Abstract
Most connective tissue cells (e.g. tendon and ligament cells) attach to extracellular matrix (ECM) and exert so-called cell traction forces (CTFs) to the ECM. CTFs are essential for many cellular functions such as maintenance of cell shape, cell motility, and cell communication. Therefore, many techniques have been developed over the years to measure CTFs in order to better understand tissue physiology and pathology. This article provides a brief review of CTF in terms of its generation and transmission and also CTF measurement techniques, with a focus on cell traction force microscopy (CTFM). Examples of using CTFM to determine CTFs are given to illustrate various applications of CTFM. Finally, the potential applications of CTFM in musculoskeletal research are suggested.
1. Introduction
The human body is constantly exposed to a mechanical environment. The musculoskeletal, respiratory, and cardiovascular systems are well known examples of organs that are subjected to mechanical forces in various forms such as tension, compression, and shear stress [1]. The cells in these systems sense mechanical forces and transduce them into biochemical signals by so-called mechanotransduction mechanisms; as a result, cells alter their ECM gene and protein expression to maintain normal tissue structure and function or cause pathological changes of the tissues. Besides the external forces acting on organs and tissues and eventually on cells, cells also generate their own mechanical forces that act on the extracellular matrix (ECM). The cellular forces are referred to as cell traction forces (CTFs). Cells use traction forces to perform various tasks, including maintaining cell shape, migrating within tissues, reorganizing ECM, and communicating with neighboring cells. As such, CTFs play a critical role in a number of fundamental biological processes such as embryogenesis, angiogenesis, inflammation, wound healing, and metastasis.
The generation of CTFs involves interaction between the actin cytoskeleton and motor protein myosin-II [2], which results in intracellular tension. Both the actin cytoskeleton and myosin are involved in numerous biological activities of a cell [3]. The intracellular tension is transmitted to the ECM through focal adhesions (FAs), which link the actin cytoskeleton to the ECM. In addition to transmitting intracellular tension, FAs act as a mini-force sensors and transducers [4]. As the generation and transmission of CTFs involve the actin cytoskeleton, motor proteins, and FAs, any changes in one or more of the three components due to the action of biological factors (growth factors and cytokines), biochemical agents, and external mechanical forces will lead to changes in CTFs (Fig. 1). On the other hand, CTFs deform the ECM, which in turn modulates cellular function, including DNA synthesis, ECM protein secretion, and cell differentiation [5]. Thus, CTFs are important biophysical markers which can be used to characterize the biological state of a cell, and thus, measurement of CTFs will aid in better understanding of tissue physiology and pathology. Finally, using CTF measurement techniques to discern the extent of traction/contraction forces produced by cells may guide the development of optimal protocols for more effective treatment of tissue wound healing in clinical settings.
Fig. 1.
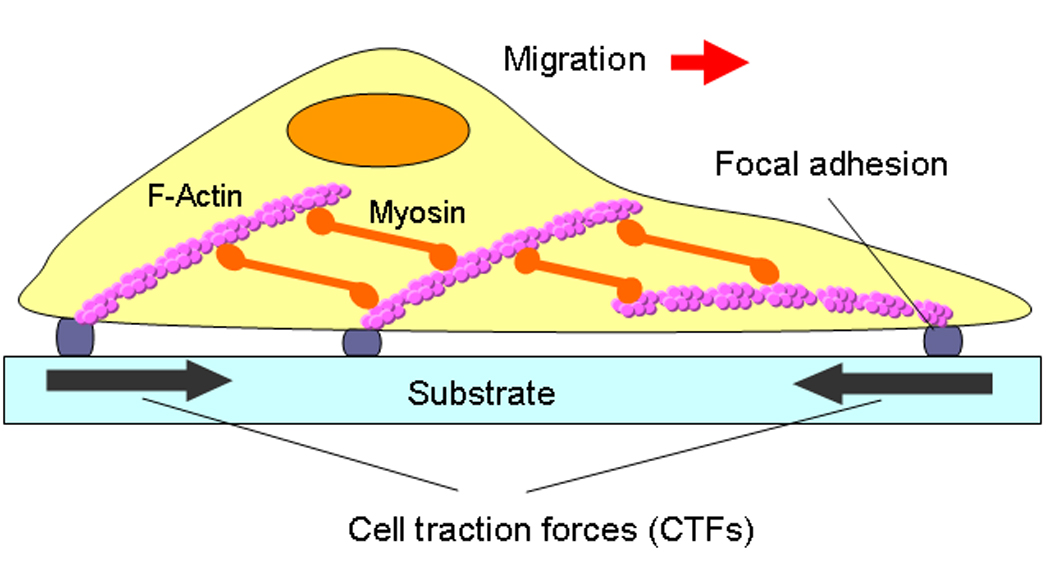
An illustration of CTFs on an adherent cell. As shown, the generation of CTFs involves actin filaments, myosin, and focal adhesions. As a result, any changes of the three components in terms of structure and/or activities will result in changes in CTFs. (modified from Fig. 1 with permission in Wang and Lin 2007, Biomech Model Mechanobiol 6: 361–371).
2. Techniques for CTF measurement
Many techniques have been developed over the years to measure CTFs. One of the earliest approaches was the use of a cell-populated collagen gel (CPCG) [6, 7]. In this approach, cells are suspended in a collagen solution by mixing, and once the collagen solution polymerizes, a gel disk containing cells is obtained. The traction forces generated by cells inside the gel disk contract the gel; consequently, the diameter and area of the gel disk decrease. The CTF is estimated by measuring changes in the diameter or area of the gel disk. Later on, the CPCG method was refined by including a force monitor to quantify the CTF magnitudes [8, 9]. However, one limitation of the CPCG approach is that it only measures the collective traction force of a group of cells, not individual cells.
To overcome this limitation, a thin silicone membrane can be used as a substrate with cells grown on its surface [10]. As the thin membrane is highly compliant, wrinkles are generated by CTFs on the membrane surface. This elegant approach enables one for the first time to “visualize” CTFs, as number of wrinkles and extent of wrinkling reflect the nature of CTFs, such as CTF magnitudes and distribution. One obvious drawback of this approach is that it only provides a qualitative, not quantitative measurement of CTFs. So later, effort was made to quantify CTFs based on wrinkles on silicone membrane [11]. However, because the wrinkling of a thin membrane is a highly nonlinear problem, there are no known mathematical models that can accurately determine a complex CTF field on the membrane.
Toward the goal of measuring CTFs quantitatively, a flexible micropost array has been developed in recent years [12–14]. The micropost array is made of an elastic silicone polymer. When a cell attaches to the microposts, the CTFs bend them. By determining the extent of the micropost bending, CTFs, including magnitude and direction at each post, can be easily calculated using beam theory.
While a silicone polymer has the advantages of non-toxicity, transparency, and high elasticity, the stiffness of this material is too high to sense small deformations produced by small CTFs. To overcome this problem, Pelham and Wang [15] introduced highly compliant polyacrylamide gel (PG) as a substrate for CTF determination using so-called cell traction force microscopy (CTFM). In CTFM, fluorescent microbeads are embedded in the PG and its surface is coated with ECM proteins such as collagen to promote cell attachment. The cells on the PG surface generate CTFs and therefore deform the gel, causing displacement of the beads underlying and surrounding a cell. Two images are obtained by photographing fluorescent beads before and after cells are removed. By comparing the two images using pattern recognition algorithms, the bead displacements can be determined. The obtained displacements can then be converted into CTFs based on elasticity theory [16, 17] (Fig. 2).
Fig. 2.
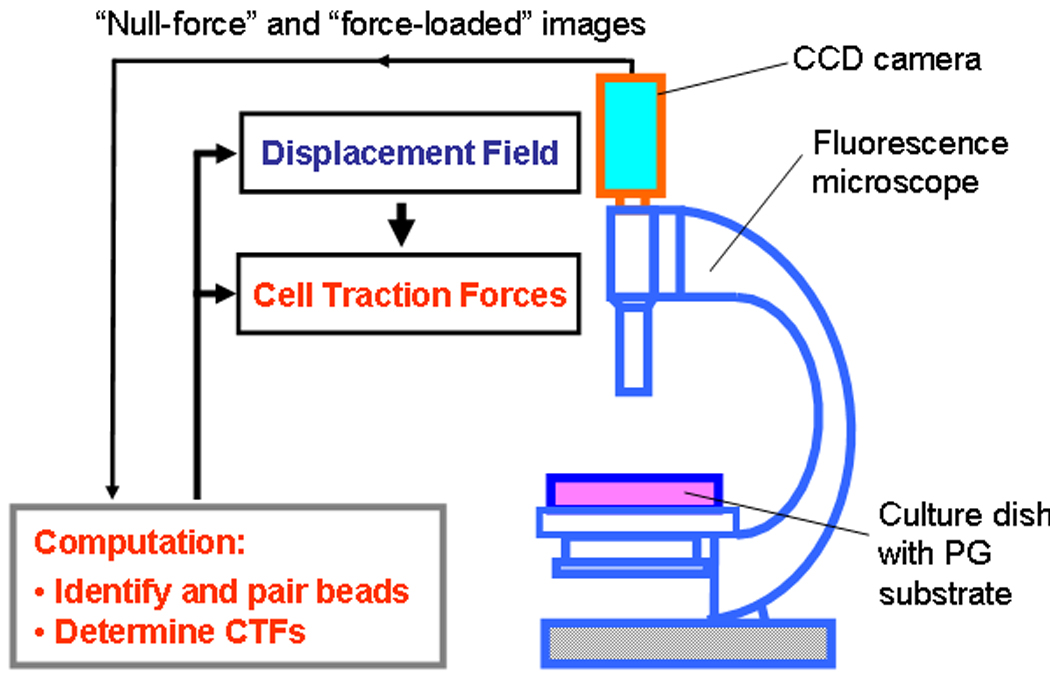
A general CTFM scheme. Two main procedures are involved in the implementation of CTFM: a) obtaining bead displacements through fluorescent microscopy; and b) converting displacements into CTFs using computation algorithms based on elasticity theory. (modified from Fig. 1 with permission in Wang et al. “Cell Traction Force Microscopy for Musculoskeletal Research” in “A Practical Manual for Musculoskeletal Research” edited by Leung, Qin, Chung, and Qin, World Scientific, 2008).
3. CTFM Applications
As demonstrated in the following application examples, CTFM can accurately determine traction forces of individual cells and grouped cells. Because the generation of CTFs involves the actin cytoskeleton, myosin, and focal adhesions, which are related to nearly all cell activities, CTFM may be used to characterize biophysical and biochemical characteristics of cells by determining CTFs.
a. Traction forces generated by human tendon fibroblasts (HPTFs)
In the CTFM experiments, we fabricated PG substrates and conjugated them with collagen type I to promote the attachment of HPTFs. After plating for 6 hrs on the gel substrate, HPTFs exhibited an elongated shape, a typical morphology for these cells. The images of “force-loaded” and “null-force” images, which correspond to before and after cells were removed, respectively, were obtained by micro-photographing and used to determine the substrate displacement field and CTFs based on the published methods [17]. The maximum displacement of the gel substrate was found to be ~1.2 μm, and the maximum traction stress ~250 Pa, with CTFs being concentrated at the front and rear of the fibroblast (Fig. 3).
Fig. 3.
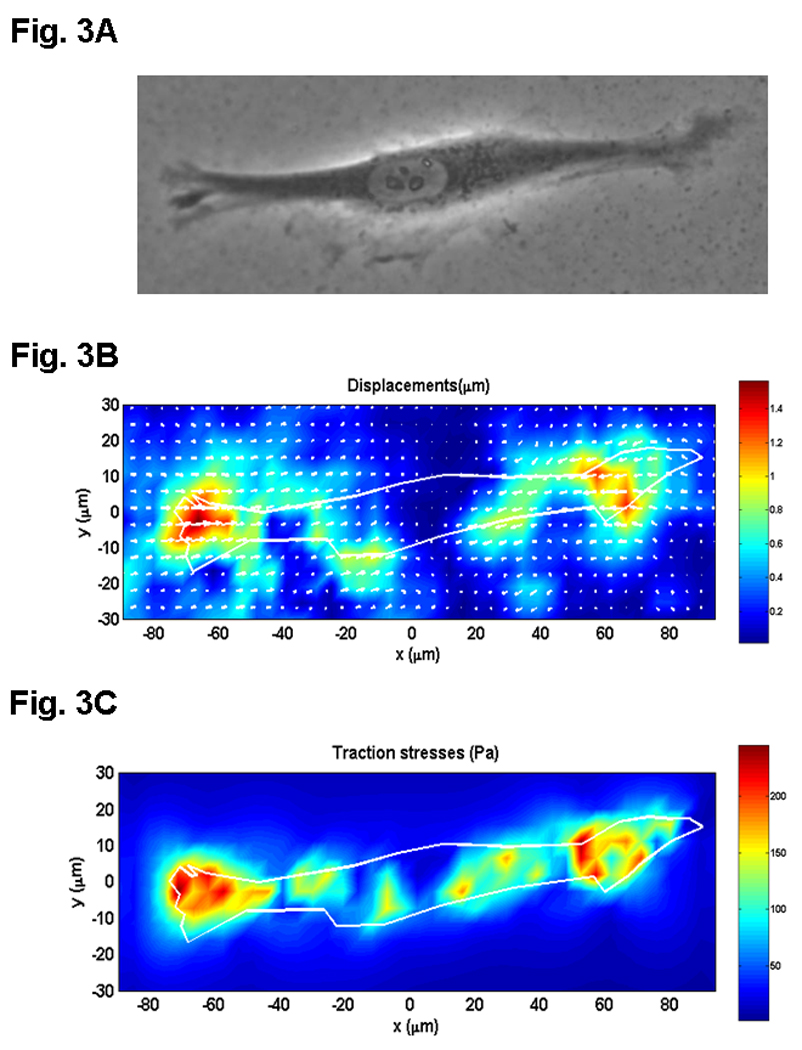
CTFM application to determine traction forces of human patellar tendon fibroblast (HPTF). A. A HPTF on PG substrate embedded with fluorescent microbeads, which are not shown on this phase contrast image. B. PG substrate displacements defined by beads’ movements near the PG surface. C. CTF field. (adopted with permission from Fig. 6 in Yang et al. 2006, J Theor Biol 242:607–616).
b. Traction forces generated by muscle cells
We used a micropatterning technique to fabricate an array of highly elongated, rectangular micro-islands (10 µm × 200 µm) of collagen on PAG dishes. After plating, muscle cells adapted to the rectangular shape during growth and differentiation. Using similar procedures described above, we then determined the displacements and traction forces of a muscle cell (Fig. 4). The results indicate that CTFM may be used as a functional tool to evaluate muscle force without the need to use animals.
Fig. 4.
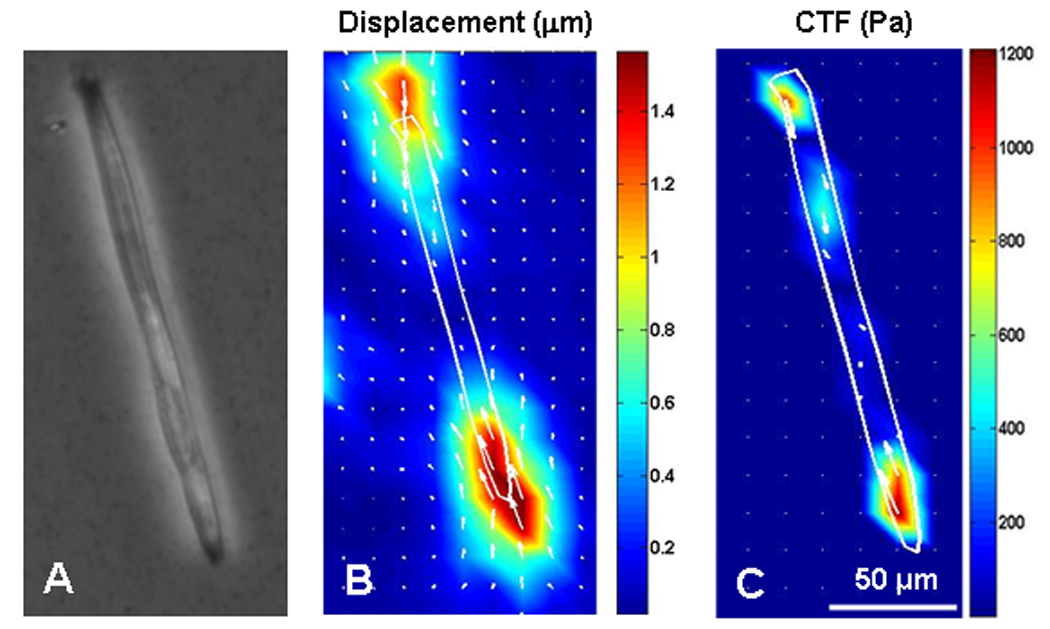
CTFM application to determine traction force of a muscle cell. A. the muscle cell. B. Substrate displacement field. C. CTF field. (adopted with permission from Fig. 4 in Li et al. 2008, J Biomech 41: 3349–3353).
c. Detection of cell differentiation by CTFM
We tested whether CTFM can detect cell differentiation. A well established culture protocol [18] was first used to induce differentiation of fibroblasts into myofibroblasts, which express a high level of α-SMA, a specific marker of myofibroblasts. Then, traction forces of individual fibroblasts and myofibroblasts were determined by CTFM and the resulting CTFs were used to construct a CTF histogram. It was found that the two populations of fibroblasts and myofibroblasts were clearly distinguishable on the CTF histogram (Fig. 5). The results suggest that CTFM may be an effective approach to detect fibroblast differentiation in wound healing.
Fig. 5.
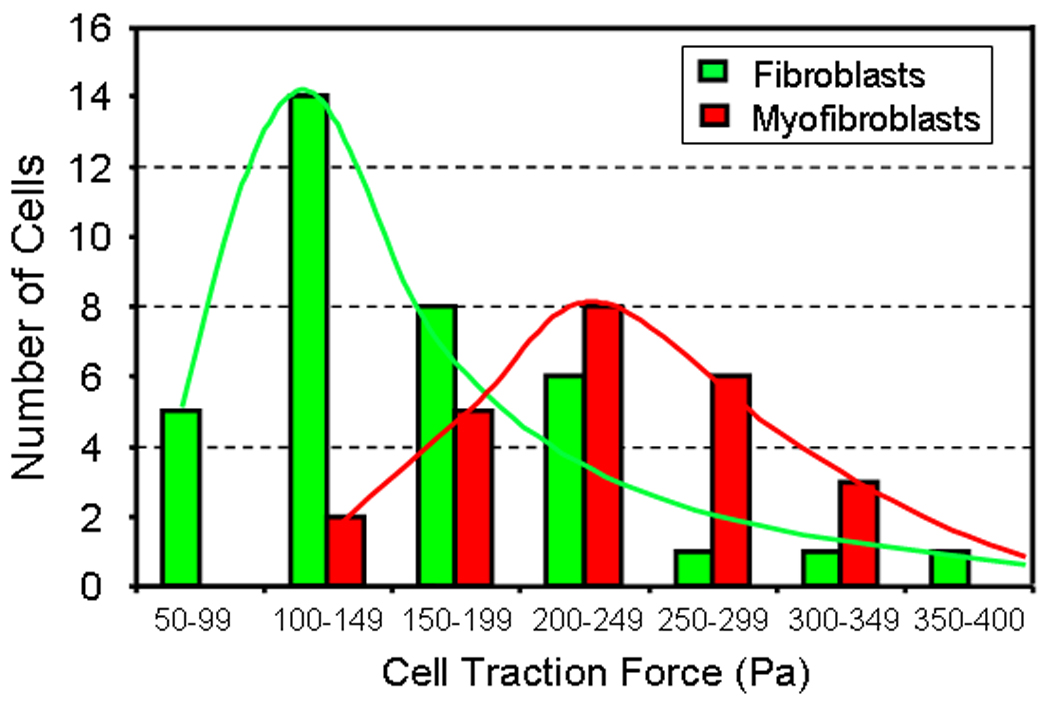
CTFM application to detect cell differentiation. After fibroblasts were differentiated into myofibroblasts, two populations of cells were clearly separated by their large differences in CTF distributions resulting from CTF measurements. (adopted from Fig. 4 with permission in Wang et al. “Cell Traction Force Microscopy for Musculoskeletal Research” in “A Practical Manual for Musculoskeletal Research” edited by Leung, Qin, Chung, and Qin, World Scientific, 2008)
d. Traction forces of a group of cells
In this test, we first fabricated fibronectin-coated, square micro-islands on PG substrates using microcontact printing technology [19]. We then plated cells on the micro-islands. Finally, we applied CTFM to determine the CTFs of the square cell island. It was found that CTFs were not evenly distributed on the cell island; instead, large traction forces were concentrated at the corners and around the edges of the cell island (Fig. 6).
Fig. 6.
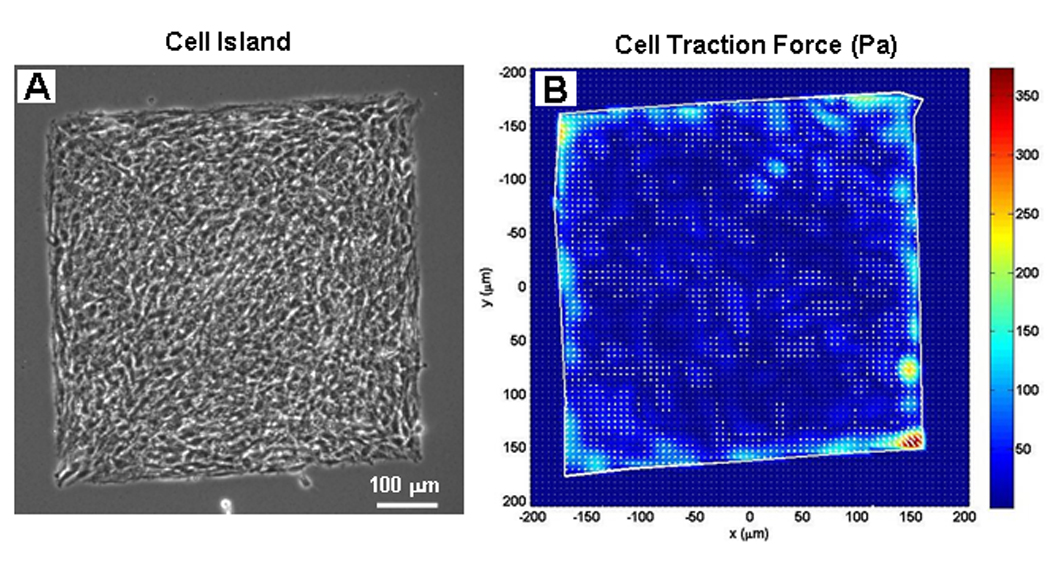
CTFM application on a group of cells. A. A group of cells on a square micro-island. B. CTF distribution of the cell island. (adopted from Fig. 5 with permission in Wang et al. “Cell Traction Force Microscopy for Musculoskeletal Research” in “A Practical Manual for Musculoskeletal Research” edited by Leung, Qin, Chung, and Qin, World Scientific, 2008)
4. Concluding Remarks
The generation of CTFs involves the intracellular components (e.g. actin cytoskeleton) and extracellular matrix components (e.g. focal adhesions). These cellular components are related to nearly all cell activities. As a result, any biological, biochemical, or biomechanical stimuli acting on cells likely cause changes in CTFs. Therefore, determination of CTFs enables one to have a better understanding of tissue physiology and pathology at the cellular level. Among those techniques that have been developed to measure CTFs, CTFM is one of the most widely used that can accurately determine traction/contraction forces of individual cells and groups of cells. This technique has many potential applications in orthopaedic research. For example, fibroblasts differentiate into myofibroblasts during tissue wound healing. CTFM may be used to detect the myofibroblasts by measuring their large contraction forces and, by doing so, help determine when myofibroblasts are present in the wound area and when they disappear after wounds are healed. This information may help devise means of reducing tissue scarring, which is a common orthopaedic problem after tissue healing. Another application of CTFM is evaluation of muscle function (i.e. the ability to generate muscular forces) by measuring contraction forces of a single muscle fiber, therefore eliminating the need to use animals. Finally, as CTFM is an individual cell-based technology, it can be used to characterize differentiation states of individual adult stem cells from bone, tendons, ligaments, etc.
ACKNOWLEDGEMENT
The funding support of NIH grants AR049921 and AR049921-S2 is gratefully acknowledged (JHW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Wang JH, Thampatty BP. An introductory review of cell mechanobiology. Biomech Model Mechanobiol. 2006;5(1):1–16. doi: 10.1007/s10237-005-0012-z. [DOI] [PubMed] [Google Scholar]
- 2.Wang JH, Lin JS. Cell traction force and measurement methods. Biomech Model Mechanobiol. 2007 doi: 10.1007/s10237-006-0068-4. [DOI] [PubMed] [Google Scholar]
- 3.Ingber DE. Mechanosensation through integrins: cells act locally but think globally. Proc Natl Acad Sci U S A. 2003;100(4):1472–1474. doi: 10.1073/pnas.0530201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingber DE. Tensegrity: the architectural basis of cellular mechanotransduction. Annu Rev Physiol. 1997;59:575–599. doi: 10.1146/annurev.physiol.59.1.575. [DOI] [PubMed] [Google Scholar]
- 5.Harris AK, Stopak D, Wild P. Fibroblast traction as a mechanism for collagen morphogenesis. Nature. 1981;290(5803):249–251. doi: 10.1038/290249a0. [DOI] [PubMed] [Google Scholar]
- 6.Bell E, Ivarsson B, Merrill C. Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro. Proc Natl Acad Sci U S A. 1979;76(3):1274–1278. doi: 10.1073/pnas.76.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moon AG, Tranquillo RT. Fibroblast-Populated Collagen Microsphere Assay of Cell Traction Force .1. Continuum Model. Aiche Journal. 1993;39(1):163–177. doi: 10.1115/1.2795998. [DOI] [PubMed] [Google Scholar]
- 8.Delvoye P, Wiliquet P, Leveque JL, Nusgens BV, Lapiere CM. Measurement of mechanical forces generated by skin fibroblasts embedded in a three-dimensional collagen gel. J Invest Dermatol. 1991;97(5):898–902. doi: 10.1111/1523-1747.ep12491651. [DOI] [PubMed] [Google Scholar]
- 9.Campbell BH, Clark WW, Wang JH. A multi-station culture force monitor system to study cellular contractility. J Biomech. 2003;36(1):137–140. doi: 10.1016/s0021-9290(02)00325-1. [DOI] [PubMed] [Google Scholar]
- 10.Harris AK, Wild P, Stopak D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science. 1980;208(4440):177–179. doi: 10.1126/science.6987736. [DOI] [PubMed] [Google Scholar]
- 11.Burton K, Taylor DL. Traction forces of cytokinesis measured with optically modified elastic substrata. Nature. 1997;385(6615):450–454. doi: 10.1038/385450a0. [DOI] [PubMed] [Google Scholar]
- 12.du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Siberzan P, Ladoux B. Force mapping in epithelial cell migration. Proc Natl Acad Sci U S A. 2005;102(7):2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci U S A. 2003;100(4):1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Xie L, Starr ZC, Yang Z, Lin JS, Wang JH. Development of micropost force sensor array with culture experiments for determination of cell traction forces. Cell Motil Cytoskeleton. 2007 doi: 10.1002/cm.20200. [DOI] [PubMed] [Google Scholar]
- 15.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94(25):13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Traction fields, moments, and strain energy that cells exert on their surroundings. Am J Physiol Cell Physiol. 2002;282(3):C595–C605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, Lin JS, Chen J, Wang JH. Determining substrate displacement and cell traction fields--a new approach. J Theor Biol. 2006;242(3):607–616. doi: 10.1016/j.jtbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Anderson S, DiCesare L, Tan I, Leung T, SundarRaj N. Rho-mediated assembly of stress fibers is differentially regulated in corneal fibroblasts and myofibroblasts. Exp Cell Res. 2004;298(2):574–583. doi: 10.1016/j.yexcr.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]


