Abstract
The mosquito Anopheles gambiae has heteromorphic sex chromosomes, while the mosquito Aedes aegypti has homomorphic sex chromosomes. We use retrotransposed gene duplicates to show an excess of movement off the An. gambiae X chromosome only after the split with Ae. aegypti, suggesting that their ancestor had homomorphic sex chromosomes.
HETEROMORPHIC sex chromosomes, both XX/XY and ZZ/ZW systems, have evolved independently multiple times in both animals and plants (Bull 1983; Charlesworth 1996; Rice 1996). Sex chromosomes are thought to evolve from a pair of autosomes that acquire a new sex-determining locus. Theory suggests that natural selection will favor tight linkage between the newly arisen sex-determining locus and sexually antagonistic alleles (i.e., genes that are beneficial in one sex, but detrimental in the other), which favors the suppression of recombination near the sex-determining locus (Charlesworth et al. 2005). In some species, this nonrecombining region includes only a small portion of the sex chromosome (hereafter referred to as homomorphic sex chromosomes), whereas in other species, this region encompasses most of the sex chromosomes (heteromorphic sex chromosomes). In many species the nonrecombining region progressively expands from only the portion near the sex-determining locus to nearly the full extent of the sex chromosomes (Lahn and Page 1999; Lawson Handley et al. 2004; Nicolas et al. 2005). However, the broad phylogenetic distribution of homomorphic sex chromosomes suggests that this progression does not happen in every species (e.g., Matsubara et al. 2006; Tsuda et al. 2007), although why it should occur in some lineages and not in others is unknown. As noted by Gilchrist and Haldane (1947, p. 187): “It is a striking fact that this [the suppression of recombination across the sex chromosome] has not happened in many large and successful groups.”
Within the order Diptera, there are a wide variety of sex chromosomes and sex-determination mechanisms, including XY, ZW, multiple-X, and homomorphic systems, often varying within the same family (Marin and Baker 1998; Schutt and Nothiger 2000; Sanchez 2008). The mosquito Anopheles gambiae (a species in the subfamily Anophelinae) has fully differentiated heteromorphic X and Y chromosomes that show no evidence of recombination (Krzywinski et al. 2004). The mosquito Aedes aegypti (subfamily Culicinae) has a nonrecombining sex-determining region that spans only a few megabases on chromosome 1; this chromosome is homologous to chromosomes X and 2R of An. gambiae (Nene et al. 2007). An. gambiae and Ae. aegypti diverged ∼150 million years ago (Krzywinski et al. 2006).
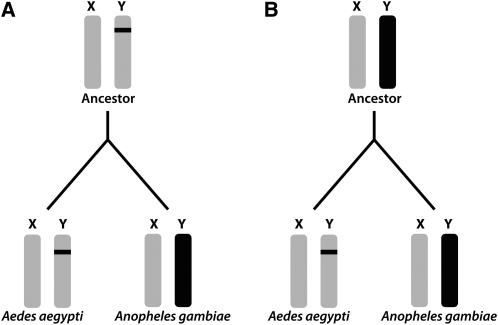
Because of the rapid turnover of sex-chromosome systems among the Diptera, it is not clear if the common ancestor of Ae. aegypti and An. gambiae had only a sex-determining region (i.e., homomorphic sex chromosomes) or fully differentiated heteromorphic sex chromosomes (Rai and Black 1999). The generally accepted model of sex-chromosome evolution, in which homomorphic sex chromosomes progressively suppress recombination and become heteromorphic, predicts that the common ancestor of Ae. aegypti and An. gambiae had homomorphic sex chromosomes (Figure 1A). This implies that evolution of heteromorphic sex chromosomes in An. gambiae occurred in a short period of time after the split between these two lineages and before the radiation of the Anophelines and that the homomorphic sex chromosomes of Ae. aegypti have been nearly static over evolutionary time. Alternatively, the common ancestor may have had nearly or fully differentiated sex chromosomes, and Ae. aegypti evolved from heteromorphic sex chromosomes to having only a small sex-determining region (Figure 1B; Rao and Rai 1987). We imagine this transition may have occurred by one of two mechanisms: either the sex-determining locus was transposed from the ancestral sex chromosome to an autosome or, in an XO sex-determination system, one of the “numerator” genes located on the X chromosome sustained an inactivating mutation, effectively making a karyotypic XX individual into a genetically male XO individual. (The precise mechanism of sex determination in Ae. aegypti is not known.)
Figure 1.—
Hypotheses for sex-chromosome evolution in Anopheles gambiae and Aedes aegypti. (A) The ancestor of An. gambiae and Ae. aegypti had homomorphic sex chromosomes and heteromorphism evolved along the Anopheline lineage. (B) The ancestor of An. gambiae and Ae. aegypti had heteromorphic chromosomes and homomorphism evolved along the Culicine lineage.
To determine the state of the mosquito common ancestor, we examined genes duplicated by retrotransposition in the An. gambiae genome. Several organisms with heteromorphic sex chromosomes, including mammals and Drosophila, have an excess of retrotransposed genes moving from the X chromosome to autosomes compared to genes moving between autosomes or from the autosomes to the X (Betran et al. 2002; Emerson et al. 2004; Vinckenbosch et al. 2006; Meisel et al. 2009). This pattern is further found to be strongly associated with the origin of new X chromosomes in both mammals and Drosophila (Potrzebowski et al. 2008; Meisel et al. 2009), although it continues long after X chromosomes arise. While there are many hypotheses for the evolutionary forces that drive gene movement off X chromosomes—including sexual antagonism and meiotic sex-chromosome inactivation (e.g., Hense et al. 2007)—it is likely that all of these forces also act in mosquitoes, implying excess movement off the heteromorphic X in this clade as well. We reasoned that if the common ancestor of Ae. aegypti and An. gambiae had homomorphic sex chromosomes (Figure 1A), there should be an excess of retrogene movement off the X chromosome in An. gambiae only after the divergence of the two lineages (i.e., since An. gambiae evolved a differentiated X chromosome). In contrast, if the common ancestor had fully heteromorphic chromosomes (Figure 1B), then our prediction is that there will be an excess of gene movement off the An. gambiae X on both the shared ancestral branch and the Anopheles-specific branch after the split with Aedes. (Note that the Ae. aegypti genome is largely not assembled onto chromosomes, precluding a similar analysis in this species.)
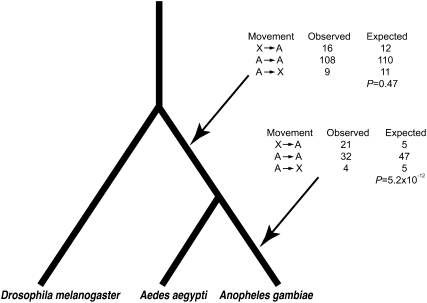
We collected data on all functional, intact duplicates in the An. gambiae genome and all orthologs between An. gambiae and Ae. aegypti from Ensembl version 54. When genes are retrotransposed there will be introns in the parental copy, but no introns in the daughter copy, allowing us to polarize gene movement. Although introns may be lost—and more rarely gained—over time, the rate of such changes is quite low (Coulombe-Huntington and Majewski 2007). Nevertheless, unless a parental gene loses all of its introns and the daughter gene gains introns, such changes will merely cause us to miss events rather than to assign them to an incorrect chromosome. Using gene-tree/species-tree reconciliation (Goodman et al. 1979), we identified retrotransposition events in the An. gambiae genome that have occurred since the split with Drosophila melanogaster and assigned them to a branch on the basis of the timing of the inferred duplication event in the gene tree. Calculating the expected number of movements on the basis of the equations presented in Betran et al. (2002), we find that an excess of movement off the X chromosome has in fact occurred since the split with D. melanogaster (χ2 = 23.83, d.f. = 2, P = 6.7 × 10−6). We then divided the retrotransposition events into those that occurred before the divergence of An. gambiae and Ae. aegypti and those that occurred only in An. gambiae since the split. We determined that there is a 400% excess of retrotransposition events off the X chromosome since the An. gambiae and Ae. aegypti split (Figure 2: χ2 = 51.97, d.f. = 2, P = 5.2 × 10−12). However, there is no excess of retrotransposition off the X chromosome prior to the split between An. gambiae and Ae. aegypti (Figure 2: χ2 = 1.51, d.f. = 2, P = 0.47). This strongly suggests a recent origin of fully differentiated heteromorphic sex chromosomes in An. gambiae.
Figure 2.—
Retroposition events off the X chromosome. There is an excess of genes moving off the X chromosome on the An. gambiae-specific lineage, but not on the branch leading to the common ancestor of An. gambiae and Ae. aegypti.
The deepest split between species within the subfamily Anophelinae—all of which have fully differentiated sex chromosomes—occurs soon after the split with the Culicinae (Krzywinski et al. 2006). This implies that the evolution of heteromorphic sex chromosomes must have occurred very soon after the split with Ae. aegypti. To determine whether there was a burst of retrotransposition off the X following this split, we examined the amino acid sequence identity between X-to-autosome retrotransposed proteins and their parental paralogs. A comparison of these distributions indicates that there is no difference in the percentage of identity of genes retrotransposed off the An. gambiae X chromosome and one-to-one orthologs between An. gambiae and Ae. aegypti (71.1% vs. 70.7%, t-test, P = 0.92; JTT amino acid distances, 0.508 vs. 0.436, t-test, P = 0.57). Given the fact that functional retrotransposed genes have been found to evolve more rapidly than single-copy genes (Betran et al. 2002), these results support the idea that these duplication events occurred soon after the split between An. gambiae and Ae. aegypti.
Our results have important implications for two further areas of research. First, a recent article (Moyle et al. 2010) proposed that X-to-autosome duplication events could be partly responsible for the large X-effect—the disproportionate effect of the X chromosome on reproductive isolation (Coyne and Orr 2004). This is because gene movement between chromosomes can itself cause reproductive isolation (e.g., Masly et al. 2006), and any excess movement involving the X will lead to an excess of reproductive isolation loci mapping to this chromosome. One prediction of this model is that species showing the large X-effect should also show an excess of X-to-autosome gene movement. As An. gambiae does in fact exhibit patterns consistent with the large X-effect (Slotman et al. 2005), our demonstration of an excess of movement off the X supports this model.
Second, it has been proposed that the excess movement off the X in Drosophila is the cause of the deficit of male-biased genes on the X in the same species (e.g., Vibranovski et al. 2009), although the number of retrotransposed genes is much smaller than the number of missing male-biased genes (Betran et al. 2002; Parisi et al. 2003). We have previously shown that there is no deficit of male-biased genes on the An. gambiae X chromosome, at any significance level (Hahn and Lanzaro 2005). Given the observed excess of gene movement off the X presented here, we therefore find little support for a causal link between movement and genome-wide patterns of male-biased gene expression.
Our results suggest that retrogene movement is a general feature of sex-chromosome evolution and support the hypothesis that the common ancestor of An. gambiae and Ae. aegypti had homomorphic sex chromosomes. It appears that the nonrecombining region around the sex-determining locus in An. gambiae expanded rapidly after the divergence with Ae. aegypti. Further investigation into the causes of the rapid expansion in the An. gambiae lineage and the long-term stasis in the Ae. aegypti lineage is clearly warranted.
Acknowledgments
We thank Charlotte Jeffries for help during the early stages of this project. This work is supported by National Science Foundation grant DBI-0845494 to M.W.H.
References
- Betran, E., K. Thornton and M. Long, 2002. Retroposed new genes out of the X in Drosophila. Genome Res. 12 1854–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull, J. J., 1983. Evolution of Sex Determining Mechanisms. Benjamin Cummings, Menlo Park, CA.
- Charlesworth, B., 1996. The evolution of chromosomal sex determination and dosage compensation. Curr. Biol. 6 149–162. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., B. Charlesworth and G. Marais, 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95 118–128. [DOI] [PubMed] [Google Scholar]
- Coulombe-Huntington, J., and J. Majewski, 2007. Intron loss and gain in Drosophila. Mol. Biol. Evol. 24 2842–2850. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Emerson, J. J., H. Kaessmann, E. Betran and M. Y. Long, 2004. Extensive gene traffic on the mammalian X chromosome. Science 303 537–540. [DOI] [PubMed] [Google Scholar]
- Gilchrist, B. M., and J. B. S. Haldane, 1947. Sex linkage and sex determination in a mosquito, Culex molestus. Hereditas 33 175–190. [Google Scholar]
- Goodman, M., J. Czelusniak, G. W. Moore, A. E. Romero-Herrera and G. Matsuda, 1979. Fitting the gene lineage into its species lineage, a parsimony strategy illustrated by cladograms constructed from globin sequences. Syst. Zool. 28 132–163. [Google Scholar]
- Hahn, M. W., and G. C. Lanzaro, 2005. Female-biased gene expression in the malaria mosquito Anopheles gambiae. Curr. Biol. 15 R192–R193. [DOI] [PubMed] [Google Scholar]
- Hense, W., J. F. Baines and J. Parsch, 2007. X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol. 5: e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski, J., D. R. Nusskern, M. K. Kern and N. J. Besansky, 2004. Isolation and characterization of Y chromosome sequences from the African malaria mosquito Anopheles gambiae. Genetics 166 1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski, J., O. G. Grushko and N. J. Besansky, 2006. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol. Phylogenet. Evol. 39 417–423. [DOI] [PubMed] [Google Scholar]
- Lahn, B. T., and D. C. Page, 1999. Four evolutionary strata on the human X chromosome. Science 286 964–967. [DOI] [PubMed] [Google Scholar]
- Lawson Handley, L.-J., H. Ceplitis and H. Ellegren, 2004. Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics 167 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin, I., and B. S. Baker, 1998. The evolutionary dynamics of sex determination. Science 281 1990–1994. [DOI] [PubMed] [Google Scholar]
- Masly, J. P., C. D. Jones, M. A. F. Noor, J. Locke and H. A. Orr, 2006. Gene transposition as a cause of hybrid sterility in Drosophila. Science 313 1448–1450. [DOI] [PubMed] [Google Scholar]
- Matsubara, K., H. Tarui, M. Toriba, K. Yamada, C. Nishida-Umehara et al., 2006. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc. Natl. Acad. Sci. USA 103 18190–18195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel, R. P., M. V. Han and M. W. Hahn, 2009. A complex suite of forces drive gene traffic from Drosophila X chromosomes. Genome Biol. Evol. 1 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle, L. C., C. D. Muir, M. V. Han and M. W. Hahn, 2010. The contribution of gene movement to the “Two Rules of Speciation”. Evolution 64 1541–1557. [DOI] [PubMed] [Google Scholar]
- Nene, V., J. R. Wortman, D. Lawson, B. Haas, C. Kodira et al., 2007. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 316 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, M., G. Marais, V. Hykelova, B. Janousek, V. Laporte et al., 2005. A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol. 3 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M., R. Nuttall, D. Naiman, G. Bouffard, J. Malley et al., 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrzebowski, L., N. Vinckenbosch, A. C. Marques, F. Chalmel, B. Jegou et al., 2008. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 6 709–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, K. S., and W. C. Black, 1999. Mosquito genomes: structure, organization, and evolution. Adv. Genet. 41(41): 1–33. [DOI] [PubMed] [Google Scholar]
- Rao, P. N., and K. S. Rai, 1987. Comparative karyotypes and chromosomal evolution in some genera of nematocerous (Diptera, Nematocera) families. Ann. Entomol. Soc. Am. 80 321–332. [Google Scholar]
- Rice, W. R., 1996. Evolution of the Y sex chromosome in animals. Bioscience 46 331–343. [Google Scholar]
- Sanchez, L., 2008. Sex-determining mechanisms in insects. Int. J. Dev. Biol. 52 837–856. [DOI] [PubMed] [Google Scholar]
- Schutt, C., and R. Nothiger, 2000. Structure, function and evolution of sex-determining systems in Dipteran insects. Development 127 667–677. [DOI] [PubMed] [Google Scholar]
- Slotman, M., A. Della Torre and J. R. Powell, 2005. Female sterility in hybrids between Anopheles gambiae and A. arabiensis, and the causes of Haldane's rule. Evolution 59 1016–1026. [PubMed] [Google Scholar]
- Tsuda, Y., C. Nishida-Umehara, J. Ishijima, K. Yamada and Y. Matsuda, 2007. Comparison of the Z and W sex chromosomal architectures in elegant crested tinamou (Eudromia elegans) and ostrich (Struthio camelus) and the process of sex chromosome differentiation in palaeognathous birds. Chromosoma 116 159–173. [DOI] [PubMed] [Google Scholar]
- Vibranovski, M. D., Y. Zhang and M. Y. Long, 2009. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 19 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinckenbosch, N., I. Dupanloup and H. Kaessmann, 2006. Evolutionary fate of retroposed gene copies in the human genome. Proc. Natl. Acad. Sci. USA 103 3220–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]