et al. DNA polymerase λ, a novel DNA repair enzyme in human cells. J. Biol. Chem. 2002;277:13184–13191. doi: 10.1074/jbc.M111601200. [DOI] [PubMed] [Google Scholar]
- Garcia-Ortiz MV, Ariza RR, Roldan-Arjona T. An OGG1 orthologue encoding a functional 8-oxoguanine DNA glycosylase/lyase in Arabidopsis thaliana. Plant Mol. Biol. 2001;47:795–804. doi: 10.1023/a:1013644026132. [DOI] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman BL, Niyogi KK. Evidence for base excision repair of oxidative DNA Damage in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 2009;284:17006–17012. doi: 10.1074/jbc.M109.008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiwanger BM, Matsumoto Y, Nicolas E, Dianov GL, Bohr VA, Taraschi TF. DNA base excision repair in human malaria parasites is predominantly by a long-patch pathway. Biochemistry. 2000;39:763–772. doi: 10.1021/bi9923151. [DOI] [PubMed] [Google Scholar]
- Hays JB. Arabidopsis thaliana, a versatile model system for study of eukaryotic genome-maintenance functions. DNA Repair. 2002;1:579–600. doi: 10.1016/s1568-7864(02)00093-9. [DOI] [PubMed] [Google Scholar]
- Kavli B, Otterlei M, Slupphaug G, Krokan HE. Uracil in DNA – General mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst) 2007;6:505–516. doi: 10.1016/j.dnarep.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Kelley MR, Kow YW, Wilson DM., III Disparity between DNA base excision repair in yeast and mammals: translational implications. Cancer Res. 2003;63:549–554. [PubMed] [Google Scholar]
- Kimura S, Sakaguchi K. DNA repair in plants. Chem. Rev. 2006;106:753–766. doi: 10.1021/cr040482n. [DOI] [PubMed] [Google Scholar]
- Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- Kunz BA, Anderson HJ, Osmond MJ, Vonarx EJ. Components of nucleotide excision repair and DNA damage tolerance in Arabidopsis thaliana. Environ. Mol. Mutagen. 2005;45:115–127. doi: 10.1002/em.20094. [DOI] [PubMed] [Google Scholar]
- Levin JD, Demple B. Analysis of class II (hydrolytic) and class I (beta-lyase) apurinic/apyrimidinic endonucleases with a synthetic DNA substrate. Nucleic Acids Res. 1990;18:5069–5075. doi: 10.1093/nar/18.17.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Schuermann D, Gallego F, Kovalchuk I, Tinland B. Repair of damaged DNA by Arabidopsis cell extract. Plant Cell. 2002;14:263–273. doi: 10.1105/tpc.010258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Satoh MS, Poirier GG, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem. Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Bogenhagen DF. Repair of a synthetic abasic site in DNA in a Xenopus laevis oocyte extract. Mol. Cell. Biol. 1989;9:3750–3757. doi: 10.1128/mcb.9.9.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Bogenhagen DF. Repair of a synthetic abasic site involves concerted reactions of DNA synthesis followed by excision and ligation. Mol. Cell. Biol. 1991;11:4441–4447. doi: 10.1128/mcb.11.9.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Murray JA. Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 2002;30:203–212. doi: 10.1046/j.1365-313x.2002.01274.x. [DOI] [PubMed] [Google Scholar]
- Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, Martinez-Macias MI, Ariza RR, Roldan-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl Acad. Sci. USA. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholl ID, Nealon K, Kenny MK. Reconstitution of human base excision repair with purified proteins. Biochemistry. 1997;36:7557–7566. doi: 10.1021/bi962950w. [DOI] [PubMed] [Google Scholar]
- Ohtsubo T, Matsuda O, Iba K, Terashima I, Sekiguchi M, Nakabeppu Y. Molecular cloning of AtMMH, an Arabidopsis thaliana ortholog of the Escherichia coli mutM gene, and analysis of functional domains of its product. Mol. Gen. Genet. 1998;259:577–590. doi: 10.1007/s004380050851. [DOI] [PubMed] [Google Scholar]
- Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldan-Arjona T. Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 2008;67:671–681. doi: 10.1007/s11103-008-9346-0. [DOI] [PubMed] [Google Scholar]
- Parlanti E, Fortini P, Macpherson P, Laval J, Dogliotti E. Base excision repair of adenine/8-oxoguanine mispairs by an aphidicolin-sensitive DNA polymerase in human cell extracts. Oncogene. 2002;21:5204–5212. doi: 10.1038/sj.onc.1205561. [DOI] [PubMed] [Google Scholar]
- Parlanti E, Pascucci B, Terrados G, Blanco L, Dogliotti E. Aphidicolin-resistant and -sensitive base excision repair in wild-type and DNA polymerase β-defective mouse cells. DNA Repair (Amst) 2004;3:703–710. doi: 10.1016/j.dnarep.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Pascucci B, Stucki M, Jonsson ZO, Dogliotti E, Hubscher U. Long patch base excision repair with purified human proteins. DNA ligase I as patch size mediator for DNA polymerases δ and ε. J. Biol. Chem. 1999;274:33696–33702. doi: 10.1074/jbc.274.47.33696. [DOI] [PubMed] [Google Scholar]
- Petrucco S, Volpi G, Bolchi A, Rivetti C, Ottonello S. A nick-sensing DNA 3′-repair enzyme from Arabidopsis. J. Biol. Chem. 2002;277:23675–23683. doi: 10.1074/jbc.M201411200. [DOI] [PubMed] [Google Scholar]
- Prasad R, Dianov GL, Bohr VA, Wilson SH. FEN1 stimulation of DNA polymerase β mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem. 2000;275:4460–4466. doi: 10.1074/jbc.275.6.4460. [DOI] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Roldan-Arjona T, Ariza RR. Repair and tolerance of oxidative DNA damage in plants. Mutat. Res. 2009;681:169–179. doi: 10.1016/j.mrrev.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Roldan-Arjona T, Anselmino C, Lindahl T. Molecular cloning and functional analysis of a Schizosaccharomyces pombe homologue of Escherichia coli endonuclease III. Nucleic Acids Res. 1996;24:3307–3312. doi: 10.1093/nar/24.17.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl Acad. Sci. USA. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan-Arjona T, Garcia-Ortiz MV, Ruiz-Rubio M, Ariza RR. cDNA cloning, expression and functional characterization of an Arabidopsis thaliana homologue of the Escherichia coli DNA repair enzyme endonuclease III. Plant Mol. Biol. 2000;44:43–52. doi: 10.1023/a:1006429114451. [DOI] [PubMed] [Google Scholar]
- Sarkar SN, Bakshi S, Mokkapati SK, Roy S, Sengupta DN. Dideoxynucleoside triphosphate-sensitive DNA polymerase from rice is involved in base excision repair and immunologically similar to mammalian DNA pol β. Biochem. Biophys. Res. Commun. 2004;320:145–155. doi: 10.1016/j.bbrc.2004.05.152. [DOI] [PubMed] [Google Scholar]
- Shatilla A, Ramotar D. Embryonic extracts derived from the nematode Caenorhabditis elegans remove uracil from DNA by the sequential action of uracil-DNA glycosylase and AP (apurinic/apyrimidinic) endonuclease. Biochem. J. 2002;365:547–553. doi: 10.1042/BJ20020375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki N, Yoshida K, Kobayashi T, Toji S, Tamai K, Koiwai O. Over-expression of human DNA polymerase λ in E. coli and characterization of the recombinant enzyme. Genes Cells. 2002;7:639–651. doi: 10.1046/j.1365-2443.2002.00547.x. [DOI] [PubMed] [Google Scholar]
- Singhal RK, Prasad R, Wilson SH. DNA polymerase β conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J. Biol. Chem. 1995;270:949–957. doi: 10.1074/jbc.270.2.949. [DOI] [PubMed] [Google Scholar]
- Srivastava DK, Berg BJ, Prasad R, Molina JT, Beard WA, Tomkinson AE, Wilson SH. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J. Biol. Chem. 1998;273:21203–21209. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- Stierum RH, Dianov GL, Bohr VA. Single-nucleotide patch base excision repair of uracil in DNA by mitochondrial protein extracts. Nucleic Acids Res. 1999;27:3712–3719. doi: 10.1093/nar/27.18.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem. Rev. 2003;103:2729–2759. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- Uchiyama Y, Kimura S, Yamamoto T, Ishibashi T, Sakaguchi K. Plant DNA polymerase λ, a DNA repair enzyme that functions in plant meristematic and meiotic tissues. Eur. J. Biochem. 2004;271:2799–2807. doi: 10.1111/j.1432-1033.2004.04214.x. [DOI] [PubMed] [Google Scholar]
- Uchiyama Y, Takeuchi R, Kodera H, Sakaguchi K. Distribution and roles of X-family DNA polymerases in eukaryotes. Biochimie. 2008;91:165–170. doi: 10.1016/j.biochi.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Visnes T, Akbari M, Hagen L, Slupphaug G, Krokan HE. The rate of base excision repair of uracil is controlled by the initiating glycosylase. DNA Repair (Amst) 2008;7:1869–1881. doi: 10.1016/j.dnarep.2008.07.012. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wu X, Friedberg EC. DNA repair synthesis during base excision repair in vitro is catalyzed by DNA polymerase ε and is influenced by DNA polymerases α and δ in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:1051–1058. doi: 10.1128/mcb.13.2.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser T, Gassmann M, Thommes P, Ferrari E, Hafkemeyer P, Hubscher U. Biochemical and functional comparison of DNA polymerases α, δ, and ε from calf thymus. J. Biol. Chem. 1991;266:10420–10428. [PubMed] [Google Scholar]
- Wiederhold L, Leppard JB, Kedar P, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Zeitlin SG, Patel S, Kavli B, Slupphaug G. Xenopus CENP-A assembly into chromatin requires base excision repair proteins. DNA Repair (Amst) 2005;4:760–772. doi: 10.1016/j.dnarep.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Zharkov DO, Grollman AP. The DNA trackwalkers: principles of lesion search and recognition by DNA glycosylases. Mutat. Res. 2005;577:24–54. doi: 10.1016/j.mrfmmm.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Zharkov DO, Ishchenko AA, Douglas KT, Nevinsky GA. Recognition of damaged DNA by Escherichia coli Fpg protein: insights from structural and kinetic data. Mutat. Res. 2003;531:141–156. doi: 10.1016/j.mrfmmm.2003.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.