Abstract
Centromeres control chromosome inheritance in eukaryotes, yet their DNA structure and primary sequence are hypervariable. Most animals and plants have megabases of tandem repeats at their centromeres, unlike yeast with unique centromere sequences. Centromere function requires the centromere-specific histone CENH3 (CENP-A in human), which replaces histone H3 in centromeric nucleosomes. CENH3 evolves rapidly, particularly in its N-terminal tail domain. A portion of the CENH3 histone-fold domain, the CENP-A targeting domain (CATD), has been previously shown to confer kinetochore localization and centromere function when swapped into human H3. Furthermore, CENP-A in human cells can be functionally replaced by CENH3 from distantly related organisms including Saccharomyces cerevisiae. We have used cenh3-1 (a null mutant in Arabidopsis thaliana) to replace endogenous CENH3 with GFP-tagged variants. A H3.3 tail domain–CENH3 histone-fold domain chimera rescued viability of cenh3-1, but CENH3's lacking a tail domain were nonfunctional. In contrast to human results, H3 containing the A. thaliana CATD cannot complement cenh3-1. GFP–CENH3 from the sister species A. arenosa functionally replaces A. thaliana CENH3. GFP–CENH3 from the close relative Brassica rapa was targeted to centromeres, but did not complement cenh3-1, indicating that kinetochore localization and centromere function can be uncoupled. We conclude that CENH3 function in A. thaliana, an organism with large tandem repeat centromeres, has stringent requirements for functional complementation in mitosis.
CENTROMERES are essential for chromosome inheritance, because they nucleate kinetochores, the protein complexes on eukaryotic chromosomes that attach to spindle microtubules. Despite the essential requirement for centromeres in chromosome segregation, their DNA sequences and the sequences of kinetochore proteins are highly variable. Kinetochores in Saccharomyces cerevisiae and related budding yeasts assemble on small, unique centromere DNAs (125 bp in S. cerevisiae) (Meraldi et al. 2006). Centromere DNAs in the fission yeast Schizosaccharomyces pombe are larger, consisting of a central core sequence of 4–5 kb, which binds kinetochore proteins, flanked by large inverted repeats whose heterochromatic nature is important for centromere function (the total size of the S. pombe centromere DNA is 35–110 kb). At the other extreme from small yeast centromeres are holocentric organisms, such as Caenorhabditis elegans, in which kinetochore proteins bind along the entire length of mitotic chromosomes (Dernburg 2001). Most plants and animals have extremely large centromere DNA tracts consisting of megabases of simple tandem repeats. The repeat sequence evolves extremely rapidly, and only a small fraction of the repeat array is likely to be bound by kinetochore proteins. Furthermore, kinetochores can be nucleated by noncentromeric DNA sequences in plant and animal cells (Amor and Choo 2002; Nagaki et al. 2004; Nasuda et al. 2005; Heun et al. 2006; Wade et al. 2009). Despite these findings, the maintenance of massive centromere repeat arrays in both animal and plant taxa suggests that repeats are a central feature of centromere biology in these organisms.
Although centromere DNAs are extremely diverse, all eukaryote kinetochores contain the centromere-specific histone H3 variant CENH3 (originally described as CENP-A in human) (Henikoff and Dalal 2005; Black and Bassett 2008). CENH3 replaces conventional H3 specifically in a subset of centromere nucleosomes. It is essential for kinetochore function in all eukaryotes where this requirement has been tested. Conventional histones are among the most conserved proteins in eukaryote genomes. In contrast, CENH3 is rapidly evolving. The C-terminal histone-fold domain, which complexes with other histones to form the globular nucleosome core, can be aligned with conventional H3's but evolves rapidly and shows signatures of adaptive evolution in some residues (Malik and Henikoff 2001; Talbert et al. 2002; Cooper and Henikoff 2004). The N-terminal tail domain of conventional histone H3 protrudes from the nucleosome core and is not resolved in the structure solved by X-ray crystallography (Luger et al. 1997). In CENH3, the tail domain evolves so rapidly that its sequence can barely be aligned between closely related species.
Experiments in yeast and in animals have delineated functionally important regions within CENH3. S. cerevisiae kinetochores contain only a single CENH3/Cse4p nucleosome (Furuyama and Biggins 2007). In S. cerevisiae Cse4p, amino acid residues required for normal function are distributed throughout the histone-fold domain (Keith et al. 1999). The N-terminal tail of Cse4p contains an essential region termed the END domain, but overexpression of a Cse4p lacking the tail altogether can rescue a cse4 deletion mutant (Chen et al. 2000; Morey et al. 2004). In Drosophila melanogaster cells, CENH3/Cid from the distantly related D. bipectinata did not localize to kinetochores unless a specific region of the histone-fold domain, loop 1, was swapped with the corresponding region from D. melanogaster CENH3/Cid (Vermaak et al. 2002). In human, the histone-fold domain is important for centromere targeting (Sullivan et al. 1994). The functionally important region within the histone-fold domain was further defined by inserting loop 1 and the α-2 helix from CENH3/CENP-A (termed the CENP-A targeting domain, or CATD) into conventional H3 (Black et al. 2004). H3 containing the CATD acquires several functions of CENP-A when expressed in human cells. It localizes to kinetochores, binds the kinetochore protein CENP-N, has a rigid secondary structure when assembled into nucleosomes, and can restore normal chromosome segregation in cells depleted for CENP-A using RNA interference (RNAi) (Black et al. 2004, 2007a,b; Carroll et al. 2009).
Despite these extensive studies, questions about structure–function relationships within CENH3 remain. CENH3 function may differ between small yeast centromeres and the large tandem repeat centromeres of animals and plants, particularly because larger centromere DNAs are likely to contain many more CENH3 nucleosomes and may require a higher level of organization. Experiments in D. melanogaster and in human cells have used RNAi to downregulate the endogenous protein, and a conditional knockout has been made in chicken DT-40 cells (Blower and Karpen 2001; Goshima et al. 2003; Regnier et al. 2005; Black et al. 2007b). These experiments are challenging because CENH3 is very stable. If preexisting CENH3 is partitioned equally between duplicated sister centromeres, its amount will be approximately halved at each cell division. Therefore the protein may persist for many cell divisions after induction of RNAi, as shown by Western blots indicating that ∼10% of endogenous CENH3 remains in human cells subjected to two rounds of RNAi (Black et al. 2007b).
We have chosen to study CENH3 in the model plant A. thaliana, which combines facile genetics and transgenesis with centromere DNA structure that is similar to most plants and animals (megabases of tandem repeats with a repeating unit of 178 bp) (Murata et al. 1994; Copenhaver et al. 1999). Although Drosophila and mouse CENH3 knockout mutants have been characterized (Howman et al. 2000; Blower et al. 2006), a large-scale structure–function analysis of CENH3 has not been attempted in these organisms. A cenh3 null mutant in A. thaliana allows us to completely replace the endogenous protein with transgenic variants (Ravi and Chan 2010). Here we report four major conclusions regarding CENH3 function in A. thaliana: (1) CENH3 function requires an N-terminal histone tail domain, although either the CENH3 tail or the H3 tail can support mitotic chromosome segregation. (2) Inserting the CENP-A targeting domain of CENH3 into H3 does not confer CENH3 function. (3) Complementation of cenh3 by heterologous CENH3 requires that the species of origin be closely related to A. thaliana. (4) Localization of a heterologous CENH3 protein to kinetochores in the presence of native CENH3 does not necessarily indicate that it can complement a cenh3 mutant. Overall, our results indicate that requirements for CENH3 function in A. thaliana are more stringent that those obtained in human cells. They underscore the usefulness of comparative studies of centromere function using genetically tractable experimental organisms.
MATERIALS AND METHODS
Plant materials:
Plants were transformed by the Agrobacterium floral dip method using standard protocols. Plants were grown under 16 hr of light/8 hr of dark at 20°.
Cloning of CENH3 transgenes:
The CENH3 genomic fragment used to complement cenh3-1 was amplified by PCR and cloned into pCAMBIA1300 or pCAMBIA3300, using the endogenous HindIII site at nucleotide −1489 (relative to ATG = +1) and a BamHI site added to a PCR primer at nucleotide −2200.
To express CENH3 variants from the CENH3 promoter with an N-terminal GFP tag, we first constructed a cassette vector, CP93. This vector is based on pCAMBIA1300 and has an insert from EcoRI to HindIII that contains the following elements: the endogenous CENH3 promoter beginning at nucleotide −1489, a KpnI site, the GFP open reading frame without a STOP codon (Fang and Spector 2005), a hexaglycine linker, SalI and XbaI sites, and finally the CENH3 transcriptional terminator from the STOP codon until nucleotide 2200. The CENH3 coding region including introns was cloned into CP93 from SalI to XbaI to give GFP–CENH3.
Chimeric transgenes combining H3.3 (encoded by At1g13370) and CENH3 were constructed by overlapping PCR and cloned into CP93 from SalI to XbaI. First, the entire H3.3 coding region including introns was cloned to give GFP–H3.3. Second, we fused the tail domain of CENH3 (nucleotides 1–706) to the histone-fold domain of H3.3 (nucleotides 133–597). Third, we fused the tail domain of H3.3 (nucleotides 1–132) to the histone-fold domain of CENH3 (nucleotides 707–1629) to give GFP–tailswap. Fourth, to introduce the CATD of CENH3 into H3.3, we fused H3.3 (nucleotides 1–306) to CENH3 (nucleotides 1167–1419) and again to H3.3 (nucleotides 532–597).
To delete the tail domain of CENH3 in a GFP-tagged protein, we PCR amplified nucleotides 707–1629 of CENH3 and cloned them into CP93 from SalI to XbaI. To delete the tail domain of CENH3 in an untagged protein, we first constructed a cassette vector, CP225. Cloned from EcoRI to PstI within pCAMBIA1300, this contained the endogenous CENH3 promoter beginning at nucleotide −1489, MluI, SalI, and XbaI sites, and finally the CENH3 transcriptional terminator from the STOP codon until nucleotide 2200. We then cloned nucleotides 707–1629 of CENH3 into CP225 from SalI to XbaI.
To express heterologous CENH3 variants from the CENH3 promoter with an N-terminal GFP tag, we cloned them from SalI to XbaI into CP93. A. arenosa CENH3, Napa cabbage (Brassica rapa subspecies pekinensis) CENH3, and S. cerevisiae CSE4 (which lacks introns) were PCR amplified from genomic DNA. Napa cabbage was purchased from the Davis Farmer's Market. Maize (Zea mays) CENH3, human CENP-A, and C. elegans HCP-3 were PCR amplified from cDNA. Human cDNA was from HeLa cells.
DNA extraction and genotyping:
Genomic DNA preparation and PCR genotyping were performed using standard methods. cenh3-1 was genotyped with dCAPS primers. In lines containing GFP-tagged transgenic proteins, we were able to distinguish the endogenous CENH3 gene from the transgene by placing one primer upstream of the ATG in the 5′-UTR. To genotype endogenous CENH3 in lines containing an untagged CENH3 transgene, we first performed a PCR reaction with a primer outside the genomic DNA fragment present in the transgene. This PCR product was purified and then used as the substrate in a dCAPS genotyping reaction. Primer sequences are available on request.
Embryo analysis:
Differential interference contrast microscopy of developing seeds was performed as described (Aida et al. 1997).
Fluorescence microscopy:
Kinetochore localization of GFP-tagged proteins was assayed in floral tissues (anthers and petals) using a DeltaVision deconvolution microscope. Images were captured with a ×60 oil immersion lens and exposure time of 0.5 s. Z-stacks containing several cell layers with a step size of 0.2 um were captured. For cell cycle localization of wild-type (WT) GFP–CENH3, deconvolved Z-stacks were analyzed using a volume-rendering function in Imaris (Bitplane). For localization of heterologous CENH3s, raw images were scaled identically using SoftWoRx software (Applied Precision). Z-stacks were converted to two-dimensional (2D) projections in SoftWoRx and saved as TIF files to be processed using Adobe Photoshop and Illustrator.
Aneuploidy measurements:
We assayed the fidelity of chromosome segregation by counting chromosomes in somatic anther cells. Chromosome spreads were prepared according to standard protocols. Centromere DNA was labeled by fluorescence in situ hybridization (FISH), using a fluorescein-labeled probe. FISH analysis was performed as described (Comai et al. 2003).
S. cerevisiae genetics and cloning:
We created a cse4Δ∷kanMX4 deletion in a diploid of the SK1 strain using standard PCR-based transformation methods. CSE4 was cloned from −575 to 1059 (relative to ATG = +1) into the high-copy 2-μm plasmid YEp351, using the endogenous HindIII site at −575 and a BamHI site added to a PCR primer. We constructed cse4–tailswap and cse4Δ129 (Morey et al. 2004) constructs using the endogenous CSE4 promoter (−575 to 0) by overlapping PCR and cloned them into YEp351 from HindIII to BamHI. These plasmids were transformed into cse4Δ∷kanMX4/CSE4 diploid strains, which were then sporulated. Tetrad dissection was used to determine whether cse4Δ∷kanMX4 haploid cells containing plasmid-borne CSE4 variants were viable.
Sequence analysis:
To calculate percentage of identity between CENH3 proteins, we used BLAST to make pairwise alignments between A. thaliana CENH3 and proteins from other species. BLAST alignments were edited by hand. We used the shorter of the two protein sequences as the denominator. Multiple sequence alignment was done with ClustalW.
RESULTS
Isolation of a cenh3 null mutation from a tetraploid A. thaliana population:
CENH3 in A. thaliana is encoded by the HTR12 gene (Talbert et al. 2002). We refer to the gene as CENH3 in accordance with the nomenclature used in other plant species and in several unicellular eukaryotes. Public databases did not list any T-DNA insertions within the CENH3 ORF, and a set of 22 point mutations in the gene created by TILLING did not contain an obvious null allele such as a premature STOP codon (7 created nonsynonymous changes in exons) (http://www.arabidopsis.org). As CENH3 is an essential protein, we were concerned that a complete loss-of-function mutation might be lethal to both male and female gametophytes. Therefore, we utilized a TILLING population created by mutagenizing tetraploid A. thaliana with ethylmethanesulfonate (L. Comai, V. Sundaresan, and R. K. Tran unpublished results). Gametophytes in tetraploid plants are diploid and therefore a gametophyte lethal mutation can be complemented by a wild-type gene.
cenh3-1 is a G-to-A transition at nucleotide 161 relative to ATG = +1 and changes the conserved splice acceptor sequence AG to AA in the second intron (Figure 1A). This mutation is predicted to disrupt normal splicing of CENH3. We performed RT–PCR from plants heterozygous for cenh3-1 using PCR primers that bracket the mutation. One predominant novel splice form was seen in cenh3-1 heterozygotes that was not observed in wild type (other splice variants may also exist, but were not obvious when the RT–PCR product was directly sequenced). In this cDNA, exon 1 splices incorrectly to nucleotide 140 in the middle of intron 2. Notably, nucleotides surrounding the cryptic splice site used in cenh3-1 are identical to that of the regular splice junction (TAGAT, where the fourth A in the sequence is the beginning of the exon). The incorrectly spliced cDNA is predicted to encode the first 18 amino acids of CENH3, followed by a frameshift and STOP codon after 46 amino acids (Figure 1A). The CENH3 histone-fold domain contains sequences essential for centromere localization and for kinetochore function (Sullivan et al. 1994; Keith et al. 1999; Vermaak et al. 2002; Lermontova et al. 2006; Black et al. 2007b). As the histone-fold domain of A. thaliana CENH3 begins at amino acid 82, we believe that cenh3-1 is a null mutation.
Figure 1.—
A cenh3 null allele in Arabidopsis is embryo lethal. (A) cenh3-1 is predicted to encode a nonfunctional truncated protein. The cenh3-1 gene splices incorrectly, produced a protein that is truncated before the essential histone-fold domain. White boxes, 5′-UTR; dark gray boxes, coding sequence; Light gray boxes, wild-type protein sequence; black box, frameshifted protein sequence resulting from incorrect splicing. (B) Genetic scheme for converting a tetraploid TILLING mutant into a heterozygous diploid. If an allele can be passed from a triploid parent to diploid offspring, it must be transmitted through a haploid gamete. (C) Siliques from wild-type and cenh3-1/CENH3 plants are split open to reveal developing seeds. White seeds indicate embryonic lethality in cenh3-1 homozygous progeny. (D) Genotyping of progeny from cenh3-1/CENH3. The ∼1:2 ratio of CENH3/CENH3 to cenh3-1/CENH3 progeny is consistent with embryonic lethality of cenh3-1. (E) cenh3-1/cenh3-1 embryos arrest at the midglobular stage. Developing seeds in the top and bottom rows were both from cenh3-1/CENH3 siliques. Embryos for each column were fixed at equivalent times after fertilization (labels describe the wild-type developmental stages).
We isolated cenh3-1 in tetraploid plants, which can be converted into diploids via two sequential genetic crosses (Figure 1B) (Henry et al. 2009). Crossing a tetraploid to a diploid yields triploid offspring. Meiosis in triploids produces gametes with a range of chromosome numbers, ranging from haploid (5 in A. thaliana) to diploid (10 in A. thaliana). Crossing a triploid to a diploid gives progeny with a range of chromosome numbers, but ∼30% are true diploids. If a mutant allele can be passed from a triploid parent to a diploid offspring in such a cross, it must be transmitted through a haploid gametophyte. We created cenh3-1/CENH3 heterozygous diploids by crossing a cenh3-1–containing triploid to a wild-type diploid in both directions. Therefore, the cenh3-1 null allele can be transmitted through both male and female gametophyte lineages, indicating that either gametophyte can function without a wild-type CENH3 gene.
Plants heterozygous for the cenh3-1 allele were phenotypically indistinguishable from wild type. However, siliques from heterozygous cenh3-1/CENH3 plants contained 1/4 dead seeds (124/508 dead seeds, from 11 siliques) (Figure 1C), and progeny of these plants contained ∼1/3 CENH3/CENH3 homozygotes and 2/3 cenh3-1/CENH3 heterozygotes (Figure 1D). cenh3-1 homozygotes were never isolated. Taken together, the above observations indicated that cenh3-1 causes zygotic lethality. To understand the nature of embryo lethality, we examined the time course of embryogenesis by optical clearing of fertilized ovules from a heterozygous plant. cenh3-1 homozygous embryos did not develop beyond the midglobular stage and arrested at this stage when presumed CENH3/CENH3 and cenh3-1/CENH3 seeds from the same silique had progressed to later stages of embryogenesis (Figure 1E). This is a common arrest point for many A. thaliana embryo-lethal mutations. Several cell divisions must take place after fertilization in cenh3-1/cenh3-1 homozygous mutant embryos, as they arrest at the multicellular midglobular stage. However, persistence of either maternal CENH3 mRNA or CENH3 protein are likely to provide residual kinetochore function until failure of chromosome segregation results in the observed lethality. CENH3 loaded into paternal chromosomes has been shown to persist through multiple cell divisions in the endosperm after fertilization (Ingouff et al. 2007).
To confirm that embryonic lethality was caused by the cenh3-1 mutation, we transformed cenh3-1/CENH3 heterozygotes with a genomic DNA fragment spanning nucleotides −1489 to 2200 of CENH3. In the T1 generation, 13/59 transformed plants were homozygous for the cenh3-1 mutation, indicating that embryo lethality can be rescued by a CENH3 transgene (Table 1). In summary, A. thaliana cenh3-1 is a presumed null mutation that allows us to completely replace endogenous CENH3 in an organism with large tandem repeat-based centromeres.
TABLE 1.
Complementation of cenh3-1 by CENH3, GFP–CENH3, and GFP–tailswap transgenes
| Transgene | Total no. of plants | CENH3/CENH3 | cenh3-1/CENH3 | cenh3-1/cenh3-1 | P-value |
|---|---|---|---|---|---|
| CENH3 (Basta) | 32 T1 | 10 | 14 | 8 | |
| CENH3 (Hygromycin) | 27 T1 | 9 | 13 | 5 | |
| Total CENH3 | 59 T1 | 19 | 27 | 13 | 0.4396 |
| GFP–CENH3 | 70 T1 | 22 | 45 | 3 | 0.0003 |
| GFP–tailswap | 33 T1 | 6 | 27 | 0 | 0.0039 |
| GFP–tailswapa | 61 T2 | 15 | 38 | 8 | 0.1757 |
Transformed plants born from cenh3-1/CENH3 T0 parents were genotyped at the endogenous CENH3 locus in the T1 generation. CENH3 transgenics were selected using either Basta (pCAMBIA3300) or Hygromycin (pCAMBIA1300). cenh3-1/CENH3 GFP–tailswap/+ plants that yielded cenh3-1 homozygous progeny in the T2 generation were selected on the basis of their lower seed lethality in the T1 generation. P-values were calculated using the χ2 distribution, with complementation as the null hypothesis.
These plants were the progeny of T1 cenh3-1/CENH3 GFP–tailswap/+ plants that showed 6–25% seed lethality.
GFP-tagged CENH3 can complement a cenh3-1 null mutant, although not all transformed lines show complementation:
Previous studies have shown that CENH3 proteins GFP tagged at their C termini are localized to kinetochores in A. thaliana (Fang and Spector 2005; Lermontova et al. 2006), but the lack of a cenh3 mutant meant that complementation could not be assayed. One such CENH3–GFP transgene (Fang and Spector 2005) did not complement the embryo-lethal phenotype of cenh3-1 when crossed into the mutant background (data not shown). We tagged CENH3 at its N terminus with GFP, using the endogenous CENH3 promoter and terminator to control transgene expression (Ravi and Chan 2010). When cenh3-1/CENH3 plants are transformed with a GFP–CENH3 transgene, 3/70 transformed T1 plants were cenh3-1 homozygotes (Table 1). Genotyping results from T1 plants showed statistically significant differences from expected values for full complementation, using the χ2 test. In cenh3-1/CENH3 T1s expressing a GFP–CENH3 transgene, embryo lethality in the siliques would be expected to range from 25% (signifying lack of complementation in the T2 generation) to 6.25% (consistent with full complementation in the T2 generation by a single locus insertion; multiple loci may further reduce embryo lethality). Some cenh3-1/CENH3 GFP–CENH3/+ transgenic plants (2/9 T1 individuals) showed seed lethality close to 25%, indicating that these transgenes failed to complement.
Reduced complementation frequency by GFP–CENH3 relative to CENH3 shows that the tag may affect centromere function in a subtle manner. This is consistent with chromosome missegregation seen when chromosomes containing GFP-tagged CENH3 are mixed with wild-type chromosomes in a GFP–CENH3 × wild-type cross. The entire GFP–CENH3 derived genome is eliminated from the zygote in some F1 individuals, and ∼40% of the developing seeds from the cross show abortion (Ravi and Chan 2010). Despite these observations, cenh3-1 homozygotes rescued by GFP–CENH3 were phenotypically indistinguishable from WT and had a full seed set on self-fertilization (Figure 2, A and C). One such GFP–CENH3 complemented line has been stably propagated at least for three successive generations without any observable phenotype. GFP fluorescence at kinetochores was observed in all phases of the cell cycle in these plants (Figure 2B). Ten kinetochores were labeled in interphase, whereas 20 individual kinetochores could be visualized in mitotic cells.
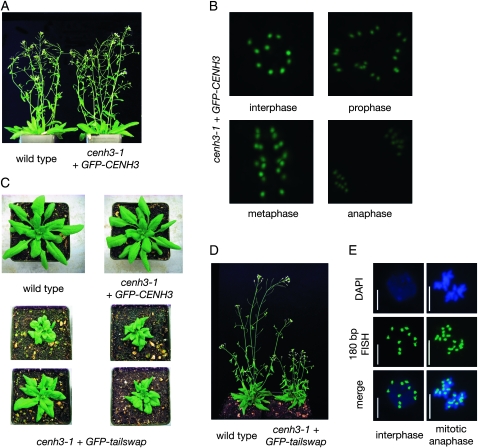
Figure 2.—
Morphological phenotype of cenh3-1 plants complemented by GFP–CENH3 and GFP–tailswap. (A) GFP–CENH3 complemented plants are indistinguishable from wild type. (B) GFP fluorescence labels kinetochores in interphase and mitotic cells from anther squashes. Three dimensional images were captured by deconvolution microscopy. (C) GFP–tailswap complemented plants have a variable vegetative phenotype. (D) GFP–tailswap complemented plants have short stature, and are infertile. (E) Centromeres in GFP–tailswap complemented plants are visualized by FISH in somatic nuclei using a 180-bp centromere DNA repeat probe. We did not detect somatic aneuploidy after inspecting >500 nuclei.
Mouse CENP-A/CENH3 tagged with GFP at its C terminus is only partially functional, because a cenpa−/− mouse rescued by Cenpa–GFP survives only until embryonic day 10.5 (an N-terminal GFP tag on CENP-A was not tested in mouse) (Kalitsis et al. 2003). A. thaliana cenh3-1 GFP–CENH3 plants have a wild-type growth phenotype. Therefore, we can use an N-terminal GFP tag to assay kinetochore localization without grossly disrupting the ability of CENH3 to complement a null mutation.
The CENH3 histone-fold domain controls kinetochore localization, but rescue of cenh3-1 mitotic chromosome segregation by CENH3 transgenes requires an N-terminal histone tail:
To determine which regions of CENH3 confer centromere localization and kinetochore function, we generated chimeric proteins that combined domains of CENH3 and H3.3 (we chose H3.3 because it can be incorporated into chromatin independently of DNA replication and have not tested chimeras between CENH3 and H3.1) (Ahmad and Henikoff 2002). These proteins were expressed as fusion proteins with an N-terminal GFP using the endogenous CENH3 promoter and terminator and transformed into cenh3-1/CENH3 plants (Figure 3). We assayed kinetochore localization in T1 transformants by GFP fluorescence and genotyped for the presence of cenh3-1 homozygotes among transformed plants. If cenh3-1 homozygotes were not detected, we counted the frequency of seed lethality in cenh3-1/CENH3 T1 plants that were hemizygous for the CENH3 transgene, to determine whether the transgene could complement embryonic lethality of cenh3-1.
Figure 3.—
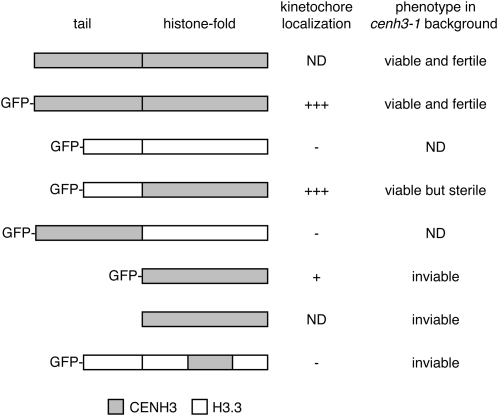
Kinetochore localization of CENH3/H3.3 chimeras and complementation of cenh3-1 embryonic lethality. CENH3/H3.3 chimeras were tagged with GFP at their N termini and cloned under the control of the endogenous CENH3 promoter. The indicated transgenic proteins were introduced into cenh3-1/CENH3 T0 plants. Transformed T1 plants were assayed for GFP fluorescence at kinetochores. Complementation of cenh3-1 embryonic lethality (i.e., viability) was assayed by counting seed lethality in siliques from at least eight cenh3-1/CENH3 plants in the T1 generation. ND, not determined; +++, strong GFP fluorescence at kinetochores; +, weaker GFP fluorescence at kinetochores; -, no GFP fluorescence at kinetochores.
Previous results have indicated that the CENH3 histone-fold domain controls kinetochore localization (Sullivan et al. 1994; Lermontova et al. 2006; Black et al. 2007b). We confirmed that the CENH3 N-terminal tail domain fused to the H3.3 C-terminal histone-fold domain did not localize to kinetochores (Figure 3). In contrast, a protein containing the H3.3 tail domain and the CENH3 histone-fold domain (termed GFP–tailswap) showed kinetochore GFP fluorescence. GFP–tailswap rescued the embryo-lethal phenotype of cenh3-1 (previously described in Ravi and Chan (2010), although complemented cenh3-1 homozygotes were not isolated in the T1 generation, and only a subset of cenh3-1/CENH3 transformants showed complementation when T2 progeny were analyzed. GFP–tailswap complemented plants had misshapen rosette leaves and shorter internodes (Figure 2, C and D). This vegetative phenotype varied in penetrance and expressivity. Despite the altered vegetative growth of cenh3-1 GFP–tailswap plants, we inspected a total of >500 somatic nuclei from floral tissues (from three independent transformants) using centromere DNA FISH and did not detect aneuploidy (Figure 2E). In the absence of obvious aneuploidy, developmental perturbations in cenh3-1 GFP–tailswap may arise from altered cell cycle timing or from defects in asymmetric cell division. Although chromosome segregation in GFP–tailswap is accurate, the protein must be significantly less functional than native CENH3, because it induces a dramatic level of chromosome missegregation and seed abortion when GFP–tailswap containing chromosomes mix with wild-type chromosomes after a cross (Ravi and Chan 2010). Interestingly, cenh3-1 GFP–tailswap plants were mostly sterile (irrespective of the penetrance of their vegetative phenotype) suggesting that this transgene is functional in mitosis, but not in meiosis (Figure 2D). We are currently investigating the nature of sterility caused by GFP–tailswap.
As the N-terminal tail of CENH3 can be replaced with the H3.3 tail, we deleted the tail in the context of a GFP-tagged transgenic protein, and in a transgene without a GFP tag (Figure 3). The GFP-tagged protein localized to kinetochores, albeit more weakly than full-length GFP-tagged CENH3. Neither transgene rescued embryonic lethality of cenh3-1, based on the percentage of seed lethality (∼25%) in T1 plants. Furthermore, we genotyped 24 T2 progeny from a cenh3-1/CENH3 individual containing a GFP-tagged tail-deleted CENH3 transgene and did not find any cenh3-1/cenh3-1 progeny. This shows that CENH3 requires a tail domain of some kind—either the CENH3 tail or the H3.3 tail—to nucleate a functional kinetochore in mitosis.
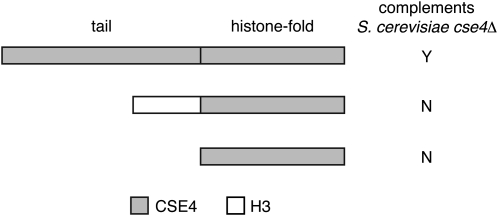
In S. cerevisiae, the N-terminal tail of Cse4p/CENH3 contains an essential domain, but a Cse4 protein completely lacking the tail (cse4Δ129) can complement a deletion mutant if overexpressed from the GAL1 promoter in an integrated transgene (Morey et al. 2004). We created a “tailswap” Cse4p by replacing the endogenous tail domain with the S. cerevisiae H3 tail (Figure 4). Wild-type CSE4 complemented a cse4Δ mutant when expressed from a high-copy 2-μm plasmid, but cse4–tailswap and cse4Δ129 did not (in contrast to previous studies, we expressed cse4Δ129 from the native CSE4 promoter, albeit from a high-copy plasmid). This result shows that S. cerevisiae Cse4p differs from A. thaliana CENH3, in that the essential function of the tail domain cannot be replaced by the H3 tail.
Figure 4.—
Tailswap CSE4p fails to complement cse4 in S. cerevisiae. High-copy 2-μm plasmids containing the indicated proteins controlled by the CSE4 promoter were introduced into a S. cerevisiae cse4/CSE4 strain. Following sporulation, tetrads contained either four viable colonies (complementation) or two viable colonies (noncomplementation).
Introducing the CENP-A targeting domain (CATD) into H3.3 does not confer kinetochore localization or complementation of cenh3-1 in Arabidopsis:
In humans, a small region of the histone-fold domain of CENH3 (named the CENP-A targeting domain or CATD) can induce kinetochore localization when introduced into H3 (Black et al. 2004). The CATD comprises loop 1 and α-helix 2 of the histone-fold domain (Figure 5). CENH3 function may also be acquired in this domain swap, because H3 containing the CATD can restore a normal chromosome segregation phenotype in cells where endogenous CENP-A has been depleted with RNAi (Black et al. 2007b). We introduced the CATD of A. thaliana CENH3 (amino acids 114–155) into H3.3, tagged the protein with an N-terminal GFP, and expressed it from the endogenous CENH3 promoter (Figure 3). This chimeric protein did not localize to kinetochores or rescue seed lethality of cenh3-1. Our results show that the ability of the previously defined CATD to convert H3 into CENH3 is not conserved in all eukaryotes with large tandem repeat centromeres. The boundaries within the histone-fold domain that determine CENH3 function may differ between animals and plants.
Figure 5.—
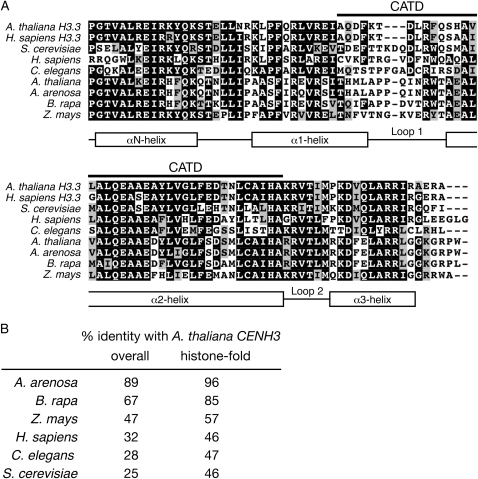
Histone-fold domain multiple sequence alignment of heterologous CENH3 proteins expressed in A. thaliana. (A) Histone-fold domain multiple sequence alignment of Arabidopsis thaliana and human (Homo sapiens) conventional H3.3 proteins and CENH3 proteins from Saccharomyces cerevisiae, human, Caenorhabditis elegans, Arabidopsis thaliana, A. arenosa, Brassica rapa, and maize (Zea mays). (B) Percentage of identity of heterologous CENH3 proteins with A. thaliana CENH3. The histone-fold domain was defined as beginning at amino acid 82 in A. thaliana CENH3, the start of the α-N helix.
Heterologous CENH3 must be from a close relative to complement A. thaliana cenh3-1.
Although CENH3 is very fast evolving relative to conventional H3's, experiments in human cells have suggested that CENH3 proteins from distantly related organisms (even S. cerevisiae) can functionally replace CENP-A (Wieland et al. 2004). However, these experiments relied on RNAi to reduce endogenous protein levels, and the stability of the CENP-A protein means that cells subjected to gene silencing may have contained significant levels of residual CENP-A.
We attempted to complement A. thaliana cenh3-1 with several heterologous CENH3's at varying degrees of evolutionary divergence (Figure 5). CENH3 ORFs were fused to an N-terminal GFP and expressed from the endogenous A. thaliana CENH3 promoter (cDNA sequences were used for CENH3 genes outside the Brassicaceae, except S. cerevisiae CSE4, which does not have introns). These transgenes were transformed into cenh3-1/CENH3 heterozygotes. Kinetochore localization of GFP-tagged proteins (Figure 6) and complementation of cenh3-1 embryonic lethality (Figure 7) were assayed as described above for CENH3/H3.3 chimeric protein transgenes.
Figure 6.—
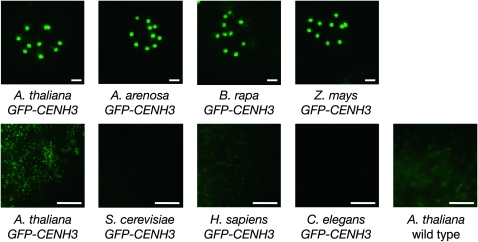
Kinetochore localization of heterologous GFP–CENH3 proteins expressed in A. thaliana. GFP fluorescence microscopy of anther squashes from cenh3/CENH3 plants expressing GFP–CENH3 proteins. Arabidopsis thaliana, A. arenosa, Brassica rapa, maize (Zea mays), human (Homo sapiens), Caenorhabditis elegans, and Saccharomyces cerevisiae GFP–CENH3 transgenics are shown. All images are scaled to the intensity distribution of A. thaliana GFP–CENH3 to normalize for background and autofluorescence. Uppermost images were deconvolved using SoftWoRx (Applied Precision) and enlarged to show centromere localization in individual nuclei. Bar for upper row, 1 μm; for lower row, 10 μm.
Figure 7.—
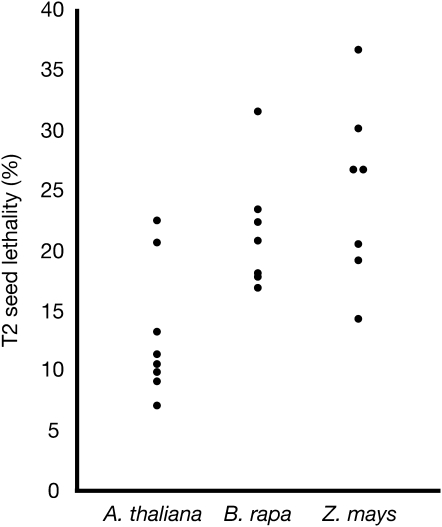
B. rapa and Z. mays GFP–CENH3 transgenes do not complement cenh3-1. A. thaliana, B. rapa, and Z. mays GFP–CENH3's were cloned under the control of the A. thaliana CENH3 promoter and introduced into cenh3-1/CENH3 T0 plants. Transformed plants were genotyped, and seed lethality was measured in at least five siliques from individual cenh3-1/CENH3 T1 plants. Each dot represents an independent T1 transformant. Noncomplementation is indicated by 25% lethality, while full complementation is indicated by 6.25% seed lethality (or lower). T2 progeny from B. rapa and Z. mays lines showing the lowest seed lethality were planted and genotyped at the endogenous CENH3 locus. We did not recover homozygotes among 24 T2 plants for each line.
GFP–CENH3 from the close relative A. arenosa complemented A. thaliana cenh3-1, as complemented plants homozygous for cenh3-1 were isolated in the T1 generation. T2 plants homozygous for the A. arenosa GFP–CENH3 transgene had a full seed set. A. arenosa GFP–CENH3 showed kinetochore localization similar to A. thaliana GFP–CENH3, which is consistent with the previous observation that A. thaliana CENH3 localizes to both A. thaliana and A. arenosa chromosomes in the allopolyploid species A. suecica (Talbert et al. 2002). B. rapa (Napa cabbage) is more distantly related to A. thaliana, but still within the Brassicaceae family. Transgenics expressing B. rapa GFP–CENH3 showed kinetochore GFP fluorescence comparable to that seen with A. thaliana GFP–CENH3 (Figure 6). However, we did not find cenh3-1 homozygotes among transformed plants, and seed lethality in cenh3-1/CENH3 T1 plants indicated that this protein did not rescue embryonic lethality (Figure 7). This result is illuminating because it clearly uncouples kinetochore localization (in the presence of wild-type endogenous CENH3) from the ability to support chromosome segregation in the absence of endogenous CENH3. As the N-terminal tail of CENH3 can be replaced with the H3.3 tail, histone-fold domain changes are likely to explain why B. rapa CENH3 is functionally different from A. thaliana and A. arenosa CENH3's. There are 13 amino acids in the histone-fold domain at which B. rapa CENH3 differs from both Arabidopsis CENH3's, of which 9 are in the CATD.
The more distantly related CENH3 from maize (Z. mays) localized to A. thaliana kinetochores when tagged with GFP (Figure 6). As expected, on the basis of B. rapa results, maize GFP–CENH3 failed to rescue embryonic lethality (Figure 7). A previous report stated that rice CENH3 did not localize to A. thaliana kinetochores when tagged with a C-terminal GFP and expressed in cultured cells (Nagaki et al. 2010). However, our results suggest that CENH3's from some grass species may be able to target correctly in the presence of the native A. thaliana CENH3, but are not functionally competent in the absence of native CENH3. GFP–CENH3 proteins from human (CENP-A), S. cerevisiae (Cse4p), and C. elegans (HCP-3) failed to localize to kinetochores in A. thaliana (Figure 6) and were not tested for complementation. We infer that animal and fungal CENH3's cannot be recruited to A. thaliana kinetochores by the endogenous cellular machinery or are unable to be maintained there.
DISCUSSION
With the isolation of cenh3-1, we have established A. thaliana as a genetic model for studying CENH3 in the context of large tandem repeat centromeres. Some of our results differ from published data in other experimental systems. These differences may result from organism-specific features of CENH3 biology, particularly because yeast centromeres are much smaller than those in animals and plants. However, differences between our findings and those in animal cells may also reflect the ability of A. thaliana genetics to completely replace native CENH3 with transgenic proteins expressed using the endogenous CENH3 promoter.
We have previously shown that GFP–CENH3 is not functionally equivalent to native CENH3, because a GFP–CENH3 × wild-type cross induces chromosome missegregation when the two chromosome sets mix in the fertilized zygote (Ravi and Chan 2010). Complementation assays presented here support this conclusion, although we do not know why GFP interferes with normal CENH3 function. It is also unclear why some GFP–CENH3 transformants can complement cenh3-1, yet others cannot. Differences in expression level caused by position effects are generally minor in A. thaliana, but may prove significant if the GFP tag significantly affects CENH3 function. Alternatively, epigenetic factors may affect the ability of a given GFP–CENH3 line to form functional kinetochores when endogenous CENH3 is removed. As complemented cenh3-1 GFP–CENH3 plants have completely normal growth and fertility, there may be relatively little difference between a GFP–CENH3 based kinetochore that is nonfunctional and one that supports accurate chromosome segregation. In general, GFP-tagging of CENH3 and other kinetochore proteins may cause subtle differences in function that are only revealed by careful genetic analysis.
The function of the fast-evolving N-terminal tail domain of CENH3 is enigmatic. A truncated tailless CENH3 is localized to kinetochores but cannot rescue embryo lethality of cenh3-1. However, the CENH3 tail may be replaced with the H3.3 tail (which has a virtually unrelated sequence) during mitosis. The hypervariable nature of the tail domain might suggest coevolution with rapidly changing centromere DNA sequences, although demonstrating such an interaction biochemically is challenging within the context of CENH3 nucleosomes. Our finding that a histone tail domain of some type is required for CENH3 mitotic function is analogous to results from S. cerevisiae, in which the tail of Cse4p is essential at native expression levels (although we have shown here that the Cse4p tail cannot be replaced by the H3 tail). Although mitosis can be rescued in cenh3-1 plants expressing GFP–tailswap, these plants are sterile. The specific role of the CENH3 tail domain in meiosis will be an intriguing topic for future study.
A major conclusion from our work is that CENH3 from a heterologous species must be closely related to A. thaliana CENH3 for functional complementation, although CENH3 from somewhat more distantly related species is targeted to kinetochores. This has been shown for fungal CENH3's (Baker and Rogers 2006), but it is important to confirm this property for an organism with large tandem repeat centromeres that are likely specified epigenetically, rather than by sequence-specific DNA binding proteins. A previous study using plant tissue culture cells found that CENH3 from tobacco (Nicotiana tabacum) could localize to A. thaliana kinetochores when tagged at its C termini with GFP and expressed from the strong constitutive 35S promoter (Nagaki et al. 2010). Similar results were seen when A. thaliana CENH3 fusion proteins were expressed in cultured tobacco cells (Kurihara et al. 2008; Nagaki et al. 2010). Importantly, our results show that kinetochore localization in the presence of endogenous CENH3 (such as that seen with B. rapa GFP–CENH3) does not necessarily reflect functional complementation. B. rapa GFP–CENH3 may have difficulty localizing to kinetochores when endogenous CENH3 is removed or may fail to interact with A. thaliana kinetochore proteins even if it localizes correctly. Our results emphasize that genetic analysis in a cenh3 mutant background is important for truly assessing whether a given CENH3 protein can function in a heterologous organism. We were surprised by the finding that the CATD swap into H3 did not confer CENH3 function and by the result that a B. rapa GFP–CENH3 did not complement cenh3-1. These data may reflect a greater overall stringency in the A. thaliana cellular machinery that recruits CENH3 to kinetochores and/or its function in nucleating assembly of kinetochore proteins. Alternatively, the residual wild-type CENH3 present in human cell experiments may have been functionally significant in allowing H3–CATD and CENH3's from distantly related organisms to restore normal chromosome segregation.
Rapid evolution of CENH3 has been suggested to compensate for changes in centromere DNA that might result in unequal binding to spindle microtubules of centromere DNA variants (Henikoff et al. 2001). The fact that complementation of A. thaliana cenh3-1 requires a CENH3 protein from a closely related species makes the plant ideal for studying the functional implications of rapid CENH3 evolution both in mitosis and in meiosis.
Acknowledgments
We thank Jong Park, Rima Woods, and Helen Tsai for technical assistance and Joanne Engebrecht, Marty Privalsky, Neelima Sinha, Brad Townsley, Shannon Ceballos, Hsuan-Chung Ho, and Liza Conrad for reagents. We also thank Daniel Chu and Sean Burgess for generous help with S. cerevisiae genetics. This study was funded by a Basil O'Connor Starter Scholar Research Award from the March of Dimes (to S.C.), a grant from the Hellman Family foundation (to S.C.), National Science Foundation (NSF) grant IOS-0745167 (to V.S.), and grant DBI-0822383 from NSF Plant Genome (to L.C.).
References
- Ahmad, K., and S. Henikoff, 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9 1191–1200. [DOI] [PubMed] [Google Scholar]
- Aida, M., T. Ishida, H. Fukaki, H. Fujisawa and M. Tasaka, 1997. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor, D. J., and K. H. Choo, 2002. Neocentromeres: role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71 695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, R. E., and K. Rogers, 2006. Phylogenetic analysis of fungal centromere H3 proteins. Genetics 174 1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, B. E., and E. A. Bassett, 2008. The histone variant CENP-A and centromere specification. Curr. Opin. Cell Biol. 20 91–100. [DOI] [PubMed] [Google Scholar]
- Black, B. E., D. R. Foltz, S. Chakravarthy, K. Luger, V. L. Woods, Jr. et al., 2004. Structural determinants for generating centromeric chromatin. Nature 430 578–582. [DOI] [PubMed] [Google Scholar]
- Black, B. E., M. A. Brock, S. Bedard, V. L. Woods, Jr. and D. W. Cleveland, 2007. a An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. USA 104 5008–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, B. E., L. E. Jansen, P. S. Maddox, D. R. Foltz, A. B. Desai et al., 2007. b Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell 25 309–322. [DOI] [PubMed] [Google Scholar]
- Blower, M. D., and G. H. Karpen, 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M. D., T. Daigle, T. Kaufman and G. H. Karpen, 2006. Drosophila CENP-A mutations cause a BubR1-dependent early mitotic delay without normal localization of kinetochore components. PLoS Genet. 2 e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, C. W., M. C. Silva, K. M. Godek, L. E. Jansen and A. F. Straight, 2009. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 11 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., R. E. Baker, K. C. Keith, K. Harris, S. Stoler et al., 2000. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell Biol. 20 7037–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai, L., A. P. Tyagi and M. A. Lysak, 2003. FISH analysis of meiosis in Arabidopsis allopolyploids. Chromosome Res. 11 217–226. [DOI] [PubMed] [Google Scholar]
- Cooper, J. L., and S. Henikoff, 2004. Adaptive evolution of the histone fold domain in centromeric histones. Mol. Biol. Evol. 21 1712–1718. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G. P., K. Nickel, T. Kuromori, M. I. Benito, S. Kaul et al., 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286 2468–2474. [DOI] [PubMed] [Google Scholar]
- Dernburg, A. F., 2001. Here, there, and everywhere: kinetochore function on holocentric chromosomes. J. Cell Biol. 153 F33–F38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y., and D. L. Spector, 2005. Centromere positioning and dynamics in living Arabidopsis plants. Mol. Biol. Cell 16 5710–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama, S., and S. Biggins, 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA 104 14706–14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., T. Kiyomitsu, K. Yoda and M. Yanagida, 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., and Y. Dalal, 2005. Centromeric chromatin: what makes it unique? Curr. Opin. Genet. Dev. 15 177–184. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., K. Ahmad and H. S. Malik, 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293 1098–1102. [DOI] [PubMed] [Google Scholar]
- Henry, I. M., B. P. Dilkes, A. P. Tyagi, H. Y. Lin and L. Comai, 2009. Dosage and parent-of-origin effects shaping aneuploid swarms in A. thaliana. Heredity 103 458–468. [DOI] [PubMed] [Google Scholar]
- Heun, P., S. Erhardt, M. D. Blower, S. Weiss, A. D. Skora et al., 2006. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell 10 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howman, E. V., K. J. Fowler, A. J. Newson, S. Redward, A. C. MacDonald et al., 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 97 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff, M., Y. Hamamura, M. Gourgues, T. Higashiyama and F. Berger, 2007. Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr. Biol. 17 1032–1037. [DOI] [PubMed] [Google Scholar]
- Kalitsis, P., K. J. Fowler, E. Earle, B. Griffiths, E. Howman et al., 2003. Partially functional Cenpa-GFP fusion protein causes increased chromosome missegregation and apoptosis during mouse embryogenesis. Chromosome Res. 11 345–357. [DOI] [PubMed] [Google Scholar]
- Keith, K. C., R. E. Baker, Y. Chen, K. Harris, S. Stoler et al., 1999. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol. Cell Biol. 19 6130–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara, D., S. Matsunaga, S. Uchiyama and K. Fukui, 2008. Live cell imaging reveals plant aurora kinase has dual roles during mitosis. Plant Cell Physiol. 49 1256–1261. [DOI] [PubMed] [Google Scholar]
- Lermontova, I., V. Schubert, J. Fuchs, S. Klatte, J. Macas et al., 2006. Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18 2443–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent and T. J. Richmond, 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389 251–260. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., and S. Henikoff, 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi, P., A. D. McAinsh, E. Rheinbay and P. K. Sorger, 2006. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7 R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey, L., K. Barnes, Y. Chen, M. Fitzgerald-Hayes and R. E. Baker, 2004. The histone fold domain of Cse4 is sufficient for CEN targeting and propagation of active centromeres in budding yeast. Eukaryot. Cell 3 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, M., Y. Ogura and F. Motoyoshi, 1994. Centromeric repetitive sequences in Arabidopsis thaliana. Jpn. J. Genet. 69 361–370. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Z. Cheng, S. Ouyang, P. B. Talbert, M. Kim et al., 2004. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36 138–145. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., K. Terada, M. Wakimoto, K. Kashihara and M. Murata, 2010. Centromere targeting of alien CENH3s in Arabidopsis and tobacco cells. Chromosome Res. 18 203–211. [DOI] [PubMed] [Google Scholar]
- Nasuda, S., S. Hudakova, I. Schubert, A. Houben and T. R. Endo, 2005. Stable barley chromosomes without centromeric repeats. Proc. Natl. Acad. Sci. USA 102 9842–9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi, M., and S. W. Chan, 2010. Haploid plants produced by centromere-mediated genome elimination. Nature 464 615–618. [DOI] [PubMed] [Google Scholar]
- Regnier, V., P. Vagnarelli, T. Fukagawa, T. Zerjal, E. Burns et al., 2005. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 25 3967–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, K. F., M. Hechenberger and K. Masri, 1994. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 127 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P. B., R. Masuelli, A. P. Tyagi, L. Comai and S. Henikoff, 2002. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak, D., H. S. Hayden and S. Henikoff, 2002. Centromere targeting element within the histone fold domain of Cid. Mol. Cell. Biol. 22 7553–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, C. M., E. Giulotto, S. Sigurdsson, M. Zoli, S. Gnerre et al., 2009. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland, G., S. Orthaus, S. Ohndorf, S. Diekmann and P. Hemmerich, 2004. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae. Mol. Cell. Biol. 24 6620–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]