Abstract
We recently demonstrated that antibiotic administration has a reproducible effect on the community structure of the indigenous gastrointestinal microbiota of mice. In this addendum we report on additional experimentation using the antibiotic vancomycin. In accord with our previous findings, vancomycin administration results in consistent alteration of the microbiota of the cecal contents and the cecal mucosa. These alterations are largely reversed by a three-week period of recovery without antibiotics. In contrast to our previous results using other antibiotics, the alterations in community structure associated with vancomycin occured without a significant decrease in the overall bacterial biomass. These results indicate that different antibiotics have specific effects on the gut microbiota. This points the way towards targeted, therapeutic alteration of the gut bacterial community as a whole.
Key words: microbiota, microbial community, community structure, vancomycin, 16S rRNA
Introduction
The recent start of the Human Microbiome Project by the NIH signals a growing acceptance of the role of complex microbial communities in human health and disease.1 It is important to note however, that the idea that the indigenous microbiota can influence health is not new. The concept of probiotics or beneficial microbes, was put forth over a century ago by Elie Metchnikoff not long after the widespread acceptance of Koch's postulates for disease causation by microbes.2
More recently, the emergence of antibiotic associated diarrhea and colitis due to Clostridium difficile raised general awareness that it was possible for the disease to arise secondary to alterations in the indigenous microbiota.3 Shortly after Koch's postulates were fulfilled for C. difficile and antibiotic associated colitis,4 it was demonstrated that the normal gut microbiota could prevent this disease in an experimental animal model.5 Only recently however have we developed the scientific tools that allow us to profile the community structure of the indigenous microbiota and follow its dynamics.6
Antibiotic Administration Modifies the Gut Microbiota
Culture-independent methods for the profiling of complex microbial communities, such as construction and analysis of 16S rRNA-encoding gene clone libraries, were initially developed by investigators studying environmental consortia of bacteria.7,8 The efforts of the Human Microbiome Project are the direct descendents of these earlier works. We have used these techniques to begin exploration of the underlying role of the indigenous microbiota and antibiotic associated diarrhea both related to and independent of C. difficile infection. We recently reported that patients with recurrent C. difficile infection possessed a microbiota that was characterized by a markedly reduced overall diversity compared to controls and patients with an initial episode of disease.9 In an attempt to understand how long-lasting changes in the community structure of the indigenous gut microbiota could arise we undertook a series of experiments involving antibiotic treatment of mice to follow community dynamics under this ecologic stress.
In an article published recently in Infection and Immunity, we presented the results of our study that temporally followed changes in the structure of the indigenous gut microbiota after antibiotic administration.10 In this study, two separate antibiotic regimens were employed to perturb the gut microbiota. Treatment of mice with a combination of amoxicillin, metronidazole and bismuth resulted in significant changes in the overall abundance and community composition of the gut microbiota. However, following withdrawal of this antibiotic combination, there was a significant recovery of the gut community towards baseline. Conversely, while treatment with the broad-spectrum antibiotic cefoperazone also resulted in significant disturbance to the community, withdrawal of this particular antibiotic resulted in longer-lasting changes in community structure. In particular, animals that were treated with cefoperazone possessed gut microbial communities that were still depressed in terms of overall diversity despite recovery of overall bacterial biomass six weeks after the withdrawal of the antibiotic. Interestingly, introduction of a non-treated animal into the cage housing cefoperazone-treated animals after withdrawal of the antibiotic resulted in normalization of the microbial community, presumably via corprophagy. This latter observation has parallels in clinical medicine in that transplantation of normal human feces has been used for the treatment of recurrent C. difficile infection. A recent publication from another group demonstrated that in a single patient who underwent this treatment, the reduced overall diversity of the gut bacteria community was reversed by treatment with donor stool.11
We are grateful for the reception our work has received from the scientific community at large since publication. Other investigators have had a number of questions regarding our study and we will address one of these more common questions in this communication as well as present some additional data we have generated. The most common question we have received regards the rationale for the selection of the two antibiotic regimens that were reported. Our initial interest in the microbial ecology of the gastrointestinal tract stems from an interest we have in inflammatory bowel disease (IBD). We utilize a murine model of IBD in which gastrointestinal inflammation is triggered in interleukin 10-deficient mice by challenge with the bacterium Helicobacter hepaticus.12,13 We are interested in determining what role the indigenous gut microbiota has in the modulation of IBD triggered by H. hepaticus. The two antibiotic regimens that we chose were selected because of our interest in this murine model of IBD. The combination of amoxicillin, metronidazole and bismuth is available in a commercial formulation with rodent chow and was designed for control of Helicobacter infection in mouse colonies.14,15 Conversely, the antibiotic cefoperazone is used in selective media designed to isolate H. hepaticus from murine fecal pellets and tissue.13,16 Therefore, our experiments were designed as control experiments to determine what effect these two antibiotic regimens would have on the baseline gut microbiota.
Effects of Vancomycin Administration on the Indigenous Gut Microbiota
We have extended our analysis of the effect of antibiotics on the indigenous gut microbiota by using additional regimens with distinct antimicrobial spectra. Vancomycin is a glycopeptide antibiotic with a spectrum of activity restricted to Gram-positive organisms due to its inhibitory activity against peptidoglycan synthesis.17 Wild-type C57BL/6 mice were treated with vancomycin at a concentration of 100 mg per liter in sterile water for 10 days. Two mice were sacrificed at the end of this ten-day treatment with antibiotics while three mice were transferred to untreated sterile water for a period of three weeks prior to sacrifice. Cecal tissue was collected and processed for microbial ecology analysis as in our recent publication. In addition to the cecal tissue, which contained the mucosa-associated microbiota, we also collected luminal contents for analysis. The comparison was made to four untreated littermates. As opposed to the pyrosequencing approach that was utilized in our prior study, standard 16S rRNA-encoding gene clone libraries were constructed for analysis of community structure as published previously.9 A total of 1,576 clones were retrieved from the samples, with a range of 78–95 clones per sample. Although construction of clone libraries does not sample as deeply as pyrosequencing, it is a technique that can reveal significant shifts in community structure.
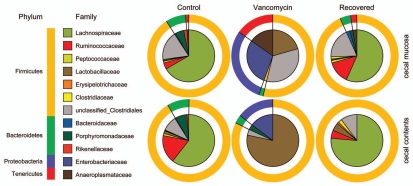
OTU-based analyses with the program mothur revealed that vancomycin reduced the richness of the mucosal and luminal communities (Fig. 1 and Table 1). Furthermore, communities exposed to vancomycin are different in structure when compared to control communities (Fig. 2). Examination of the classifications of the partial SSU gene sequences revealed that the microbial community in control animals was, as expected, dominated by Firmicutes and Bacteroidetes (Fig. 3). Among the Firmicutes there was a dominance of members from the family Lachnospiraceae. At baseline, the community of mucosa-associated organisms was similar to that found in the lumen. Following administration of vancomycin as observed for the other antibiotic regimens, there was a marked shift in the membership and community structure. Members of the Proteobacteria became much more prominent in both the lumen and mucosa (p < 0.05 as determined by Metastats18). Members of the Tenericutes became more prominent in the mucosa (p < 0.05), but were not detected in the luminal contents. In the mucosa, there was an overall decrease in the proportion of the 16S rRNA-encoding gene sequences that were retrieved from members of the Firmicutes (p < 0.05), but this proportion remained similar in the lumen. While the Firmicutes in the lumen and mucosa of control animals were dominated by members of the Lachnospiraceae, this distribution changed significantly following vancomycin administration. In the lumen and mucosa members of the Lactobacillaceae increased (p < 0.05 and p < 0.01, respectively), while members of the Lachnospiraceae decreased (both p < 0.01). These results are completely consistent with the known antibiotic spectrum of vancomycin. As Gram-negative organisms, members of the Proteobacteria would not be expected to be susceptible to vancomycin because of their outer membrane, which impedes the ability of the antibiotic to encounter its target, the peptidoglycan precursor UDP-N-acetlymuramylpentapeptide.17 Likewise, most members of the Tenericutes, including those detected in this study, are likely not susceptible to vancomycin, in this case due to the lack of a cell wall which would be comprised of peptidoglycan.19 Although they are Gram-positive organisms, lactobacilli are often phenotypically resistant to the action of vancomycin.
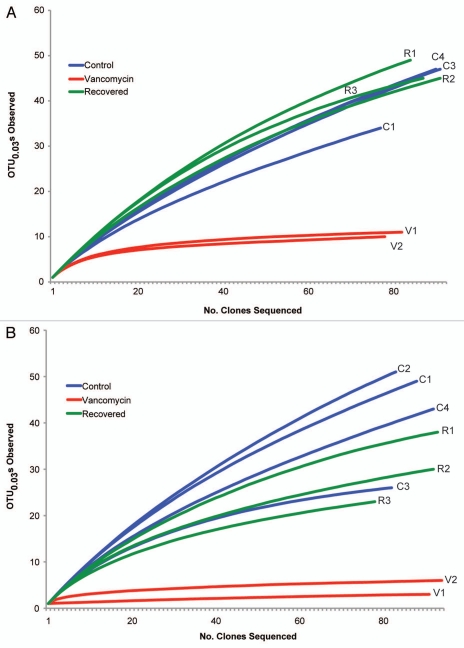
Figure 1.
Reduction of richness in gut microbiota following vancomycin administration. Partial 16S rRNA-encoding gene sequences were obtained from the cecal mucosa (A) and cecal contents (B) from individual mice treated with 10 days of vancomycin (Vancomycin,V), animals that received 10 days of vancomycin followed by a three-week recovery period without antibiotic (Recovered, R) and animals that received no antibiotics (Control, C). Rarefaction curves were generated using mothur20 and OTUs were defined at the 0.03 cutoff (97% similarity).
Table 1.
Changes in the ecologic diversity of the cecal microbiota in vancomycin treated mice
| Observed richness | Chao1 richness estimator (95% c.i.) | Shannon diversity index (95% c.i.) | ||
| Cecal mucosa | Control | 107 | 151 (128–198) | 3.90 (3.75–4.05) |
| Vancomycin | 12 | 15 (12–35) | 1.87 (1.74–1.99)* | |
| Recovered | 83 | 96 (88–117) | 4.04 (3.92–4.15) | |
| Cecal contents | Control | 117 | 168 (142–220) | 4.26 (4.14–4.38) |
| Vancomycin | 6 | 7 (6–20) | 0.74 (0.59–0.88)* | |
| Recovered | 61 | 86 (70–129) | 3.48 (3.34–3.62) |
significantly different when compared to control and recovered groups, p < 0.01; unpaired t-test.
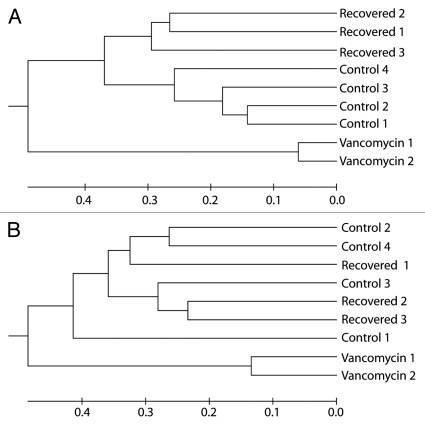
Figure 2.
Vancomycin alters community structure and recovery results in a return to near-baseline structure. UPGMA clustering of communities from the cecal mucosa (A) and cecal contents (B) from individual mice treated vancomycin (Vancomycin), vancomycin followed by a three-week recovery period without antibiotic (Recovered) and animals that received no antibiotics (Control). Dendrogram was generated using distances reflecting community structure dissimilarity (1-similarity). Community structure similarity was calculated in mothur20 using the Yue and Clayton (θ) index at OTU0.03.
Figure 3.
Distribution of phylogentic groups shift following vancomycin administration. Classification of partial 16S rRNA-encoding gene sequences obtained from the cecal contents and cecal mucosa from mice treated with vancomycin (vancomycin) and vancomycin followed by a three-week recovery period without antibiotic (recovered). Comparisons were made to the communities detected in untreated mice (control). Sequences were analyzed and classified to the level of phylum (outer ring) and family (inner pie chart) using the RDP Classifier.21
Similar to what was noted in our previous study of amoxicillin, metronidazole and bismuth treatment,10 following a three-week period without antibiotics, the community richness and structure of the cecal microbiota in vancomycin-treated animals returned to near baseline status (Figs. 1–3). The Firmicutes were once again the dominant phylum encountered and amongst these the Lachnospiraceae were the dominant family. Although the Bacteroidetes did not recover in the lumen to levels that could be detected, despite returning to the mucosa, the difference between the abundances of luminal populations in control and recovered groups was not statistically significant (p = 0.43). This is a situation where pyrosequencing of the communities, as was done in the previous study, may have resulted in a more complete understanding of recovery dynamics in this system. When overall diversity between the three groups of animals was compared using an OTU-based approach (Table 1), it is clear that the vancomycin treatment reduced overall diversity, but the three week period of recovery without antibiotics resulted in an increase of diversity that was similar to that of the gut microbiota in the untreated animals.
One significant difference between treament with vancomycin compared to the previous two antibiotic regimens, is that although vancomycin treatment resulted in large shifts in community composition there was not a major decrease in bacterial biomass. As in the previous study, we used a quantitative PCR assay targeting the 16S rRNA-encoding gene to determine relative changes in bacterial biomass. We have previously noted that treatment with the antibiotic combination of amoxicillin, metronidazole and bismuth reduced overall bacterial load in the cecal mucosa by approximately 2.5 orders of magnitude and cefoperazone treatment resulted in a three orders of magnitude decrease. Conversely, vancomycin treatment did not significantly change overall bacterial load in the mucosa (fold change mucosa = 0.318 ± 0.267, p = 0.24; Kruskal-Wallis).
Conclusions
In order to understand the role that the indigenous microbiota can play in health and disease within the gastrointestinal tract, it is important to be able to understand the dynamics of this community. Understanding how the community reacts to a given ecologic stressor will give us insight into how the microbiota assembles and maintains community structure. Our work with various antibiotic treatments and the effect that they have on the gut microbiota has demonstrated that while these effects are reproducible for a given treatment, the specific effects will vary. Presumably, this could lead to different changes in the function of the gut microbial community. Now that we have a better understanding of the specific changes that various antibiotic treatments have on the gut microbes, we are currently conducting experiments that determine if these alterations and community structure modify the inflammatory response in murine models of inflammatory bowel diseases. It is our hope that the insight from these studies will lead the way to novel methods for treatment of IBD.
Acknowledgements
We thank Dr. Dion Antonopoulos for his work on the original manuscript and continued insight. We thank Judith Opp for technical assistance. This work was funded in part by NIH DK070875 and DK070875-03S1 (Vincent B. Young).
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/12614
Financial Disclosure
This work was supported by NIH grants R01DK070875 (Vincent B. Young) and R01DK070875-03S1 (Vincent B. Young and Courtney J. Robinson).
References
- 1.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaufmann SH. Elie Metchnikoff's and Paul Ehrlich's impact on infection biology. Microbes Infect. 2008;10:1417–1419. doi: 10.1016/j.micinf.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Wilson KH. The microecology of Clostridium difficile. Clin Infect Dis. 1993;16:214–218. doi: 10.1093/clinids/16.supplement_4.s214. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett JG, Onderdonk AB, Cisneros RL, Kasper DL. Clindamycin-associated colitis due to a toxin-producing species of Clostridium in hamsters. J Infect Dis. 1977;136:701–705. doi: 10.1093/infdis/136.5.701. [DOI] [PubMed] [Google Scholar]
- 5.Wilson KH, Silva J, Fekety FR. Suppression of Clostridium difficile by normal hamster cecal flora and prevention of antibiotic-associated cecitis. Infect Immun. 1981;34:626–628. doi: 10.1128/iai.34.2.626-628.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tringe SG, Hugenholtz P. A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol. 2008;11:442–446. doi: 10.1016/j.mib.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 8.Pace NR, Stahl DA, Lane DJ, Olsen GJ. Analyzing natural microbial populations by rRNA sequences. ASM News. 1985;51:4–12. [Google Scholar]
- 9.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 10.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2009 doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 12.Cahill RJ, Foltz CJ, Fox JG, Dangler CA, Powrie F, Schauer DB. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun. 1997;65:3126–3131. doi: 10.1128/iai.65.8.3126-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young VB, Knox KA, Pratt JS, Cortez JS, Mansfield LS, Rogers AB, et al. In vitro and in vivo characterization of Helicobacter hepaticus cytolethal distending toxin mutants. Infect Immun. 2004;72:2521–2527. doi: 10.1128/IAI.72.5.2521-2527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foltz CJ, Fox JG, Yan L, Shames B. Evaluation of various oral antimicrobial formulations for eradication of Helicobacter hepaticus. Lab Anim Sci. 1996;46:193–197. [PubMed] [Google Scholar]
- 15.Kerton A, Warden P. Review of successful treatment for Helicobacter species in laboratory mice. Lab Anim. 2006;40:115–122. doi: 10.1258/002367706776319033. [DOI] [PubMed] [Google Scholar]
- 16.Shames B, Fox JG, Dewhirst F, Yan L, Shen Z, Taylor NS. Identification of widespread Helicobacter hepaticus infection in feces in commercial mouse colonies by culture and PCR assay. J Clin Microbiol. 1995;33:2968–2972. doi: 10.1128/jcm.33.11.2968-2972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barna J, Williams D. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu Rev Microbiol. 1984;38:339–357. doi: 10.1146/annurev.mi.38.100184.002011. [DOI] [PubMed] [Google Scholar]
- 18.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5:1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trachtenberg S. Mollicutes—Wall-less bacteria with internal cytoskeletons. J Struct Biol. 1998;124:244–256. doi: 10.1006/jsbi.1998.4063. [DOI] [PubMed] [Google Scholar]
- 20.Schloss P, Westcott S, Ryabin T, Hall J, Hartmann M, Hollister E, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and environmental microbiology. 2009;75:7537. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]