Abstract
Admixture between genetically divergent populations facilitates genomic studies of the mechanisms involved in adaptation, reproductive isolation, and speciation, including mapping of the loci involved in these phenomena. Little is known about how pre- and postzygotic barriers will affect the prospects of “admixture mapping” in wild species. We have studied 93 mapped genetic markers (microsatellites, indels, and sequence polymorphisms, ∼60,000 data points) to address this topic in hybrid zones of Populus alba and P. tremula, two widespread, ecologically important forest trees. Using genotype and linkage information and recently developed analytical tools we show that (1) reproductive isolation between these species is much stronger than previously assumed but this cannot prevent the introgression of neutral or advantageous alleles, (2) unexpected genotypic gaps exist between recombinant hybrids and their parental taxa, (3) these conspicuous genotypic patterns are due to assortative mating and strong postzygotic barriers, rather than recent population history. We discuss possible evolutionary trajectories of hybrid lineages between these species and outline strategies for admixture mapping in hybrid zones between highly divergent populations. Datasets such as this one are still rare in studies of natural hybrid zones but should soon become more common as high throughput genotyping and resequencing become feasible in nonmodel species.
ADMIXTURE or hybrid zones between genetically divergent populations are increasingly being explored for their use in studies of adaptation, reproductive isolation, and speciation (Rieseberg et al. 1999; Martinsen et al. 2001; Wu 2001; Vines et al. 2003; Payseur et al. 2004; reviewed by Coyne and Orr 2004), especially for their potential in identifying recombinants for gene mapping (otherwise known as “admixture mapping”; Chakraborty and Weiss 1988; Briscoe et al. 1994; Rieseberg et al. 1999; Reich et al. 2005; Slate 2005; Zhu et al. 2005; Lexer et al. 2007; Nolte et al. 2009). In many taxa of animals and plants, recombinants are created by admixture between divergent populations or species in hybrid zones or ecotones (Buerkle and Lexer 2008; Gompert and Buerkle 2009). The growing interest of evolutionary geneticists in admixture has its roots in both basic evolutionary genetics and breeding.
With respect to evolutionary genetics, admixed populations have been viewed as important resources for studying the genetics of adaptation and speciation, since the discovery that by fitting geographical clines of allele frequencies across hybrid zones, the strength of intrinsic and extrinsic (ecological) barriers to gene flow can be estimated (Barton and Hewitt 1985; Barton and Gale 1993). More recently, the genomics era has taken these concepts to a new level by providing genetic or physical genome maps for many species so that clines or introgression patterns of individual loci can be compared to their genomic background (see below; Falush et al. 2003; Gompert and Buerkle 2009). Thus, hybrid zones permit the identification and study of quantitative trait loci (QTL), genes, or other genetic elements involved in reproductive isolation and speciation in situ, directly in natural populations, if sufficient genetic recombination has occurred (Rieseberg and Buerkle 2002). In applied genetics, studies of hybrid zones yield information on the genomic architecture of barriers to introgression, which is of great interest to breeders concerned with the establishment of pedigrees for tree selection and domestication (Stettler et al. 1996).
Most animal or plant hybrid zones studied to date involve hybridization between parental populations that are much more divergent than the admixed human populations that have been used successfully for gene mapping in human medical genetics (e.g., Reich et al. 2005; Zhu et al. 2005). Little experience exists with interpreting genomic patterns of ancestry and admixture in such highly divergent, nonhuman populations. Early genomic work on hybrid zones, based on dominant genetic markers, suggested the feasibility of mapping genome regions involved in reproductive isolation and speciation (Rieseberg et al. 1999; Rogers et al. 2001), but these studies did not allow tests for selection on genotypes at single loci in different genomic backgrounds. This became possible only recently due to the development of novel analytical tools suited to large numbers of codominant markers, especially linkage models of Bayesian admixture analysis (Falush et al. 2003, 2007) and methods to fit “genomic clines” of codominant marker genotypes across complete genomic admixture gradients (Lexer et al. 2007; Gompert and Buerkle 2009; Nolte et al. 2009; Teeter et al. 2010). Great advances also have been made in interpreting single-locus estimates of genetic divergence between populations and species (Beaumont 2005; Foll and Gaggiotti 2008; Excoffier et al. 2009a). Here, we bring these approaches together to yield novel insights into genomic patterns of reproductive isolation and mating in hybrid zones of two widespread and important members of the “model tree” genus Populus. Our goal was to infer patterns of reproductive isolation and the likely evolutionary trajectories of hybrid populations and to develop strategies for genetic mapping in admixed populations.
Populus alba (white poplar) and P. tremula (European aspen) are ecologically divergent (floodplain vs. upland habitat) hybridizing tree species related to P. trichocarpa, the first completely sequenced forest tree (Tuskan et al. 2006). The two species are highly differentiated for neutral DNA-based markers (Lexer et al. 2007) and numerous phenotypic and ecological traits (Lexer et al. 2009). Mosaic hybrid zones between these species often form in riparian habitats (Lexer et al. 2005; hybrids sometimes referred to as P. × canescens) and have been proposed as potential “mapping populations” for identifying QTL and genes of interest in evolutionary biology (Lexer et al. 2007; Buerkle and Lexer 2008) and breeding (Fossati et al. 2004; Lexer et al. 2004). Previous studies of these hybrid zones were conducted with a relatively small number of genetic markers and without making use of linkage information; the genomic composition of hybrid zones between these species has never been studied with a genomewide panel of codominant markers with known linkage relationships. Specifically, we address the following questions in this contribution:
(1) What does an analysis of admixture and differentiation based on a genome-wide panel of mapped markers tell us about patterns of reproductive isolation and mating in hybrid zones of European Populus species? (2) What are the likely roles of pre- and postzygotic barriers vs. recent, localized historical factors in generating the observed genomic patterns? (3) What are the practical implications for admixture mapping in hybrid zones between highly divergent populations? We showcase where the genetic peculiarities of hybrid zones will limit their use for gene mapping and where they suggest new approaches that were perhaps not foreseen by geneticists with a focus on human medical applications.
MATERIALS AND METHODS
Sampling of admixture zones:
Three interspecific “mosaic” hybrid zones and adjacent parental populations of P. alba and P. tremula were sampled. These are defined by the three river drainage systems in which they are situated: Danube (Northeastern Austria), Ticino (Northwestern Italy), and Tisza (Northeastern Hungary) (Table 1). The three hybrid or admixture zones were previously discovered with the help of genetic markers (Bartha 1991; Fossati et al. 2004; Lexer et al. 2005). The main focus of this article is on the Danube hybrid zone; the Ticino and Tisza populations were used as additional “replicates” to check whether genotypic patterns observed for the Danube could be generalized to other localities.
TABLE 1.
Interspecific hybrid zones of P. alba and P. tremula sampled in the present study
| Locality | Country | Coordinates | Geography | Population | N | HE | HO | FISa | As |
|---|---|---|---|---|---|---|---|---|---|
| Danube | Austria | 48.26°N, 16.23°E | ∼90 km of river valley | Hybrid zone | 192 | 0.596 | 0.489 | 0.201 | 8.581 |
| Riverine lowland forest | P. alba | 48 | 0.508 | 0.436 | 0.114 | 6.770 | |||
| Eastern Alps | P. tremula | 48 | 0.533 | 0.425 | 0.177 | 6.757 | |||
| Ticino | Italy | 45.28°N, 8.98°E | ∼95 km of river valley | Hybrid zone | 138 | 0.590 | 0.517 | 0.129 | 6.694 |
| Riverine lowland forest | P. alba | 40 | 0.428 | 0.355 | 0.169 | 4.609 | |||
| Alps/Lago Maggiore | P. tremula | 40 | 0.479 | 0.351 | 0.197 | 5.021 | |||
| Tisza | Hungary | 48.32°N, 22.26°E | ∼85 km of river valley | Hybrid zone | 106 | 0.610 | 0.470 | 0.227 | 7.331 |
| Riverine lowland forest | P. alba | 40 | 0.462 | 0.401 | 0.151 | 5.074 | |||
| Zemplen mountains | P. tremula | 40 | 0.547 | 0.450 | 0.167 | 5.568 |
Included are coordinates and additional geographic information, number of individuals sampled (N), and the following genetic diversity summary statistics estimated from 68 mapped, codominant microsatellites: expected (HE) and observed heterozygosity (HO), inbreeding coefficient FIS, and allelic richness (As).
All FIS values significant at P < 0.001.
In each zone, widely spaced trees were sampled with a minimum distance of 50 m to avoid sampling of asexually derived ramets, based on results of a previous analysis of fine-scale spatial structure (van Loo et al. 2008). Sampling was carried out without regard to morphology, and emphasis was placed on broad geographic coverage within each river valley (Table 1); sampling transects is not feasible in mosaic hybrid zones and was not required for the statistical analyses employed in this study (Gompert and Buerkle 2009). Parental reference populations were sampled adjacent to each hybrid zone in lowland forest (P. alba) and upland habitat (P. tremula), with widely spaced individuals as described by Lexer et al. (2005).
Within-genome sampling and laboratory analyses:
Ninety-three polymerase chain reaction (PCR) or DNA sequence-based molecular genetic markers were typed in the Danube hybrid zone and its parental reference populations. These included 83 microsatellites, eight sequence-based markers, and two insertion–deletion polymorphisms (supporting information, Table S1). Seventy-six of the markers were developed from genomic libraries or directly from the P. trichocarpa genome assembly v.1 (http://www.ornl.gov/sci/ipgc; van der Schoot et al. 2000; Smulders et al. 2001; Tuskan et al. 2004), and 17 were developed from expressed sequence tags (ESTs) (Joseph and Lexer 2008; De Carvalho et al. 2010). The markers were distributed across all 19 chromosomes of the Populus genome with an average of 5 ± 1 (SE) markers per linkage group (Table S1). Different sets of markers were used at various stages of the work, depending on the requirements of each statistical method (see below).
To facilitate marker genotyping, genomic DNA was extracted from silica-dried leaves with the Dneasy plant mini kit (QIAGEN). Subsequently, all microsatellites and indel polymorphisms were PCR amplified using the protocols described previously by Lexer et al. (2005) and precisely sized using an Applied Biosystems (ABI) 3100 Genetic Analyzer and associated fluorescent dyes and software. All sequence-based polymorphisms were examined by PCR amplification and direct sequencing, using a Biomek NX S8 Laboratory Automated Workstation (Beckman Coulter) and an ABI 3730 Genetic Analyzer with accompanying chemistry, following precisely the protocols of Joseph and Lexer (2008).
Statistical analyses:
Descriptive statistics:
Since European hybrid zones of P. alba and P. tremula were characterized for their genetic diversity and structure elsewhere (Fossati et al. 2004; Lexer et al. 2005), descriptive population genetic analyses were kept to a minimum here: populations were characterized for their gene diversity equivalent to expected (HE) heterozygosity, observed (HO) heterozygosity (for fully codominant markers), inbreeding coefficient (FIS) and allelic richness corrected for sample size by rarefaction, and the following measures of genetic divergence: FST, Hedrick's (2005) G′ST, and the allele frequency differential (δ). These calculations were carried out using the FSTAT software (Goudet 1995) or manually on the basis of the output of this program.
Bayesian admixture analysis with linkage model:
Bayesian admixture analysis of the focal hybrid zone (Danube/Austria) was carried out with Structure 2.2, using a linkage model (Falush et al. 2003) that uses genotype and linkage information for each marker to estimate admixture parameters. Admixture analysis can reveal information regarding patterns of reproductive isolation (RI) and mating in recent generations. For example, mating events involving F1's may be rare or absent (Milne et al. 2003), or F1's may mate frequently with one or both parental species (Minder et al. 2007). Alternatively, mating may happen preferentially among F1's or other early hybrid classes, potentially leading to partial or complete reproductive isolation between hybrids and their parents (Rieseberg et al. 2003; Jiggins et al. 2008). These aspects are of great interest in both speciation genetics and admixture mapping, and each scenario leaves a characteristic signature in admixture coefficients obtained by a linkage model.
Since 12 of the 80 microsatellites used at this step were scored as band present/absent (Table S1; dominant markers), we used the Structure module of Falush et al. (2007) that is able to resolve incomplete or ambiguous marker information. The analysis was carried out for a K = 2 gene pool model based on previous results for this hybrid zone (Lexer et al. 2005; 2007) using a burn-in of 50,000 and 100,000 subsequent iterations, assuming unphased genotypic data. Then, the site-by-site output of Structure 2.2 was used to estimate the probabilities of interspecific heterozygosity or homozygosity for each locus in each individual. With unphased data, Structure estimates two probabilities informative regarding heterozygosity (ss2 and ss3; assignment probabilities of each allele at an unphased locus in a diploid individual), and two probabilities informative regarding homozygosity (ss1 and ss4; probabilities of assignment of both alleles of a locus to one or the other parental species). The following linear combinations of these probabilities were used to plot and explore hetero- and homozygosity in the Danube hybrid zone: (ss2 + ss3)−(ss1 + ss4), which quantifies evidence for interspecific heterozygosity for each locus and individual, with values ranging from −1 to +1; and ss1-ss4, which summarizes the evidence for interspecific homozgosity for alleles from P. tremula (−1) vs. P. alba (+1).
Effect of null alleles:
Allele nonamplification is a potential source of error when interpreting homo- and heterozygosity of PCR-based markers. Commonly used methods for estimating null allele frequencies are not easily applicable to hybrid zones. Thus, we assessed the role of null alleles by comparing interspecific heterozygosities from Structure to the actual marker heterozygosities in the raw data. Interspecific heterozygosity as examined by Structure refers to the ancestry of alleles in each individual, not to actual marker heterozygosities. For example, an individual may present a heterozygous marker genotype at a focal locus but may nevertheless be classified as an interspecific homozygote by Structure, because both alleles of that locus are derived from the same parental species. This scenario should be encountered fairly frequently for highly variable microsatellites. It can be used to rule out null alleles as the cause of interspecific homozygosity for the individuals in question.
Genome scan for interspecific divergence:
Admixture analysis can only reveal the effects of very recent gene flow across hybrid zones because admixture proportions Q from the donor species will decrease by approximately one-half with each new backcross generation. Thus, gene flow further back in the past is better captured by estimating divergence between hybridizing populations. A genome scan for interspecific divergence was carried out for the two parental populations of P. alba and P. tremula sampled adjacent to the focal (Danube) hybrid zone, using all 93 marker loci (Table S1). Interspecific divergence was estimated using FST and Hedrick's (2005) G′ST. Wright's FST offers the advantage that plenty of experience is available for it (including potentials and pitfalls; Jost 2008) and that it can be estimated for different kinds of markers within an analysis-of-variance framework (Weir and Cockerham 1984). Hedrick's (2005) G′ST, on the other hand, is useful because it takes within-population heterozygosity into account. Thus, a comparative analysis of FST and G′ST for the same set of loci should be informative regarding the amount of interspecific differentiation at each locus considering its within-population diversity.
To complete the interspecific divergence analysis, a standard neutrality test was carried out that compares FST to neutral coalescent simulations, using the frequentist approach of Beaumont and Nichols (1996), also discussed by Beaumont and Balding (2004). This test is often used to detect candidate loci for divergent selection, i.e., loci that are more divergent between two populations or species than expected under neutrality (Beaumont 2005). Many aspects of population demography can affect the outcome of this type of test, e.g., hierarchical population structure (Excoffier et al. 2009a), thus highly divergent “outlier” loci from FST-based tests need not necessarily indicate the action of selection (Foll and Gaggiotti 2008; Excoffier et al. 2009a). Nevertheless, the test allowed us to compare the pattern of divergence between our study species to published work on other hybridizing taxa. It was carried out with two rounds of simulations, one to obtain “neutral” FST and a second round to identify outliers. This was done for two datasets, one including all 93 loci genotyped for the Danube hybrid zone, including microsatellites, indels, and sequence polymorphisms and the other one comprising microsatellites only. Since the distributions were very similar in shape, only the results of the full analysis are discussed below.
Heterozygosity vs. hybrid index in observed and simulated data:
Our interest in speciation genetics and admixture mapping led us to explore patterns of reproductive isolation (RI) and mating in the Danube hybrid zone more deeply. This was achieved by examining interspecific heterozygosity vs. genomic admixture for each individual, using the 68 fully codominant microsatellites typed for the Danube hybrid zone (Table S3). We were also interested in checking the generality of our results, and thus we analyzed two additional replicate hybrid zones, Ticino (Italy) and Tisza (Hungary), for 18 widely spaced, fully codominant microsatellites (Table S3). Our rationale was as follows: if patterns of RI and mating are primarily determined by ecological differences among localities or by local history (e.g., human influence), then we may expect genotypic patterns to vary across localities. Conversely, if RI and mating are determined primarily by intrinsic organismal features shared by all populations, then we would expect genotypic patterns to be similar across localities.
For all three zones, microsatellite alleles were first combined into two allelic classes (converted into biallelic markers) with frequency differentials between species equal to the observed differential based on all alleles, following Gompert and Buerkle (2009). These biallelic data were then used to estimate interclass heterozygosity, which corresponds to a measure of interspecific heterozygosity, similar to but simpler than that obtained by Structure (above). This was plotted against genomic admixture estimated by a maximum likelihood hybrid index, all calculations carried out with the R package Introgress (Gompert and Buerkle 2010). This two-dimensional representation of genomic composition for each individual (heterozygosity vs. hybrid index) was then compared to simulated data for parental genotypes and F1's. Our rationale was to check whether observed, genetically intermediate hybrid genotypes in nature were more compatible with the simulated F1's (maximum heterozygosity with intermediate hybrid index) or with recombinant hybrids of subsequent generations (reduced interspecific heterozygosity compared to simulated F1's). In simulations, 500 individuals were sampled from the allele frequency distributions for each locus for each genotypic class (parentals and F1's). Note that this analysis follows similar principles to the method by Anderson and Thompson (2002) implemented in their NewHybrids software. Compared to their approach, ours makes fewer assumptions, because we do not attempt to assign hybrids to artificial categories such as F1, F2, backcross (BC), or “parental-like.”
Tests for epistatic interactions:
The presence of genetic discontinuities between recombinant hybrids and their parental species (steep clines despite fertile F1's) can be explained by several competing hypotheses, some of which can be addressed by available data for these species. A plausible hypothesis for which no data are currently available involves epistasis, i.e., negative genomic interactions among loci (Gavrilets 1997). Thus we used a recently developed method (Teeter et al. 2010) to test for epistatic interactions in the Danube hybrid zone, the focal population with the best genomic coverage. To keep the number of pairwise tests manageable, the analysis was restricted to those 29 fully codominant microsatellites with the greatest differentiation between the parental species (δ ≥ 0.8).
In a first step, single locus genomic clines of genotype frequencies at each locus were fitted against genomic admixture gradients (i.e., against the genetic background) using multinomial logistic regression, as in Lexer et al. (2007) but using the methodological improvements by Gompert and Buerkle (2009). Comparisons of observed clines against simulated neutral expectations allow the detection of under- or overrepresentation of interspecific homo- and heterozygotes, respectively (Gompert and Buerkle 2009). However, those were not at the center of attention here. Instead, two-locus interactions were explored by adding a second predictor locus to each regression model, and the two models for each focal locus (with or without the predictor) were compared using the Aikake information criterion (AIC = −2 L + 2p, where L is the log-likelihood of the model and p is the number of parameters in the model). Since smaller AICs represent higher likelihoods and better fits, ΔAIC was calculated as AICbasic model − AICwith predictor, so that large positive values of ΔAIC indicate loci for which the model fit is greatly improved by including the predictor locus, consistent with epistatic interactions. Since differences in AIC are used for interpretation of the results, rather than formal statistical tests of null hypotheses, there is no adjustment for multiple tests. The results can be interpreted on the basis of the magnitudes of ΔAIC for each pairwise locus combination. Particular pairwise interactions may be studied further in the future, on the basis of candidate gene information, the magnitude of ΔAIC, or other analyses.
Use of different types of genetic markers in statistical analyses:
All 93 marker loci were used for estimating interspecific genomic divergence (FST and G′ST) between P. alba and P. tremula in the focal hybrid zone (Danube/Austria; Table S1), and a subset of 80 microsatellites was used for Bayesian admixture analysis in Structure 2.2 (Table S2). These 80 loci comprised all those for which information on genomic location was regarded adequate for analysis on the basis of a linkage model. The 80 loci included 12 microsatellites for which allele calling was difficult and that were thus scored conservatively via the presence/absence of a well-defined focal band. The 68 remaining, easily scorable and fully codominant microsatellites were used for analyzing heterozygosity vs. hybrid index and for epistasis tests in the Danube population. Out of these, 18 widely spaced, codominant markers were used to assess the generality of the results using the replicate localities Ticino (Italy) and Tisza (Hungary) (Table S3).
RESULTS
Genetic diversity in admixture zones:
All three hybrid zones exhibited increased diversity compared to the parental species, measured in the form of expected (HE) and observed (HO) heterozygosity and allelic richness (Table 1). Inbreeding coefficients (FIS) were significantly positive, consistent with previously observed Wahlund effects arising from wide local sampling of the parental reference populations (De Carvalho et al. 2010) and widespread departures from random mating due to assortative mating in hybrid zones (van Loo et al. 2008).
Patterns of genomic ancestry inferred by linkage-based Bayesian analysis:
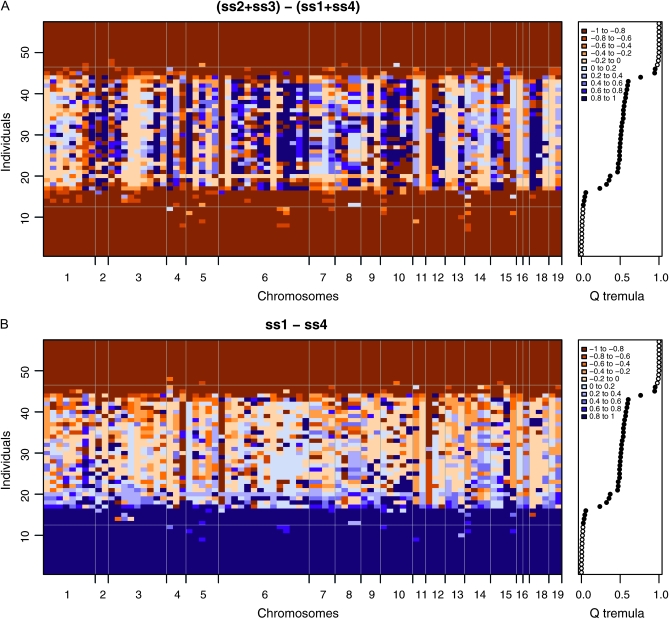
Bayesian-based admixture analysis based on 80 mapped microsatellites indicated an admixed ancestry for 31 (16%) individuals (0.05 < Q < 0.95) out of 192 sampled from the Danube hybrid zone of P. alba and P. tremula. Locus-by-locus analysis of these intermediate genotypes revealed “genomic mosaics” consisting of heterozygous loci and loci homozygous for alleles derived from one or another of the parental species (Figure 1, A and B), as expected for advanced recombinant hybrids derived from crosses of F1's. Pronounced gaps in the genotype distribution were uncovered between these recombinant hybrids and the two parental species (Figure 1, A and B; genotype data in File S1). The results are unlikely to be systematically biased by null alleles, as on average 44% of “interspecific homozygotes” classified by Structure were indeed true heterozygous marker genotypes in the raw data. Thus, classification as interspecific homozygotes at most loci was due to allelic ancestry rather than null alleles mimicking homozygosity.
Figure 1.—
Representative set of genetically intermediate and some parental individuals (along the vertical axis) from the Austrian Danube hybrid zone of P. alba and P. tremula, analyzed with 80 mapped microsatellites distributed across all 19 chromosomes of the Populus genome (horizontal axis). A linkage model (Falush et al. 2003) was used to infer probabilities of interspecific hetero- and homozygosity for each locus in each individual. (A) Evidence for interspecific heterozygosity, calculated from Structure site-by-site output as described in the text. High positive values for interspecific heterozygosity (ss2 + ss3 > ss1 + ss4) are indicated by blue, whereas brown colors indicate evidence for homozygosity for alleles from one or the other parental species (ss2 + ss3 < ss1 + ss4). (B) Evidence for interspecific homozygosity, calculated as described in the text, corresponding to the evidence (ss1 − ss4) that an individual is homozygous for alleles from P. tremula (brown color, ss4) vs. P. alba (blue color, ss1) at each locus. The recombinant, mosaic-like nature of genetically intermediate individuals is clearly visible, as are some introgressed individuals of P. alba. Overall admixture proportions (Q) of each tree are shown to the right. See text and Table S1, Table S2, and Table S3 for details.
Genome scan for species divergence:
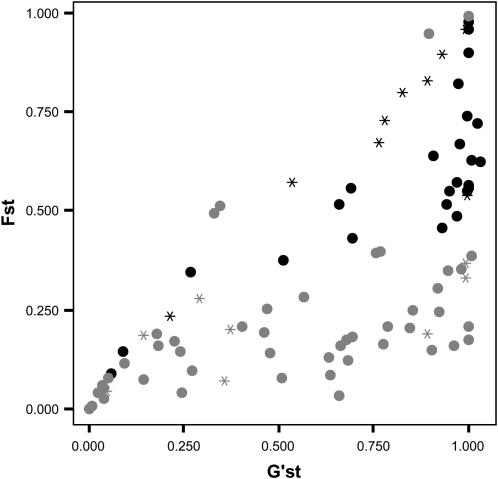
Between-locus variation for genetic divergence (FST) between the parental populations of the Danube hybrid zone was great (0.369 ± 0.289 SD), and the same was true for G′ST, which takes within-population variation into account (0.632 ± 0.354 SD) (Figure 2). At the 99% level, 33 loci (35%) were more divergent than expected under a neutral model following Beaumont and nichols (1996). This number is not readily interpretable in terms of divergent natural selection maintaining the species barrier, but it facilitates comparisons to recent studies of other groups of hybridizing species. The large proportion of loci with low interspecific divergence (low FST or G′ST; lower left corner of Figure 2) indicates extensive allele sharing, which is most plausibly attributed to past gene flow between these ecologically divergent, parapatric species (discussed below).
Figure 2.—
Results of a genomic scan for interspecific divergence between P. alba and P. tremula for the Austrian Danube hybrid zone, based on 93 DNA-based genetic markers. Hedrick's (2005) G′ST is shown on the x-axis and FST on the y-axis. Marker loci with greater-than-expected FST (99% level) relative to neutral expectations are solid; all other markers are shaded. Circles indicate markers derived from genomic DNA sequence (genomic libraries or P. trichocarpa shotgun genome sequence), stars stand for markers sourced from expressed sequence tags (ESTs). Marker loci with maximum interspecific divergence for their heterozygosity are visible along the diagonal.
Genomic composition of replicate hybrid zones:
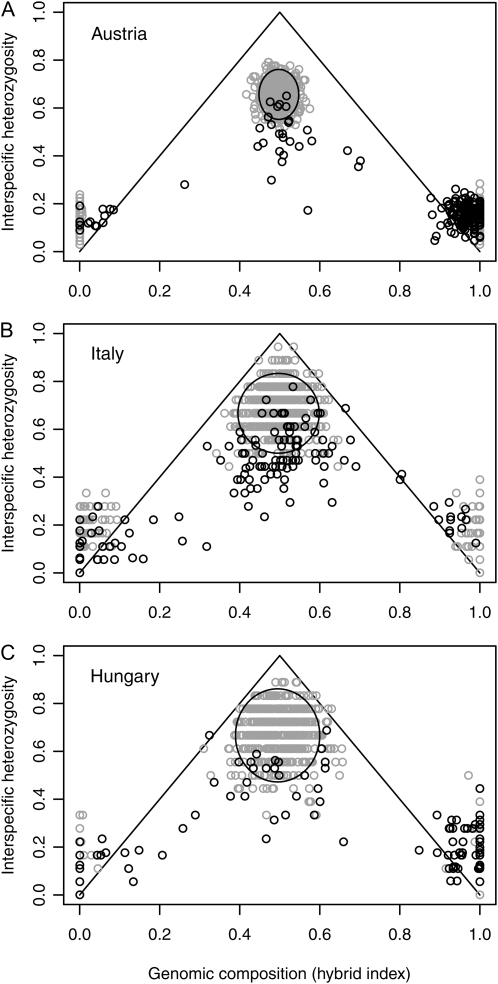
Analysis of interspecific heterozygosity vs. hybrid index in the focal (Danube/Austria) and two additional replicate hybrid zones of the same two species (Ticino/Italy and Tisza/Hungary) revealed a similar genomic composition of all three hybrid populations (Figure 3; genotype data in File S2 and File S3). In all three localities, most hybrids had intermediate hybrid index (horizontal axis) with interspecific heterozygosities (vertical axis) that fell below the 95% confidence intervals of simulated F1's (Figure 3). The increased variance in the observed and simulated data for the Ticino and Tisza populations stems from the lower number of loci genotyped for these localities (N = 18 loci) compared to the Danube (N = 68 codominantly scored loci in this analysis; see materials and methods). The reduced interspecific heterozygosity in hybrids indicates their recombinant nature, beyond the F1 generation, as also visible in the linkage-based Structure analysis for the Danube population (above; Figure 1) and from initial inspection of the raw data.
Figure 3.—
Interspecific heterozygosity (vertical axis) vs. hybrid index (horizontal axis) in three interspecific hybrid zones of P. alba and P. tremula: Danube (Austria), Ticino (Italy), and Tisza (Hungary). Solid circles indicate observed values for individual genotypes sampled in each population; shaded circles indicate values for simulated F1's (center of each graph) and parentals (left and right end of each graph). Univariate 95% confidence intervals for simulated F1's (ellipses) allow detection of observed hybrid genotypes with reduced interspecific heterozygosity (individuals below the ellipses), indicative of recombinant hybrid generations. The graphs are based on 68 codominant microsatellites in the case of the Danube hybrid zone (Austria) and 18 codominant loci for the Ticino (Italy) and Tisza (Hungary) hybrid zones, respectively.
Conspicuous gaps in the genotype distributions between hybrids and parental species were present in all three hybrid zones (Figure 3), which was not expected from previous studies of these species and hybrids with smaller genomic coverage and less powerful analytical tools (discussed below). In each locality, many advanced generation introgressants had a hybrid index that was distinguishable from the simulated “pure” parental populations (hybrid index further away from 0.0 or 1.0 than expected of parental genotypes in the simulations). This is consistent with the locus-to-locus variation in marker ancestry seen in the Structure graph (Figure 1) and with the high proportion of weakly differentiated loci detected by FST and G′ST (Figure 2).
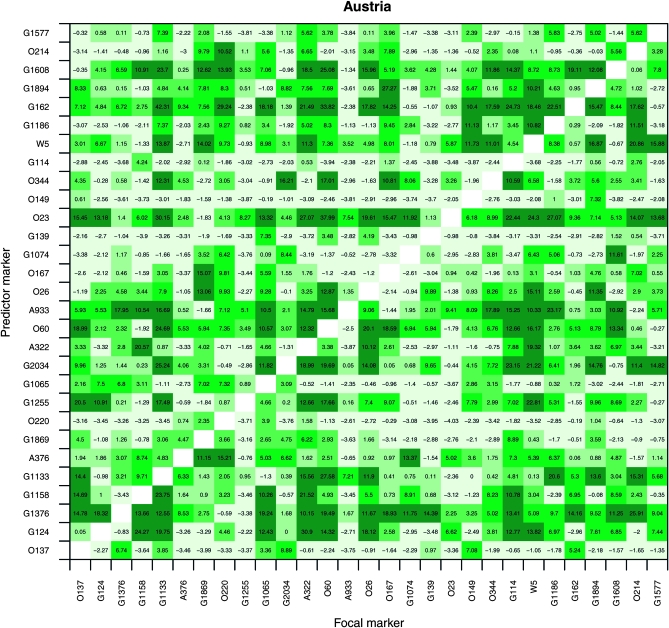
Tests for epistatic interactions:
Tests for epistatic interactions were informative regarding the possible origins of the genomic patterns seen in hybrid zones of P. alba and P. tremula (Figures 1 and 3). Among 29 loci with allele frequency differentials >0.8 in the Danube hybrid zone, a high proportion showed evidence for two-locus interactions (Figure 4). This is visible from the great number of two-locus comparisons for which an epistasis model for predicting the genotypes at a focal locus was strongly preferred over a simple one-locus model (see materials and methods for the details to these models). The results indicate that epistasis needs to be considered explicitly when discussing the origin of genotypic discontinuities between these two ecologically divergent forest trees despite the presence of fertile F1's. In this study we were interested primarily in interactions between loci, not in individual genomic clines. Nevertheless, basic cline parameters for all loci are shown in Table 2 and Table S3.
Figure 4.—
Results of tests for epistatic interactions between pairs of loci in the Danube hybrid zone, carried out for those 29 loci with the greatest interspecific divergence (allele frequency differential δ ≥ 0.8). Shown are differences between AIC values (ΔAIC) for logistic regression models that include vs. models that exclude a second predictor locus to predict genotypes at a focal locus (i.e., epistasis vs. single-locus models). Large values of ΔAIC indicate that the model fit is substantially greater for the epistasis model. The cells with the darkest green indicate a positive AIC difference of 10–50 (natural logarithmic scale) for the epistasis model relative to the one-locus model and the next shade of green an AIC difference of 5–10, and then 2–5, and then 0–2. See text for details of regression models.
TABLE 2.
Interspecific genetic divergence and results of genomic clines for 68 mapped codominant microsatellites studied in the Austrian Danube hybrid zone of P. alba and P. tremula
| Locus | Chrom | FST | G′ST | Delta | lnL ratio | P | Selectiona | ||
|---|---|---|---|---|---|---|---|---|---|
| G1719 | 1 | 0.182 | 0.694 | 0.625 | 7.486 | 0.004 | TT: + | TA: | AA: − |
| A302 | 1 | 0.070 | 0.358 | 0.448 | 1.237 | 0.648 | TT: | TA: | AA: |
| P2852 | 1 | 0.042 | 0.245 | 0.490 | 2.105 | 0.341 | TT: | TA: | AA: |
| O30_2 | 1 | 0.085 | 0.636 | 0.677 | 26.601 | 0.000 | TT: + | TA: | AA: − |
| G1568 | 1 | 0.033 | 0.660 | 0.740 | 46.820 | 0.000 | TT: + | TA: | AA: − |
| O137b | 1 | 0.488 | 0.968 | 0.969 | 20.551 | 0.000 | TT: | TA: − | AA: + |
| G124b | 1 | 0.350 | 0.947 | 0.885 | 6.276 | 0.015 | TT: | TA: | AA: |
| G1376b | 2 | 0.669 | 0.979 | 0.906 | 4.488 | 0.049 | TT: + | TA: − | AA: |
| A376b | 2 | 0.366 | 0.994 | 0.948 | 6.696 | 0.008 | TT: | TA: | AA: |
| G1158b | 2 | 0.897 | 1.001 | 0.979 | 10.200 | 0.000 | TT: | TA: | AA: |
| G1133b | 3 | 0.306 | 0.920 | 0.802 | 49.745 | 0.000 | TT: + | TA: − | AA: |
| G1629 | 3 | 0.124 | 0.681 | 0.729 | 7.098 | 0.007 | TT: + | TA: | AA: |
| O30_1 | 3 | 0.189 | 0.180 | 0.292 | 3.757 | 0.060 | TT: | TA: | AA: |
| G1416 | 3 | 0.000 | 0.000 | 0.083 | 15.482 | 0.000 | TT: | TA: | AA: + |
| O203 | 3 | 0.173 | 0.225 | 0.406 | 6.808 | 0.009 | TT: | TA: | AA: |
| G1869b | 3 | 0.174 | 1.000 | 0.979 | 5.361 | 0.031 | TT: | TA: | AA: |
| G1688 | 3 | 0.394 | 0.758 | 0.698 | 5.857 | 0.013 | TT: − | TA: | AA: − |
| O220b | 4 | 0.740 | 0.996 | 1.000 | 8.684 | 0.001 | TT: − | TA: + | AA: |
| O127 | 4 | 0.375 | 0.513 | 0.594 | 18.490 | 0.000 | TT: + | TA: − | AA: |
| G1809 | 4 | 0.431 | 0.695 | 0.677 | 13.221 | 0.000 | TT: + | TA: | AA: |
| G1255b | 5 | 0.990 | 1.000 | 1.000 | 11.093 | 0.001 | TT: | TA: + | AA: |
| G1192 | 5 | 0.146 | 0.243 | 0.458 | 5.418 | 0.018 | TT: | TA: | AA: |
| G1838 | 5 | 0.208 | 0.788 | 0.781 | 12.455 | 0.000 | TT: − | TA: + | AA: − |
| G20 | 5 | 0.558 | 0.691 | 0.615 | 8.600 | 0.001 | TT: + | TA: | AA: − |
| W15 | 5 | 0.141 | 0.479 | 0.542 | 5.944 | 0.010 | TT: − | TA: | AA: |
| G1065b | 6 | 0.628 | 1.006 | 0.938 | 31.029 | 0.000 | TT: + | TA: − | AA: |
| O369 | 6 | 0.026 | 0.039 | 0.219 | 9.428 | 0.000 | TT: | TA: + | AA: − |
| G2034b | 6 | 0.388 | 1.007 | 0.958 | 6.721 | 0.012 | TT: − | TA: | AA: |
| A322b | 6 | 0.188 | 0.892 | 0.813 | 10.775 | 0.000 | TT: | TA: | AA: + |
| O60b | 6 | 0.397 | 0.767 | 0.813 | 59.904 | 0.000 | TT: − | TA: | AA: + |
| W12 | 6 | 0.163 | 0.775 | 0.719 | 16.142 | 0.000 | TT: + | TA: − | AA: + |
| O190 | 6 | 0.638 | 0.908 | 0.792 | 4.721 | 0.025 | TT: − | TA: | AA: + |
| A933b | 6 | 0.538 | 0.995 | 0.969 | 8.354 | 0.001 | TT: − | TA: | AA: |
| G1485 | 6 | 0.061 | 0.036 | 0.083 | 27.080 | 0.000 | TT: + | TA: − | AA: |
| O26b | 6 | 0.548 | 0.950 | 0.875 | 13.139 | 0.000 | TT: + | TA: − | AA: |
| O167b | 6 | 0.959 | 1.000 | 0.990 | 13.543 | 0.002 | TT: | TA: + | AA: − |
| G1831 | 6 | 0.247 | 0.853 | 0.667 | 1.524 | 0.462 | TT: | TA: | AA: |
| G1074b | 6 | 0.978 | 0.999 | 0.938 | 7.469 | 0.004 | TT: − | TA: | AA: |
| G139b | 6 | 0.624 | 1.033 | 1.000 | 0.248 | 0.940 | TT: | TA: | AA: |
| G1260 | 7 | 0.208 | 0.404 | 0.469 | 9.752 | 0.000 | TT: + | TA: | AA: − |
| W17 | 7 | 0.194 | 0.461 | 0.521 | 15.861 | 0.000 | TT: + | TA: | AA: − |
| O312 | 7 | 0.159 | 0.663 | 0.677 | 2.663 | 0.231 | TT: | TA: | AA: |
| G2062 | 8 | 0.077 | 0.511 | 0.573 | 7.916 | 0.002 | TT: − | TA: + | AA: |
| O374 | 8 | 0.160 | 0.961 | 0.760 | 24.973 | 0.000 | TT: + | TA: | AA: − |
| O202 | 8 | 0.283 | 0.567 | 0.594 | 1.524 | 0.487 | TT: | TA: | AA: |
| O268 | 8 | 0.254 | 0.468 | 0.531 | 3.749 | 0.092 | TT: | TA: | AA: |
| G1949 | 9 | 0.096 | 0.271 | 0.479 | 7.082 | 0.010 | TT: | TA: | AA: |
| O23b | 9 | 0.209 | 1.001 | 1.000 | 12.457 | 0.000 | TT: − | TA: | AA: + |
| O21 | 9 | 0.090 | 0.058 | 0.125 | 8.391 | 0.001 | TT: | TA: | AA: |
| G2020 | 10 | 0.205 | 0.844 | 0.740 | 6.384 | 0.014 | TT: | TA: + | AA: |
| O149b | 10 | 0.548 | 0.996 | 0.958 | 11.453 | 0.000 | TT: + | TA: | AA: − |
| O344b | 10 | 0.721 | 1.022 | 1.000 | 5.124 | 0.039 | TT: | TA: + | AA: − |
| G1574 | 10 | 0.458 | 0.930 | 0.792 | 26.168 | 0.000 | TT: + | TA: − | AA: |
| G114b | 10 | 0.572 | 0.970 | 0.885 | 2.521 | 0.290 | TT: | TA: | AA: |
| G1037 | 11 | 0.000 | 0.000 | 0.073 | 10.924 | 0.000 | TT: | TA: + | AA: − |
| W05b | 12 | 0.244 | 0.923 | 0.917 | 6.368 | 0.015 | TT: | TA: | AA: |
| G1186b | 12 | 0.353 | 0.980 | 0.865 | 3.504 | 0.132 | TT: | TA: − | AA: + |
| G1353 | 13 | 0.076 | 0.145 | 0.271 | 9.181 | 0.001 | TT: | TA: | AA: |
| G162b | 13 | 0.148 | 0.902 | 0.823 | 0.629 | 0.741 | TT: | TA: | AA: |
| G1812 | 14 | 0.176 | 0.679 | 0.656 | 5.965 | 0.010 | TT: | TA: − | AA: + |
| G1894b | 15 | 0.516 | 0.941 | 0.906 | 3.770 | 0.101 | TT: | TA: | AA: |
| G1454 | 15 | 0.131 | 0.632 | 0.708 | 7.272 | 0.008 | TT: | TA: − | AA: + |
| G1608b | 15 | 0.558 | 1.000 | 0.896 | 46.011 | 0.000 | TT: | TA: − | AA: + |
| O14 | 16 | 0.041 | 0.023 | 0.052 | 6.242 | 0.001 | TT: + | TA: + | AA: − |
| O214b | 18 | 0.819 | 0.974 | 0.927 | 13.741 | 0.000 | TT: | TA: + | AA: − |
| G1577b | 18 | 0.564 | 1.000 | 0.979 | 4.121 | 0.086 | TT: | TA: | AA: |
| O28 | 18 | 0.161 | 0.183 | 0.333 | 3.382 | 0.119 | TT: | TA: | AA: |
| O206 | 19 | 0.517 | 0.659 | 0.635 | 2.032 | 0.341 | TT: | TA: | AA: |
Included are locus name, assignment to chromosomes, Weir and Cockerham's (1984) FST, Hedrick's (2005) G′ST, allele frequency differential delta and the following genomic cline parameters: ln likelihood, probability for departure from neutrality, and indications for significant over or underrepresentation (+ or −) of individual genotypic classes.
AA, interclass homozygotes for P. alba; AT, interclass heterozygotes for P. alba and P. tremula; TT, interclass homozygotes for P. tremula.
Markers included in tests for epistasis.
DISCUSSION
Despite a longstanding interest of evolutionary geneticists in using hybrid zones as natural laboratories for studying the evolutionary process (Barton and Hewitt 1985; Barton and Gale 1993; Rieseberg et al. 1999; Martinsen et al. 2001; Wu 2001; Vines et al. 2003; Coyne and Orr 2004; Payseur et al. 2004; Minder et al. 2007; Buerkle and Lexer 2008), there is little experience in the interpretation of genomic patterns of admixture and ancestry in terms of reproductive isolation and speciation (but see Payseur et al. 2004; Lexer et al. 2007; Nolte et al. 2009; Teeter et al. 2010). The results of our study allow us to address this topic and to comment on the utility of gene mapping in hybrid zones of wild species in animals and plants.
Genomic patterns of ancestry and admixture:
Analysis of a genome-wide panel of mapped markers (Figures 1–4) yields a different picture of patterns of reproductive isolation and mating in hybrid zones of P. alba and P. tremula compared to previous work based on much smaller numbers of codominant markers and less powerful analytical tools (Rajora and Dancik 1992; Fossati et al. 2004; Lexer et al. 2005). Early phenotype- and isozyme-based studies suggested that hybrid zones between these species consisted primarily of backcrosses to one parental species, namely P. alba (Bartha 1991; Rajora and Dancik 1992). Previous DNA-based studies (Lexer et al. 2005, 2007) did not allow rejection of this hypothesis, most likely due to the large error associated with individual-based analysis of a small number of markers. Here, linkage-, FST-, and genomic cline-based analyses of a genome-wide panel of mapped markers reveal the presence and positioning (in genotypic space) of unexpectedly strong reproductive barriers between these species and hybrids and suggest hypotheses on their origin and future fate.
The Bayesian analysis of genotypic ancestry (using Structure) indicates that early generation hybrids detected in the Danube hybrid zone of P. alba and P. tremula (Table 1) are genetically intermediate, recombinant hybrid genotypes (a genomic mosaic, Via and West 2008) separated from their parental species by sharp genomic discontinuities (Figure 1), equivalent to steep genomic clines sensu Gompert and Buerkle (2009) and similar to steep clines seen in simulations of hybrid zones with strong postzygotic barriers (Gavrilets 1997). Note that most trees from the Danube hybrid zone were P. alba-like (very low admixture coefficients Q; see genotypic data in File S1); only genetically intermediate individuals are shown in Figure 1, ordered by their Qs. Blocks of LD along the chromosomes are clearly visible in these genetically intermediate hybrids on the basis of the similarity of colors (probability of homo- or heterozygosity) along the horizontal axes (Figure 1, A and B). Also, occasional introgression of particular genome segments or loci across the barrier in either direction can be inferred from Figure 1. These observations are mirrored by characteristic patterns of genomic differentiation visible in our FST- and G′ST-based genome scan (Figure 2).
The genomic scan for pairwise species differentiation (Figure 2) is informative regarding gene flow in the more distant past—on the order of dozens or hundreds of generations (Whitlock 1992). Our pairwise genome scan reveals many loci with weak interspecific differentiation (FST or G′ST; Figure 2), despite the fact that the proportion of highly differentiated loci (FST greater than expected in neutral coalescent simulations) was much greater in our study (35%) than in studies of a different pair of outcrossing forest trees (12% in Quercus; Scotti-Saintagne et al. 2004) and a pair of self-incompatible annual plants (5% in Helianthus; Yatabe et al. 2007). The numerous weakly differentiated loci in our genome scan (Figure 2; lower portion of graph) indicate occasional but persistent introgression over long time scales, despite strong barriers to gene exchange. This makes sense: we know that even strong barriers to gene flow cannot prevent the introgression of neutral or advantageous alleles (Pialek and Barton 1997; Faure et al. 2008), and emerging phylogeographic data for these species suggest repeated contact upon colonization from divergent lineages (Fussi et al. 2010).
Ancestral polymorphism is an unlikely explanation for allele sharing at the weakly differentiated loci seen in Figure 2. These species diverged at least several million years ago (Stettler et al. 1996), and P. alba and P. tremula are not even sister species in most molecular phylogenetic analyses of the genus (Hamzeh and Dayanandan 2004). Accordingly, allele frequency differentials between these taxa often reach values of 0.90 or higher (Table 2; Table S3). Also, intermediate genotypes in hybrid zones often carry blocks of neighboring markers from one or the other parental species (Figure 1), which is consistent with interspecific recombination but not ancestral polymorphism.
We did not wish to interpret the highly differentiated outlier loci in our genome scan as being under divergent selection (see Foll and Gaggiotti 2008 and Excoffier et al. 2009a for a discussion of these issues). Rather, our primary interest was in comparing the overall distribution of interspecific genomic differentiation to expectations from comparable pairs of sympatric, hybridizing taxa. Nevertheless, it is noteworthy that most EST-derived markers used in our study are on the diagonal in our FST/G′ST plot (Figure 2), in contrast to numerous other markers developed from the Populus genome sequence or genomic libraries (Figure 2; Table S1). This indicates that the EST-derived polymorphisms have attained maximum differentiation for their within-species heterozygosity (Hedrick 2005), which is consistent with their placement outside the 99% confidence interval of the neutral distribution of FST.
Repeatability of genomic patterns and mechanisms of reproductive isolation:
Genomic discontinuities in hybrid zones of the type seen in Figure 1 are highly informative regarding the genetics of species boundaries and can be explained by several competing hypotheses: (1) assortative mating, an important prezygotic barrier in both animals and plants (Coyne and Orr 2004), easily mediated by differences in flowering time in the case of plants; (2) strong postzygotic barriers affecting hybrid generations beyond the F1, i.e., nuclear genetic incompatibilities caused by Dobzhansky–Bateson–Muller (DBM)-type epistatic selection (Gavrilets 1997); and (3) localized factors affecting only particular populations, e.g., natural or human-mediated disturbance in a particular locality may have triggered a recent “wave” of hybridization, mimicking a steep cline. Our data allow us to address each of these hypotheses. Additional alternative hypotheses include postmating barriers involving uniparentally inherited genetic factors, but these will result in strongly asymmetric intrinsic barriers (Turelli and Moyle 2007) for which there is no evidence in these species to date (Lexer et al. 2005).
To rule out recent local disturbance (hypothesis 3), we studied two replicate hybrid zones (Tisza and Ticino; Table 1) for 18 widely spaced codominant microsatellites. Our comparative analysis of interspecific heterozygosity vs. hybrid genomic composition in these populations (Figure 3) indicates similar patterns of reproductive isolation in all three localities: intermediate hybrids have reduced heterozygosity compared to simulated F1's, in agreement with their recombinant nature, backcrosses are infrequent (although possible in both directions), and many parental-like genotypes exhibit hybrid indices (horizontal axes) that suggest low levels of introgression in the distant past. These observations are consistent with those from Structure-based ancestry analysis (Figure 1) and the FST-based genome scan (Figure 2). More importantly, the repeatability of the results strongly contradicts the third hypothesis: these three river drainages have very different postglacial histories (timing of recolonization and onset of human disturbance); thus it is unlikely that recent events would have affected all three of them in identical ways.
So, are genomic discontinuities in hybrid zones of these hybridizing forest trees caused by prezygotic isolation (assortative mating) or by postzygotic barriers? The two parental species differ in their peak timing of flowering by up to several weeks (Fischer et al. 2005) and appreciable genetic variation for phenological traits exists even at the within-species level (Hall et al. 2007). Thus assortative mating due to phenological differences is plausible. A study of fine-scale spatial genetic structure (SGS) of the Danube hybrid zone is consistent with this hypothesis: SGS was much stronger in hybrids compared to parental-like genotypes, even after correcting for differences in the propensity of hybrids and parentals to form clones (Van Loo et al. 2008). Also, hybrids tended to form larger clones (more ramets extending over larger areas) than the parental species, which can easily lead to preferential mating between adjacent hybrid genotypes with similar phenology, i.e., to discontinuities in spatial and temporal patterns of flowering. A molecular marker-based study of progeny arrays from natural hybrids is currently underway to test the amount of assortative mating in these populations.
With respect to postzygotic barriers, our logistic regression-based analysis of the Danube hybrid zone indicates that epistasis is widespread (Figure 4). This is consistent with theoretical predictions under DBM-type incompatibilities among nuclear genes: strong epistatic selection will keep fit recombinants at low frequency, effectively maintaining the barrier and leading to steep clines despite the presence of fertile F1's (Gavrilets 1997), which would result in genotypic patterns such as those seen in Figures 1 and 3. It is thus likely that genomic patterns of admixture in hybrid zones of P. alba and P. tremula—in particular the conspicuous genomic discontinuities between hybrids and their parental species—are due to both assortative mating and DBM incompatibilities. Note that the presence of isolated adaptive peaks (e.g., strong ecological divergence between hybridizing species) may affect the dynamics and outcome of epistatic selection, compared to hybrid zones maintained by a simple balance of selection and migration (Gavrilets 1997).
Potential evolutionary trajectories of hybrid zones:
Several possible trajectories have been proposed for the evolution of hybrid populations between diploid taxa, including swamping of the species with the smaller effective population size (Ne) by gene flow from the more abundant species (Wolf et al. 2001), massive introgression from the earlier colonizing (“native”) into the later colonizing species (Excoffier et al. 2009b), homoploid hybrid speciation (Rieseberg et al. 2003), and the transfer of adaptive traits across “porous” species boundaries (Kim and Rieseberg 1999; Barton 2001; Martin et al. 2006; Whitney et al. 2006). The first two of these (genetic swamping and massive introgression of genes from the local species) appear unlikely here: P. alba is the species with the smaller effective population size Ne (Lexer et al. 2005) and phylogeographic data indicate that it recolonized Central Europe after P. tremula (Fussi et al. 2010); yet integrity of its gene pool appears to be protected by surprisingly strong reproductive barriers (Figure 1; Figure 3; Figure 4) and possibly by sufficient gene flow at the within-species level (Nem > 3.0; Lexer et al. 2005); intraspecific gene flow in the later colonizer is expected to prevent or slow down introgression and “surfing” of genes originating from the local species (Excoffier et al. 2009b).
The third trajectory, hybrid speciation, appears already more likely, since our data suggest the build-up of partial RI between intermediate hybrids and their parents (Figure 1; Figure 3; Lexer et al. 2009). The great ecological divergence between the hybridizing species (flood plain vs. upland habitats) is also in line with expectations for conditions prevailing at the onset of hybrid speciation (Buerkle et al. 2000; Nolte and Tautz 2010). Nevertheless, opportunities for interspecific gene flow still exist (Figures 1 and 3), and interspecific heterozygosities in hybrids are higher than expected, based on models of recombinational speciation (Buerkle et al. 2000); note that drift in isolated hybrid lineages tends to reduce interspecific heterozygosity. Hybrid speciation in European Populus spp. would require range shifts leading to stronger spatial separation of P. × canescens hybrids from both parental species or the presence of multiple genes with strong positive effects on hybrid fitness (“overdominance”; Buerkle et al. 2000). The latter pattern was found for 11 out of 68 loci (16%) in our regression-based genomic cline analysis of the Danube population, compared to 19% of loci with indications of underdominance (Table 2; underrepresentation of heterozygotes). On the basis of the data, P. × canescens may be on a trajectory to hybrid speciation, but genetic data on the depth of coalescences of gene sequences are required to test this further.
We regard the fourth scenario, adaptive introgression, as the most likely evolutionary consequence of hybridization between these species: our Structure-based ancestry analysis (Figure 1), genome scan of divergence between parental species (Figure 2), and two-dimensional analysis of heterozygosity vs. hybrid genomic composition (Figure 3) all indicate low but persistent (or recurrent) introgression into both parental species, consistent with the notion of the “porous genome” in speciation genetics (Wu 2001; Scotti-Saintagne et al. 2004; Minder et al. 2007). This suggests that the transfer of traits across porous species boundaries (Kim and Rieseberg 1999, Martin et al. 2006; Whitney et al. 2006) is possible in these ecologically important forest trees, and this is corroborated by the results of recent morphometric analyses of many phenotypic characters in these species and hybrids (Lexer et al. 2009). A combination of common garden experiments and genetic mapping are currently underway to test whether introgressed morphological and chemical traits are adaptive in the recipient species.
Admixture mapping in highly divergent populations:
Admixture mapping in hybrid zones has been suggested as a means of mapping loci involved in reproductive isolation and adaptive differentiation in the wild (Rieseberg and Buerkle 2002; Buerkle and Lexer 2008). The methodological approach is analogous to the mapping of disease gene loci in admixed human populations (Reich et al. 2005; Zhu et al. 2005). Our data allow us to comment on the prospects for genetic mapping in hybrid zones stemming from admixture between highly divergent lineages.
First, it is clear from Figure 3 that coarse-scale mapping of isolating factors in highly divergent populations with strong reproductive barriers (as often encountered in evolutionary genetics) will require the screening of multiple localities for recombinant, early-generation hybrids and subsequent pooling of these individuals to form a QTL mapping population of sufficient size. The same principle was used in Homo sapiens (Reich et al. 2005; Zhu et al. 2005), so this should not be a major obstacle in organisms with weak or absent population structure such as highly outcrossing, wind-pollinated forest trees. The linear models used for gene mapping in admixed populations are readily extended to accommodate the use of multiple populations and environments (Buerkle and Lexer 2008). Sample sizes of several hundred individuals will be required for admixture mapping in early recombinant hybrid generations, as in QTL mapping experiments with early generation intercrosses.
Perhaps of even greater relevance is the potential created by ultra-high throughput genotyping and resequencing: the unstoppable movement of neutral or advantageous alleles across porous genomes (this study; Barton 2001; Martin et al. 2006; Yatabe et al. 2007; Faure et al. 2008) indicates a great potential for the use of advanced introgressed populations (rather than hybrid zones with recent admixture) for high-resolution mapping using next generation sequencing approaches (Hohenlohe et al. 2010; Turner et al. 2010). Divergent, introgressed alleles from related species should readily be identified in highly advanced introgressants using the genomic coverage afforded by these methods, and appropriate neutrality tests (Nielsen 2005) facilitate the detection of fitness-related genes among those loci that do move across the barrier. This should allow the identification of functionally important heterospecific alleles in the process of spread through positive selection or neutral introgressed genes in the process of surfing on the colonization wave of rapidly expanding populations (Excoffier et al. 2009b). The marker coverage required at this step will depend on how much time (in terms of generations) has passed since the onset of admixture, and population size requirements will depend primarily on the desired threshold frequency for the detection of introgressed variants.
Clearly, admixture mapping projects in wild species are confronted with challenges not seen in admixed human populations used for gene mapping, but they also bring novel opportunities to unravel the genetics of reproductive isolation and to identify fitness-related genes that function well in divergent genomic backgrounds. Much remains to be learned about the evolutionary consequences of the origin and breakdown of reproductive barriers from genomic studies of admixture in wild species of animals and plants.
Acknowledgments
We thank Hans Herz, Wilfried Nebenführ, Stefano Gomarasca, István Asztalos, and other colleagues for help during field work; Robyn Cowan and Ludwika Sygnarski for help in the lab; Zach Gompert for help with data analysis; and Loren Rieseberg, Roger Butlin, and Sergey Gavrilets for helpful discussions. C.L.'s research on the evolutionary genomics of species barriers in Populus is supported by grant no. NE/E016731/1 of the U.K. Natural Environment Research Council and grant no. 31003A_127059 of the Swiss National Science Foundation.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.118828/DC1.
References
- Anderson, E. C., and E. A. Thompson, 2002. A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartha, D., 1991. Hybridization and introgression in Hungarian populations of white poplar (Populus alba L.). Mitt. Dtsch. Dendrol. Ges. 80 177–180 (in German). [Google Scholar]
- Barton, N. H., 2001. The role of hybridization in evolution. Mol. Ecol. 10 551–568. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., and K. S. Gale, 1993. Genetic analysis of hybrid zones, pp. 13–45 in Hybrid Zones and the Evolutionary Process, edited by R. G. Harrison and J. Price. Oxford University Press, Oxford.
- Barton, N. H., and G. M. Hewitt, 1985. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 16 113–148. [Google Scholar]
- Beaumont, M. A., 2005. Adaptation and speciation: What can FST tell us? Trends Ecol. Evol. 20 435–440. [DOI] [PubMed] [Google Scholar]
- Beaumont, M. A., and D. J. Balding, 2004. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 13 969–980. [DOI] [PubMed] [Google Scholar]
- Beaumont, M. A., and R. A. Nichols, 1996. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. Ser. B 263 1619–1626. [Google Scholar]
- Briscoe, D., J. Stephens and S. O‘Brien, 1994. Linkage disequilibrium in admixed populations: applications in gene mapping. J. Hered. 85 59–63. [PubMed] [Google Scholar]
- Buerkle, C. A., and C. Lexer, 2008. Admixture as the basis for genetic mapping. Trends Ecol. Evol. 23 686–694. [DOI] [PubMed] [Google Scholar]
- Buerkle, C. A., R. J. Morris, M. A. Asmussen and L. H. Rieseberg, 2000. The likelihood of homoploid hybrid speciation. Heredity 84 441–451. [DOI] [PubMed] [Google Scholar]
- Chakraborty, R., and K. Weiss, 1988. Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc. Natl. Acad Sci. USA 85 9119–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- De Carvalho, D., P. K. Ingvarsson, J. Joseph, L. Suter, C. Sedivy et al., 2010. Admixture facilitates adaptation from standing variation in the European aspen (P. tremula L.), a widespread forest tree. Mol. Ecol. 19 1638–1650. [DOI] [PubMed] [Google Scholar]
- Excoffier, L., T. Hofer and M. Foll, 2009. a Detecting loci under selection in a hierarchically structured population. Heredity 103 285–298. [DOI] [PubMed] [Google Scholar]
- Excoffier, L., M. Foll and R. J. Petit, 2009. b Genetic consequences of range expansions. Annu. Rev. Ecol. Evol. Syst. 40 481–501. [Google Scholar]
- Falush, D., M. Stephens and J. K. Pritchard, 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush, D., M. Stephens and J. K. Pritchard, 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure, M. F., P. David, F. Bonhomme and N. Bierne, 2008. Genetic hitchhiking in a subdivided population of Mytilus edulis. BMC Evol. Biol. 8 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, M. A., W. Adler and K. Oswald, 2005. Excursion flora for Austria, Liechtenstein and South Tyrol. Land Oberoesterreich, Landesmuseum, Linz, Austria (in German).
- Foll, M., and O. Gaggiotti, 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180 977–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossati, T., G. Patrignani, I. Zapelli, M. Sabatti, F. Sala et al., 2004. Development of molecular markers to assess the level of introgression of P. tremula into P. alba natural populations. Plant Breeding 123 382–385. [Google Scholar]
- Fussi, B., C. Lexer and B. Heinze, 2010. Phylogeography of Populus alba (L.) and Populus tremula (L.) in Central Europe: secondary contact and hybridisation during recolonisation from disconnected refugia. Tree Genet. Genomes 6 439–450. [Google Scholar]
- Gavrilets, S., 1997. Hybrid zones with Dobzhansky-type epistatic selection. Evolution 51 1027–1035. [DOI] [PubMed] [Google Scholar]
- Gompert, Z., and C. A. Buerkle, 2009. A powerful regression-based method for admixture mapping of isolation across the genome of hybrids. Mol. Ecol. 18 1207–1224. [DOI] [PubMed] [Google Scholar]
- Gompert, Z., and C. A. Buerkle, 2010. INTROGRESS: a software package for mapping components of isolation in hybrids. Mol. Ecol. Resources 10 378–384. [DOI] [PubMed] [Google Scholar]
- Goudet, J., 1995. Fstat version 1.2: a computer program to calculate F-statistics. J. Hered. 86 485–486. [Google Scholar]
- Hall, D., V. Luquez, V. M. Garcia, K. R. St Onge, S. Jansson et al., 2007. Adaptive population differentiation in phenology across a latitudinal gradient in European Aspen (Populus tremula, L.): a comparison of neutral markers, candidate genes and phenotypic traits. Evolution 61 2849–2860. [DOI] [PubMed] [Google Scholar]
- Hamzeh, M., and S. Dayanandan, 2004. Phylogeny of Populus (Salicaceae) based on nucleotide sequences of chloroplast TRNT-TRNF region and nuclear rDNA. Am. J. Bot. 91 1398–1408. [DOI] [PubMed] [Google Scholar]
- Hedrick, P. W., 2005. A standardized genetic differentiation measure. Evolution 59 1633–1638. [PubMed] [Google Scholar]
- Hohenlohe, P. A., S. Bassham, P. D. Etter, N. Stiffler, E. A. Johnson et al., 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD Tags. Plos Genet. 6 e1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins, C. D., C. Salazar, M. Linares and J. Mavarez, 2008. Hybrid trait speciation and Heliconius butterflies. Philos. Trans. R. Soc. Lond. B 363 3047–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, J. A., and C. Lexer, 2008. A set of novel DNA polymorphisms within candidate genes potentially involved in ecological divergence between Populus alba and P. tremula, two hybridizing European forest trees. Mol. Ecol. Resour. 8 188–192. [DOI] [PubMed] [Google Scholar]
- Kim, S.-C., and L. H. Rieseberg, 1999. Genetic architecture of species differences in annual sunflowers: implications for adaptive trait introgression. Genetics 153 965–977. [DOI] [PMC free article] [PubMed]
- Jost, L., 2008. GST and its relatives do not measure differentiation. Mol. Ecol. 17 4015–4026. [DOI] [PubMed] [Google Scholar]
- Lexer, C., B. Heinze, R. Alia and L. H. Rieseberg, 2004. Hybrid zones as a tool for identifying adaptive genetic variation in outbreeding forest trees: lessons from wild annual sunflowers (Helianthus spp.). Forest Ecol. Manag. 197 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexer, C., M. Fay, J. Joseph, M.-S. Nica and B. Heinze, 2005. Barrier to gene flow between two ecologically divergent Populus species, P. alba (white poplar) and P. tremula (European aspen): the role of ecology and life history in gene introgression. Mol. Ecol. 14 1045–1057. [DOI] [PubMed] [Google Scholar]
- Lexer, C., C. A. Buerkle, J. Joseph, B. Heinze and M. Fay, 2007. Admixture in European Populus hybrid zones makes feasible the mapping of loci that contribute to reproductive isolation and trait differences. Heredity 98 74–84. [DOI] [PubMed] [Google Scholar]
- Lexer, C., J. Joseph, M. van Loo, G. Prenner, B. Heinze et al., 2009. The use of digital image-based morphometrics to study the phenotypic mosaic in taxa with porous genomes. Taxon 58 349–364. [Google Scholar]
- Martin, N. H., A. C. Bouck and M. L. Arnold, 2006. Detecting adaptive trait introgression between Iris fulva and I. brevicaulis in highly selective field conditions. Genetics 172 2481–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinsen, G. D., T. G. Whitham, R. J. Turek and P. Keim, 2001. Hybrid populations selectively filter gene introgression between species. Evolution 55 1325–1335. [DOI] [PubMed] [Google Scholar]
- Milne, R. I., S. Terzioglu and R. J. Abbott, 2003. A hybrid zone dominated by fertile F(1)s: maintenance of species barriers in Rhododendron. Mol. Ecol. 12 2719–2729. [DOI] [PubMed] [Google Scholar]
- Minder, A. M., C. Rothenbuehler and A. Widmer, 2007. Genetic structure of hybrid zones between Silene latifolia and Silene dioica (Caryophyllaceae): evidence for introgressive hybridization. Mol. Ecol. 16 2504–2516. [DOI] [PubMed] [Google Scholar]
- Nielsen, R., 2005. Molecular signatures of natural selection. Annu. Rev. Genet. 39 197–218. [DOI] [PubMed] [Google Scholar]
- Nolte, A. W., and D. Tautz, 2010. The onset of hybrid speciation. Trends Genet. 26 54–58. [DOI] [PubMed] [Google Scholar]
- Nolte, A. W., Z. Gompert and C. A. Buerkle, 2009. Variable patterns of introgression in two sculpin hybrid zones suggest that genomic isolation differs among populations. Mol. Ecol. 18 2615–2627. [DOI] [PubMed] [Google Scholar]
- Payseur, B. A., J. G. Krenz and M. W. Nachman, 2004. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58 2064–2078. [DOI] [PubMed] [Google Scholar]
- Pialek, J., and N. H. Barton, 1997. The spread of an advantageous allele across a barrier: the effects of random drift and selection against heterozygotes. Genetics 145 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajora, O. P., and B. P. Dancik, 1992. Genetic characterization and relationships of Populus alba, P. tremula, and P. x canescens, and their clones. Theor. Appl. Genet. 84 291–298. [DOI] [PubMed] [Google Scholar]
- Reich, D., N. Patterson, P. De Jager, G. McDonald, A. Waliszewska et al., 2005. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat. Genet. 37 1113–1118. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., and C. A. Buerkle, 2002. Genetic mapping in hybrid zones. Am. Nat. 159 S36–S50. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., J. Whitton and K. Gardner, 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg, L. H., O. Raymond, D. M. Rosenthal, Z. Lai, K. Livingstone et al., 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301 1211–1216. [DOI] [PubMed] [Google Scholar]
- Rogers, S., D. Campbell, S. Baird, R. Danzmann and L. Bernatchez, 2001. Combining the analyses of introgressive hybridisation and linkage mapping to investigate the genetic architecture of population divergence in the lake whitefish (Coregonus clupaeformis, Mitchell). Genetica 111 25–41. [DOI] [PubMed] [Google Scholar]
- Scotti-Saintagne, C., S. Mariette, I. Porth, P. G. Goicoechea, T. Barreneche et al., 2004. Genome scanning for interspecific differentiation between two closely related oak species. [Quercus robur L. and Q. petraea (Matt.) Liebl.] Genetics 168 1615–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slate, J., 2005. Quantitative trait locus mapping in natural populations: progress, caveats and future directions. Mol. Ecol. 14 363–379. [DOI] [PubMed] [Google Scholar]
- Smulders, M. J. M., J. Van Der Schoot, P. Arens and B. Vosman, 2001. Trinucleotide repeat microsatellite markers for black poplar (Populus nigra L.). Mol. Ecol. Notes 1 188–190. [Google Scholar]
- Stettler, R. F., H. D. Bradshaw, P. E. Heilman and T. M. Hinckley, 1996. Biology of Populus and Its Implications for Management and Conservation. NRC Research Press, Ottawa.
- Teeter, K. C., L. M. Thibodeau, Z. Gompert, C. A. Buerkle, M. W. Nachman et al., 2010. The variable genomic architecture of isolation between hybridizing species of house mice. Evolution 64 472–485. [DOI] [PubMed] [Google Scholar]
- Turelli, M., and L. C. Moyle, 2007. Asymmetric postmating isolation: Darwin's corollary to Haldane’s rule. Genetics 176 1059–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, T. L., E. C. Bourne, E. J. Von Wettberg, T. T. Hu and S. V. Nuzhdin, 2010. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat. Genet. 42 260–263. [DOI] [PubMed] [Google Scholar]
- Tuskan, G. A., L. E. Gunter, Z. K. Yang, T. M. Yin, M. M. Sewell et al., 2004. Characterization of microsatellites revealed by genomic sequencing of Populus trichocarpa. Can. J. Forest Res. 34 85–93. [Google Scholar]
- Tuskan, G. A., S. DiFazio, S. Jansson, J. Bohlmann, I. Grigoriev et al., 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604. [DOI] [PubMed] [Google Scholar]
- van der Schoot, J., M. Pospiskova, B. Vosman and M. J. M. Smulders, 2000. Development and characterization of microsatellite markers in black poplar (Populus nigra L.). Thoer. Appl. Genet. 101 317–322. [Google Scholar]
- van Loo, M., J. A. Joseph, B. Heinze, M. F. Fay and C. Lexer, 2008. Clonality and spatial genetic structure in Populus × canescens and its sympatric backcross parent P. alba in a Central European hybrid zone. New Phytol. 177 506–516. [DOI] [PubMed] [Google Scholar]
- Via, S., and J. West, 2008. The genetic mosaic suggests a new role for hitchhiking in ecological speciation. Mol. Ecol. 17 4334–4345. [DOI] [PubMed] [Google Scholar]
- Vines, T. H., S. C. Kohler, A. Thiel, I. Ghira, T. R. Sands et al., 2003. The maintenance of reproductive isolation in a mosaic hybrid zone between the fire-bellied toads Bombina bombina and B. variegata. Evolution 57 1876–1888. [DOI] [PubMed] [Google Scholar]
- Weir, B. S., and C. C. Cockerham, 1984. Estimating F-statistics for the analysis of population structure. Evolution 38 1358–1370. [DOI] [PubMed] [Google Scholar]
- Whitlock, M. C., 1992. Temporal fluctuations in demographic parameters and the genetic variance among populations. Evolution 46 608–615. [DOI] [PubMed] [Google Scholar]
- Whitney, K. D., R. A. Randell and L. H. Rieseberg, 2006. Adaptive introgression of herbivore resistance traits in the weedy sunflower Helianthus annuus. Am. Nat. 167 794–807. [DOI] [PubMed] [Google Scholar]
- Wolf, D. E., N. Takebayashi and L. H. Rieseberg, 2001. Predicting the risk of extinction through hybridization. Conserv. Biol. 15 1039–1053. [Google Scholar]
- Wu, C. I., 2001. The genic view of the process of speciation. J. Evol. Biol. 14 851–865. [Google Scholar]
- Yatabe, Y., N. C. Kane, C. Scotti-Saintagne and L. H. Rieseberg, 2007. Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris. Genetics 175 1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., A. Luke, R. Cooper, T. Quertermous, C. Hanis et al., 2005. Admixture mapping for hypertension loci with genome-scan markers. Nat. Genet. 37 177–181. [DOI] [PubMed] [Google Scholar]