Abstract
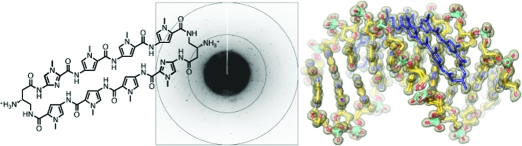
Pyrrole-imidazole polyamides are a class of small molecules that can be programmed to bind a broad repertoire of DNA sequences, disrupt transcription factor−DNA interfaces, and modulate gene expression pathways in cell culture experiments. In this paper we describe a high-resolution X-ray crystal structure of a β-amino turn-linked eight-ring cyclic Py-Im polyamide bound to the central six base pairs of the sequence d(5′-CCAGTACTGG-3′)2, revealing significant modulation of DNA shape. We compare the DNA structural perturbations induced by DNA-binding transcripton factors, androgen receptor and glucocorticoid receptor, in the major groove to those induced by cyclic polyamide binding in the minor groove. The cyclic polyamide is an allosteric modulator that perturbs the DNA structure in such a way that nuclear receptor protein binding is no longer compatible. This allosteric perturbation of the DNA helix provides a molecular basis for disruption of transcription factor−DNA interfaces by small molecules, a minimum step in chemical control of gene networks.
Introduction
Biological systems utilize allosteric modulation for integrating and responding to multiple signals.1,2 The use of allosteric modulation to bias highly dynamic protein ensembles toward conformational states favoring DNA binding provides a powerful regulatory mechanism for modulating gene activation and repression.(3) The nuclear hormone class of ligand-activated transcription factors regulates the expression of genes involved in diverse physiological processes ranging from embryonic development to adult homeostasis.4−8 Additionally, this class of transcription factors is involved in inflammatory disease and the etiology of certain cancers.5,9
Two important examples of ligand-activated nuclear transcription factors are the androgen receptor (AR) and the glucocorticoid receptor (GR).7,8 Both are structurally similar, with a high degree of conservation in their DNA-binding domains, and belong to a subset of DNA-binding receptors that includes the progesterone and mineralocorticoid receptors. This receptor subfamily contains a highly conserved three-domain architecture consisting of an N-terminal domain (NTD), a DNA-binding domain (DBD), and a C-terminal ligand-binding domain (LBD). Although pharmaceutical intervention has been targeted at the LBD, less effort has been directed toward the protein−protein or protein−DNA interface.4,5 AR and GR small-molecule modulators directed specifically to the protein−protein or protein−DNA interface would provide useful tools for understanding gene regulatory pathways and may offer alternative approaches to modulating transcription factor activity.(9) The oversupply of transcription factors can lead to dysregulated gene expression, a characteristic of many human cancers. Cell-permeable small molecules that could modulate transcription factor−DNA interfaces would allow for the chemical control of gene networks.
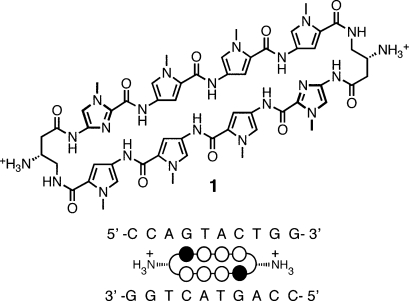
Pyrrole-imidazole (Py-Im) polyamides bind the minor groove of DNA sequence-specifically,10,11 encoded by side-by-side arrangements of N-methylpyrrole (Py) and N-methylimidazole (Im) carboxamide monomers. Im-Py pairs distinguish G·C from C·G base pairs, whereas Py-Py pairs are degenerate for T·A and A·T.10−12 Antiparallel Py-Im strands are connected by γ-aminobutyric acid (GABA) linkers to create hairpin- or cyclic-shaped oligomers. Stereocontrol of polyamide binding orientation has been achieved by the introduction of an (R)-α-amino substituent on the GABA turn, leading to increases in affinity and sequence specificity.11,13 Py-Im polyamides have been programmed to bind a broad repertoire of different DNA sequences.(14) They have been shown to permeate cell membranes,(15) access chromatin,(16) and disrupt protein−DNA interactions of medically relevant transcription factors such as HIF, AR, and GR.11,17 We have recently demonstrated that hairpin Py-Im polyamides bearing the (R)-β-amino-γ-turn, and cyclic polyamides bearing two (R)-β-amino-γ-turns, such as cycle 1 (Figure 1), possess favorable DNA-binding affinities and are useful in gene regulation studies.15g,17d−17g,18a,19
Figure 1.
Structure of cyclic polyamide 1 presented with its ball-and-stick model superimposed over the binding site on the dsDNA oligonucleotide sequence used for crystallization. Closed circles designate N-methylimidazole, open circles designate N-methylpyrrole, and half open circles substituted with ammoniums designate the β-amino-substituted γ-turn unit.
Previous studies have shown that Py-Im polyamides are able to inhibit several distinct structural classes of transcription factor−DNA complexes in vitro using electrophoretic mobility gel shift assays.11,17 Additionally, Py-Im polyamides targeted to the DNA response elements of the AR− and GR−DNA interfaces have resulted in modulation of gene expression in cell culture. Chromatin immunoprecipitation data support an inhibition model based on decreased transcription factor promoter occupancy in the presence of the match polyamide.17d,17g A structural basis for optimizing polyamide structure with regard to disruption of protein−DNA interfaces is crucial for progress in the field. Valuable insights into Py-Im polyamide binding site location and orientation have been provided by previous NMR and X-ray structural studies of polyamide−DNA complexes.,12f,12g,20 Antiparallel four-ring polyamide dimer ImPyPyPyβDp, bound to the sequence 5′-CCAGTACTGG-3′, appeared to widen the minor groove by 2 Å.(12g) Additionally, X-ray structures of hairpin polyamides bound to the nucleosome core particle (NCP) have revealed increases in minor groove widths of ∼2 Å over unliganded NCP DNA and, in addition, long-range structural changes in the NCP DNA upon polyamide binding.(16) Given the modest resolution of previous structures, there is a need for higher resolution crystallographic studies to elucidate DNA structural distortions of polyamide−DNA complexes, in particular, the influence of the turn elements. Recently, we reported the atomic resolution structure (1.18 Å resolution) of an eight-ring cyclic polyamide capped at both ends by (R)-α-amino-γ-turn units in complex with double-helical DNA.(21) The high-resolution structure of the polyamide complexed to the DNA sequence 5′-CCAGGCCTGG-3′ revealed ∼4 Å widening of the minor groove and ∼4 Å compression of the major groove along with >18° bend in the helix axis toward the major groove when compared to unliganded DNA.(21) The large perturbations to the DNA groove width suggest a molecular basis for the disruption of transcription factor−DNA interfaces by small molecules via allosteric modulation.(21) Additionally, a structural basis for the A·T base-pair sequence specificity of the α-amino-γ-turn recognition unit was elucidated, revealing a combination of several interactions. These interactions included hydrophobic packing of the β-methylene with the C2 hydrogen of adenine, specific hydrogen-bonding of the connecting amides to the groove floor before and after the turn, water-mediated hydrogen-bonding of the α-amine to the groove floor, and a conformational pucker that minimizes steric interactions of the α-amino substituent.(21) From this structure one can clearly see that the exocyclic amine of a guanine base under the turn would result in a large steric interaction with the β-methylene of the α-amino-γ-turn.(21) In light of the insight gained into polyamide turn recognition from this previous study, important structural questions remain unresolved regarding the β-amino-γ-turn, especially since these polyamides tend to be higher in affinity for certain sequences and have biological activity. Structures and models are needed for allosteric inhibition of specific transcription factor−DNA complexes, such as the AR and GR bound to their cognate DNA response elements (5′-WGWWCW-3′, with W = A or T), using molecules that have proven effective in biochemical, biophysical, and cell culture experiments.
Here we report a high-resolution X-ray structure of a cyclic polyamide 1, comprised of two antiparallel ImPyPyPy strands capped by (R)-β-amino-γ turn units, which codes for the sequence 5′-WGWWCW-3′, in complex with the 10 base-pair DNA oligonucleotide sequence 5′-CCAGTACTGG-3′, containing an ARE/GRE consensus DNA sequence (Figure 1). We observe significant structural allosteric perturbations of the DNA helix induced upon polyamide binding in the minor groove. A host of noncovalent interactions with the DNA minor groove floor and a detailed view of the β-amino-γ-turn conformation are present in the structure. The unique opportunity to observe the hydration state at this resolution reveals a network of well-ordered water molecules around the polyamide and DNA. Importantly, we are able to compare the DNA conformation of the small-molecule complex with the transcription factor AR− and GR−DNA complexes, demonstrating that structural alterations of DNA by these major-groove-binding proteins and minor-groove-binding cyclic polyamides operate in opposite directions.
Results and Discussion
Overall Structure
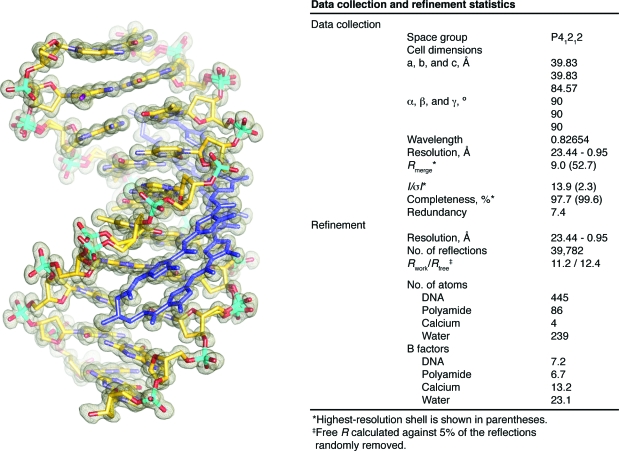
The structure of cyclic polyamide 1 in complex with d(5′-CCIAGTACTGG)2 was solved by direct methods to 0.95 Å resolution with synchrotron radiation (Figure 2).(22) One cyclic polyamide bound to a single DNA duplex is present in the asymmetric unit of the crystal in the P41212 space group. The final structure was refined anisotropically and unrestrained to Rwork = 11.2 and Rfree = 12.4 (Figure 2). The average B factors were 6.7 and 7.2 for the polyamide and DNA, respectively. The asymmetric unit contains one full polyamide-complexed DNA double-helix. In the DNA complex, the aromatic amino acids are bound with an N-to-C orientation of each ImPyPyPy strand of the cycle adjacent to the 5′-to-3′ direction of the DNA. The conformational constraints imposed by the turn unit result in ring placement that is ring-over-ring as opposed to ring-over-amide as previously seen in unlinked 2:1 binders.12f,12g,21 The substituted GABA turn appears to reinforce an antiparallel strand alignment that prevents slippage of the amide-linked heterocyclic strands, allowing less DNA-induced polyamide strand alignment. Greater than 40% of the cyclic polyamide surface area is buried, leaving only the top of the methyl groups on the heterocycles, the amide carbonyl oxygens, and the chiral β-ammonium turn solvent-exposed. Additionally, alternate phosphate conformations are observed for 7 of the 18 nucleotides of the DNA duplex, while the sugar pucker at each nucleotide remains conformationally locked.
Figure 2.
(Left) Electron density map contoured at the 1.0σ level for the X-ray crystal structure of cyclic polyamide 1 complexed to dsDNA (0.95 Å resolution). (Right) Data collection and refinement statistics.
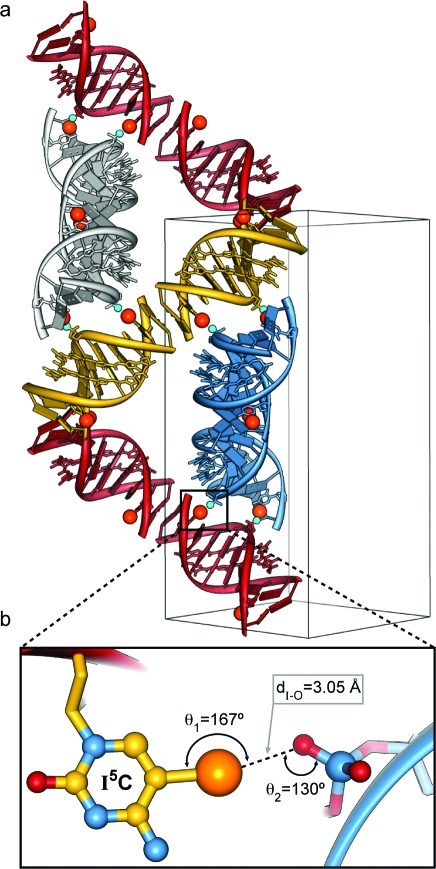
The incorporation of 5-iodocytosine in the oligonucleotide leads to a unique packing geometry in the P41212 space group where each DNA helix is stacked end-to-end with an adjacent helix, forming a pseudocontinuous 20 base-pair dimeric column of polyamide-bound duplexes, effectively desymmetrizing each end of the polyamide−DNA complex (Figure 3a). Each DNA duplex contains two 5-iodocytosine nucleobases that appear to form bridging halogen bond interactions to the phosphates of two adjacent DNA duplexes. Halogen bonding interactions have been ubiquitous in liquid crystal design; however, they are less often observed in biomolecules.(23) Recent studies from the Ho laboratory have suggested that this interaction can be used for directing macromolecular conformation.(23c) The iodine−phosphate distances are less than the sum of the van der Waals radii for I−O (3.50 Å). The first I−O halogen interaction distance is 3.05 Å, with a C−I−O angle (θ1) of 167° and a P−O−I angle (θ2) of 130° (Figure 3b). The second halogen bond interaction distance is 2.97 Å, with a C−I−O angle (θ1) of 168° and a P−O−I angle (θ2) of 137°. The electrostatic potential surface for 5-iodocytosine shown in Figure S1 (Supporting Information) reveals an electropositive crown along the C−I axis associated with the highly polarizable iodine atom, consistent with previous studies of halogen bonding interactions.(23)
Figure 3.
Outline of unit cell and 10 symmetry-related copies of the polyamide−DNA complex showing the iodine−phosphate halogen bond crystal contacts. (a) Unit cell showing the orientation and end-to-end desymmetrization of adjacent helices along with positions of halogen bonds. Halogen bonds occur between the iodine atom of 5-iodocytosine (CI) residues and the oxygen atom of phosphate groups on neighboring helices. (b) Expanded view of a CI−phosphate halogen bond interaction with an I−O distance dI−O = 3.05 Å. The sum of the I−O van der Waals radii is 3.50 Å. Iodine atoms are shown in orange. Halogen-bonded phosphate oxygen atoms are colored cyan in the top figure for clarity and red in the bottom figure.
Polyamide Conformation and Noncovalent Interactions
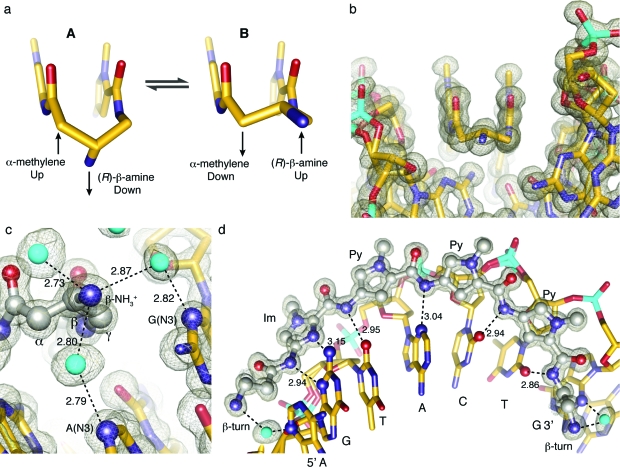
Hairpin Py-Im polyamides linked by a GABA turn substituted at the α or β position can adopt either of two possible conformations upon binding DNA. Our previous crystal strucuture of an (R)-α-amino-substituted GABA-linked cyclic polyamide shows a conformational preference where the amino group is directed up and out of the minor groove, forcing the β-methylene to the floor of the minor groove, within van der Waals contact distance of the C2 hydrogen of adenine.(21) This result left the possibility of an intrinsic preference for the alternate conformation in the absence of substitution at the α-position of the turn, allowing relief of the β-methylene interaction with the floor of the minor groove. However, in the presence of the α-amino turn, a steric clash with the minor-groove wall appears to be the dominant interaction directing turn conformation.(21) In the β-amino GABA-linked polyamide−DNA complex we observe a conformational inversion where the β position is now directed up and out of the minor-groove floor, relieving interaction with the groove floor, orienting the amino substituent along the minor groove (Figure 4a−c). Figure 4b presents a view of the complex looking down the minor groove directly at the β-amino-γ-turn, showing van der Waals interactions between the outside face of the pyrrole-imidazole strands and the walls of the minor groove.
Figure 4.
Molecular recognition details from the X-ray structure of cyclic polyamide 1. (a) Conformation of the β-amino-substituted GABA turn linkage. Conformation A (left) is the conformation observed in the previously determined α-amino-γ-turn X-ray crystal structure. Conformation B (right) shows the preferred conformation for the β-amino-γ-turn determined by X-ray crystallography in this report. The β-carbon conformational preference is puckered up and away from the DNA minor-groove floor, aligning the β-ammonium substituent along the groove floor. (b) Structural view looking down the DNA minor groove, showing the bound cyclic polyamide with electron density contoured at the 1.0σ level. (c) Geometry of the β-amino-γ-turn interacting with the adenine and guanine base pairs in the floor of the DNA minor groove through water-mediated hydrogen bonds. (d) Isolated view of one half of the polyamide (split along a plane through the long axis of the polyamide and the DNA helical axis), showing hydrogen bond distances to the DNA minor groove floor. Hydrogen-bonding interactions of the DNA−polyamide complex with electron density contoured at the 1.0σ level. Im = imidazole and Py = pyrrole.
The hydration pattern around the turn is conserved at both ends of the structure, and there are two water-mediated hydrogen bonds within 2.79−2.87 Å from the ammonium to the DNA minor-groove floor (Figure 4). The amide NH’s and imidazole lone-pairs form a continuous series of direct hydrogen bonds to the floor of the DNA minor groove. The imidazoles impart specificity for the exocyclic amine of guanine through relief of steric interaction and a G( N2-hydrogen)−Im (lone pair) hydrogen bond. The amides linking the aromatic rings and the turns contribute hydrogen bonds to the purine N3 and pyrimidine O2 lone pairs. All amides are within hydrogen-bonding distance of a DNA base (∼3.0 Å average, Figure 4). In all, there are 10 direct amide hydrogen bonds (average distance = 2.7−2.9 Å), two direct imidazole hydrogen bonds (average distance = 3.15 Å), and four (R)-β-ammonium turn water-mediated hydrogen bonds (two per turn, average distance from amine to water 2.79−2.87 Å) to the floor of the DNA minor groove. There is at least one interaction for all 12 DNA bases in the six base-pair binding site, for a total of 16 hydrogen bond interactions between the cyclic polyamide and the floor of the DNA minor groove.
Polyamide and DNA Solvation
The structure has a unit cell volume of 134 162 Å3 and a Matthews coefficient of 2.24, with a solvent content of 51%. There are 130 water molecules within 3.0 Å of the polyamide−DNA complex, with 76 of the 130 water molecules localized around the DNA phosphate backbone (Figure S2, Supporting Information). The solvent-exposed surface of the polyamide is hydrated by 22 of the 130 waters found within 3.0 Å of the complex. Most water molecules are clustered and form hydrogen-bonded networks across the carbonyl oxygens of adjacent amides linking polyamide ring pairs. Additionally, six water molecules hydrate the polyamide ammonium turns (three at each turn), with four of the six anchoring the polyamide to the floor of the DNA minor groove through bridging hydrogen bonds to the base-pair edges (Figure 4c). The major groove is also highly hydrated and contains a well-ordered seven-coordinate calcium ion in addition to other calcium ions around the duplex periphery (Figure S2).
The pattern of hydration around the β-amino-γ-turn in this structure is distinctly different from the α-amino-γ-turn in our previous structure. The β-amino-γ-turn is hydrated by three water molecules, just as in the α-amino-γ-turn where one of the water molecules forms a bridging hydrogen bond to the adenine under the polyamide turn. However, the β-amino-γ-turn contains an extra bridging hydrogen bond from one of the water molecules to the guanine of the next base pair. This observation points to the possibility of engineering turn specificity through structural modification beyond the formal 6 base-pair binding site to what would now be an 8 base-pair binding site.
Cyclic Polyamide-Induced DNA Structural Perturbations
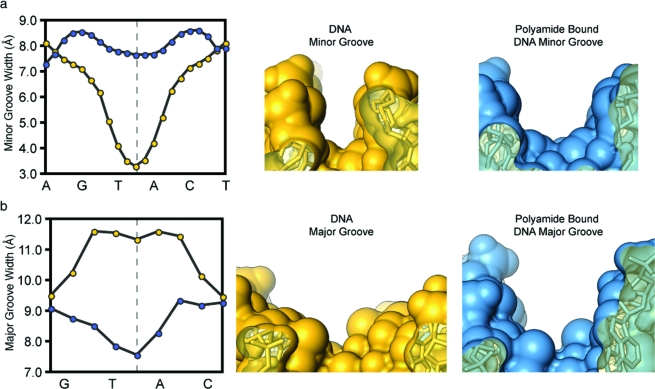
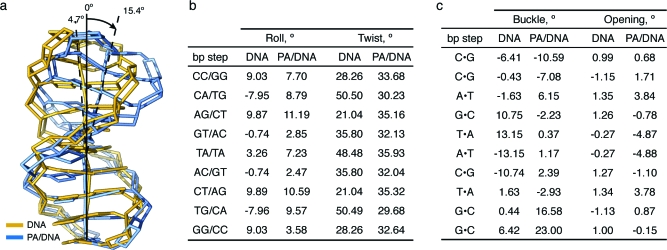
A slice through the short axis of the DNA helix, showing the minor- and major-groove geometry at the center of the polyamide binding site for uncomplexed and complexed DNA in Figure 5, reveals a >4 Å widening of the DNA minor groove upon cyclic polyamide binding and a compression of the major groove by more than 4 Å as compared to unliganded DNA.(24) Additionally, Figure 5 shows a large perturbation in the major-groove depth upon polyamide binding, converting the wide, shallow surface of the major groove from a functionally exposed protein recognition domain to a narrow, deep cleft too small to accommodate the width of a standard protein α-helical domain or β-sheet from a transcription factor. Figure 6 shows the polyamide-induced bending of the DNA helix. The helix is bent toward the major groove by >15°, resulting in major groove compression. The base-pair step parameters in Figure 6a show a large positive roll throughout the polyamide binding site, which contributes to the significant bend in the DNA helix. Additionally, polyamide binding induces a more uniform helical twist, resulting in less variability as the base-pair step changes. The helical twist values for polyamide-bound DNA range from 29 to 36°. Values for the helical twist are highly sequence dependent in native DNA and range from 21 to 50°, depending on step sequence. Major perturbations in the DNA base-pair buckle and opening are also observed upon polyamide binding (Figure 6c). At the four central base pairs of the binding site, the buckle is significantly reduced upon binding and the base pairs are opened toward the DNA major groove, with the largest variations at the central A·T base pairs. A full set of helical parameters and coordinate system definitions can be found in Figures S3−S5 (Supporting Information). The possibility that the DNA structural alterations we observe are due to the packing forces and crystal contact perturbations cannot be ruled out; however, we observe similar groove distortions and DNA bending perturbations in different sequences of DNA (5′-CCAGGCCTGG-3′) with different cyclic polyamide context (ImImPyPy) and in different space groups (P1) of DNA−polyamide complexes.(21)
Figure 5.
DNA minor- and major-groove dimensions in the absence and in the presence of polyamide 1. Native DNA structure d(5′-CCAGTACTGG-3′)2 solved by Rees and co-workers (PDB 1D8G, 0.74 Å resolution). (a) Comparison of the minor-groove width for DNA in the absence of polyamide (yellow curve and structure) and in the presence of bound polyamide (blue curve and structure). (b) Comparison of the major-groove width for DNA in the absence of polyamide (yellow curve and structure) and in the presence of bound polyamide (blue curve and structure). Polyamide has been removed from the blue complex for clarity.
Figure 6.
Allosteric distortion upon polyamide binding. (a) Significant DNA bending is observed for polyamide-bound DNA (blue structure) versus unbound DNA (yellow structure). Polyamide has been removed from the blue complex for clarity. (b) Roll and twist parameters for DNA in the absence and presence of polyamide. (c) Buckle and opening parameters for DNA in the absence and in the presence of polyamide.
Allosteric Inhibition of AR and GR Major-Groove Binding
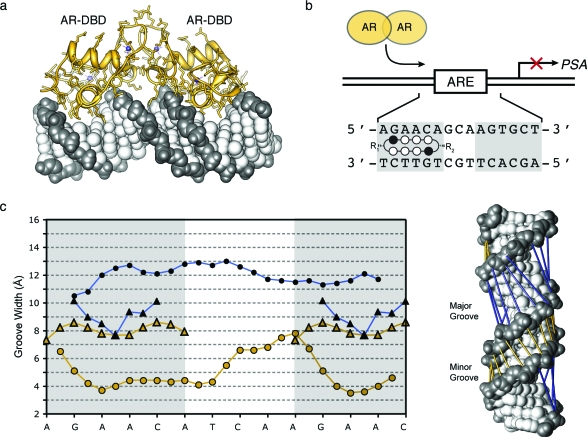
To better understand the impact of polyamide-induced DNA structural modulation on the inhibition of major-groove-binding transcription factors, we analyzed previously reported crystal structures for the androgen and glucocorticoid receptors (AR and GR) in complex with dsDNA.25,26 For the case of the androgen receptor−DNA complex, only one structure was available with a resolution limit of 3.10 Å (PDB 1R4I).(25) The comparison revealed a substantial difference in minor-groove widths for AR-bound DNA (∼4.0 Å) versus polyamide-bound DNA (∼8.0 Å). Comparison of the major-groove widths also revealed large deviations, with the AR−DNA major groove expanded to >12.0 Å and the polyamide-bound DNA major groove compressed to <9.0 Å (Figure 7).
Figure 7.
Comparison of polyamide-bound DNA to androgen receptor-bound DNA. (a) AR−DNA crystal structure (PDB 1R4I). The DNA corresponds to the androgen direct repeat response element, and the sequence used is 5′-CCAGAACATCAAGAACAG-3′. The resolution limit of the structure is 3.10 Å, and the space group is P3221. Zinc atoms are shown in blue, protein in yellow, and DNA in dark and light gray. (b) Map of the PSA-ARE site and schematic representation of a cyclic polyamide targeting the PSA-ARE site 5′-AGAACA-3′. Potential polyamide binding sites are shaded in gray. (c) Major- and minor-groove widths for the AR−DNA structure (circles) plotted with the groove widths for the cyclic polyamide−DNA structure (triangles) mapped onto the potential polyamide−DNA-binding sites of the ARE. Both potential polyamide-binding site regions are shaded in gray. (Right) Model showing groove width measurements mapped directly onto the AR−DNA crystal structure. Minor-groove widths are shown in yellow (open circles and triangles) and major-groove widths in blue (closed circles and triangles).
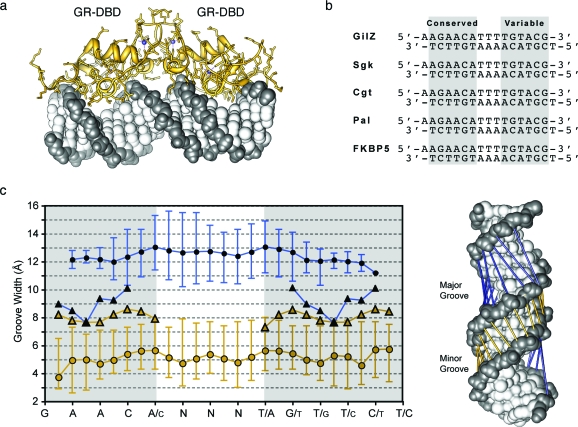
For the case of the glucocorticoid receptor, a total of 18 crystal structures were available for analysis, allowing a unique look at the impact of DNA sequence variation compared to minor- and major-groove widths.(26) The structures ranged in resolution from 1.61 to 3.00 Å, and 12 of the structures analyzed were found in the C2 space group, with the remainder found in P212121. A few of the DNA sequences are shown in Figure 8, with conserved and variable regions highlighted. A sequence alignment table with PDB numbers for all 18 structures can be found in the Supporting Information (Table S1). Our analysis of the glucocorticoid receptor shows consistency in groove distortions among different DNA sequences and different space groups, which parallel those of the androgen receptor−DNA structure irrespective of DNA sequence or space group. This analysis provides a molecular basis for the inhibition of AR− and GR−DNA binding using GABA turn-linked polyamides. The structural comparison shows that, through polyamide-induced allosteric modulation of the DNA structure, the major-groove surface geometry becomes incompatible with AR− and GR−DNA binding, an observation that is supported by previous biochemical and biophysical data.
Figure 8.
Comparison of polyamide-bound DNA to glucocorticoid receptor−DNA crystal structures. (a) GR−DNA crystal structure. Zinc atoms are shown in blue, protein in yellow, and DNA in dark and light gray. (b) DNA sequences used for each crystal structure are shown with the conserved domain highlighted. (c) Major- and minor-groove widths for the AR−DNA structure (circles) plotted with the groove widths for the cyclic polyamide−DNA structure (triangles) mapped onto the potential polyamide−DNA-binding sites of the ARE. Mean values are shown with the marker on the line, and error bars represent the minimum and maximum values for all structures. (Right) Model showing groove width measurements mapped directly onto the AR−DNA crystal structure. Minor-groove widths are shown in yellow (open circles and triangles) and major groove widths in blue (closed circles and triangles). All PDB numbers are listed in Table S1 in the Supporting Information.
Conclusion
Based on comparison of polyamide−DNA structure and protein−DNA structures, our data reveal an allosteric model for disrupting androgen and glucocorticoid receptor−DNA interfaces by small molecules. Recent data from the Yamamoto laboratory demonstrated that DNA can be thought of as an allosteric ligand for the GR, modulating the activity of the receptor to target genes based on DNA sequence.(26b) Differences at the single-base-pair level were able to affect the conformation and regulatory activity of GR. The possibility of using minor-groove DNA-binding molecules as secondary modulators of the already allosteric ligand (DNA) to further bias the conformation and regulatory activity of a transcription factor to specific target genes represents a secondary level of transcriptional control in biological systems.(26)d This study reiterates the importance of DNA shape and flexibility in protein−DNA recognition and illustrates the use of sequence-specific minor-groove ligands to modulate DNA shape as well.(27) To summarize, high-resolution structures of α-amino-γ-turn and β-amino-γ-turn cyclic polyamides can now can be compared with regard to changes in DNA shape and differences in conformation of the γ-turn unit. Regarding global DNA shape, both α- and β-amino-γ-turn cyclic polyamides are similar and able to induce large DNA structural perturbations with respect to groove width and helix bending, a critical requirement for allosteric inhibition of transcription factor binding. However, with regard to polyamide turn conformation, they are very different, with the β-amino-γ-turn adopting an inverted conformational preference compared to the α-amino-γ-turn, allowing the possibility of synthetic modifications for enhanced recognition beyond the six base-pair binding site. Importantly, the DNA structural alterations by the minor-groove synthetic cycle have been shown to be incompatible with the major-groove-binding transcription factors AR and GR. A critical next step will be to obtain high-resolution X-ray structures of hairpin polyamides and unlinked 2:1 antiparallel polyamide structures in complex with the same DNA sequence for comparison to the cyclic polyamide−DNA structures. This work is in progress and will be reported in due course.
Materials and Methods
General
Chemicals and solvents were purchased from Sigma-Aldrich and Hampton Research and were used without further purification. Water (18 MΩ) was purified using a Millipore Milli-Q purification system. Analytical HPLC analysis was conducted on a Beckman Gold instrument equipped with a Phenomenex Gemini analytical column (250 × 4.6 mm, 5 μM) and a diode array detector, and the mobile phase consisted of a gradient of acetonitrile (MeCN) in 0.1% (v/v) aqueous trifluoroacetic acid (CF3CO2H). Preparative HPLC was performed on an Agilent 1200 system equipped with a solvent degasser, a diode array detector, and a Phenomenex Gemini column (5 μm particle size, C18 110A, 250 × 21.2 mm, 5 μm). A gradient of MeCN in 0.1% (v/v) aqueous CF3CO2H was utilized as the mobile phase. UV−vis measurements were made on a Hewlett-Packard diode array spectrophotometer (model 8452 A), and polyamide concentrations were measured in 0.1% (v/v) aqueous TFA using an extinction coefficient of 69 200 M−1·cm−1 at λmax near 310 nm.
Synthesis and Purification
Cyclic polyamide 1 was synthesized as previously described and purified by reverse-phase HPLC prior to X-ray crystallography.(18a) Preparative HPLC was performed as described above. Oligonucleotides were purchased HPLC-purified from Trilink Biotechnologies (San Diego, CA). Prior to use for crystallography, oligonucleotides were desalted using a Waters Sep-Pak cartridge (5 g, C-18 sorbent).(21) The Sep-Pak was prewashed with acetonitrile (25 mL, 3×) followed by Milli-Q water (25 mL, 3×). The oligonucleotide was dissolved in 5 mL of 2.0 M NaCl and loaded directly onto the sorbent, followed by a wash with 5 mL of 2.0 M NaCl and 250 mL of Milli-Q water. Next, the oligonucleotide was eluted with acetonitrile:water (1:1) and lyophilized to dryness. Single-strand DNA and polyamide concentrations were determined by UV−vis spectroscopy on a Hewlett-Packard diode array spectrophotometer (model 8452 A).
Crystallization
Single-strand DNA was incubated with polyamide in a 2:1 ratio prior to crystallization. Crystals were obtained after 1−4 weeks from a solution of 0.5 mM duplex DNA, 0.5 mM polyamide, 23% 2-methyl-2,4-pentanediol (MPD), 35 mM calcium acetate, 10 mM Tris, pH 7.5, equilibrated in sitting drops against a reservoir of 35% MPD at 4 °C. Crystals were collected in Hampton nylon CryoLoops (10 μm, 0.1 mm) and flash-cooled to 100 K prior to data collection.
Data Collection
Polyamide−DNA crystals grew in space group P41212 with unit cell dimensions a = 39.83, b = 39.83, and c = 84.57 Å, α = 90°, β = 90°, γ = 90°, and one polyamide−dsDNA complex in the asymmetric unit. Data were collected at Stanford Synchrotron Radiation Laboratory (SSRL) beamline 12-2 with a MAR Research imaging plate detector at wavelength 0.82654 Å.
Crystal Structure Determination and Refinement
Data were processed with MOSFLM(28) and SCALA(29) from the CCP4 suite of programs.(29) The structure was solved by direct methods using the SHELX suite of programs (SHELXD).30,31 Model building and structure refinement were done with Coot(32) and REFMAC5.(33) The final polyamide−DNA complex was refined to Rwork = 11.2% and Rfree = 12.4%. Anisotropic B factors were refined in the final stages and riding hydrogens included prior to four cycles of completely unrestrained refinement.
Structural Analysis and Modeling
DNA groove analysis and helical parameters were calculated using the program Curves(34) and X3DNA.(35) Distance measurements, calculations, and preparation of structural figures were performed using UCSF Chimera.(36) All calculations reported were performed using HF/3-21G* as implemented in the Gamess program.37a−37c The electrostatic potential surface for 5-iodocytosine (CI) in Figure S1 was generated by mapping the electrostatic potential onto a surface of molecular electron density (0.002 electron/Å3) and color-coding, using the Chimera program.(36) The molecular electrostatic potential energy values range from −25 kcal/mol for values of negative potential (red) to +25 kcal/mol for values of positive potential (purple). This range was chosen to emphasize the variations in the electrostatic potential associated with the iodine atom of CI, and some regions of the electrostatic potential associated with other heteroatoms may lie beyond the 25 kcal/mol range.
Acknowledgments
We thank Douglas Rees, Jens Kaiser, and Michael Day for valuable discussions. Synchrotron data were collected at Stanford Synchrotron Radiation Laboratory (SSRL) beamline 12-2. We thank the staff of SSRL for their assistance during crystal screening and data collection. Operations at SSRL are supported by the U.S. DOE and NIH. We acknowledge the Gordon and Betty Moore Foundation for support of the Molecular Observatory at Caltech. This work was supported by the National Institutes of Health (GM27681). D.M.C. is grateful to the Kanel Foundation for a predoctoral fellowship. Coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID code 3OMJ).
Supporting Information Available
Figures S1−S5 and Table S1. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Panne D. Curr. Opin. Struct. Biol. 2008, 18, 236–242. [DOI] [PubMed] [Google Scholar]
- Panne D.; Maniatis T.; Harrison S. C. Cell 2007, 129, 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehr D. D.; Nussinov R.; Wright P. E. Nature Chem. Biol. 2009, 5, 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronemeyer H.; Gustafsson J.; Laudet V. Nat. Rev. Drug Discov. 2004, 3, 950–964. [DOI] [PubMed] [Google Scholar]
- Moore T. W.; Mayne C. G.; Katzenellenbogen J. A. Mol. Endocrinol. 2009, 24, 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. Curr. Opin. Chem. Biol. 2008, 12, 418–426. [DOI] [PubMed] [Google Scholar]
- Tsai M. J.; O’Malley B. W. Annu. Rev. Biochem. 1994, 63, 451–486. [DOI] [PubMed] [Google Scholar]
- Giguère V.; Hollenberg S. M.; Rosenfeld M. G.; Evans R. M. Cell 1986, 46, 645–652. [DOI] [PubMed] [Google Scholar]
- a Moellering R. E.; Cornejo M.; Davis T. N.; Del Bianco C.; Aster J. C.; Blacklow S. C.; Kung A. L.; Gilliland D. G.; Verdine G. L.; Bradner J. E. Nature 2009, 462, 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Henchey L. K.; Kushal S.; Dubey R.; Chapman R. N.; Olenyuk B. Z.; Arora P. S. J. Am. Chem. Soc. 2010, 132, 941–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervan P. B. Bioorg. Med. Chem. 2001, 9, 2215–2235. [DOI] [PubMed] [Google Scholar]
- Dervan P. B.; Edelson B. S. Curr. Opin. Struct. Biol. 2003, 13, 284–299. [DOI] [PubMed] [Google Scholar]
- a Pelton J. G.; Wemmer D. E. Proc. Natl. Acad. Sci. U.S.A. 1989, 86, 5723–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wade W. S.; Mrksich M.; Dervan P. B. J. Am. Chem. Soc. 1992, 114, 8783–8794. [Google Scholar]; c Mrksich M.; Wade W. S.; Dwyer T. J.; Geierstanger B. H.; Wemmer D. E.; Dervan P. B. Proc. Natl. Acad. Sci. U.S.A. 1992, 89, 7586–7590. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Trauger J. W.; Baird E. E.; Dervan P. B. Nature 1996, 382, 559–561. [DOI] [PubMed] [Google Scholar]; e White S.; Szewczyk J. W.; Turner J. M.; Baird E. E.; Dervan P. B. Nature 1998, 391, 468–470. [DOI] [PubMed] [Google Scholar]; f Kielkopf C. L.; Baird E. E.; Dervan P. B.; Rees D. C. Nat. Struct. Biol. 1998, 5, 104–109. [DOI] [PubMed] [Google Scholar]; g Kielkopf C. L.; White S.; Szewczyk J. W.; Turner J. M.; Baird E. E.; Dervan P. B.; Rees D. C. Science 1998, 282, 111–115. [DOI] [PubMed] [Google Scholar]
- Herman D. M.; Baird E. E.; Dervan P. B. J. Am. Chem. Soc. 1998, 120, 1382–1391. [Google Scholar]
- Hsu C. F.; Phillips J. W.; Trauger J. W.; Farkas M. E.; Belitsky J. M.; Heckel A.; Olenyuk B. Z.; Puckett J. W.; Wang C. C.; Dervan P. B. Tetrahedron 2007, 63, 6146–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Belitsky J. M.; Leslie S. J.; Arora P. S.; Beerman T. A.; Dervan P. B. Bioorg. Med. Chem. 2002, 10, 3313–3318. [DOI] [PubMed] [Google Scholar]; b Crowley K. S.; Phillion D. P.; Woodard S. S.; Scheitzer B. A.; Singh M.; Shabany H.; Burnette B.; Hippenmeyer P.; Heitmeier M.; Bashkin J. K. Bioorg. Med. Chem. Lett. 2003, 13, 1565–1570. [DOI] [PubMed] [Google Scholar]; c Best T. P.; Edelson B. S.; Nickols N. G.; Dervan P. B. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 12063–12068. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Edelson B. S.; Best T. P.; Olenyuk B.; Nickols N. G.; Doss R. M.; Foister S.; Heckel A.; Dervan P. B. Nucleic Acids. Res. 2004, 32, 2802–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Xiao X.; Yu P.; Lim H. S.; Sikder D.; Kodadek T. Angew. Chem., Int. Ed. 2007, 46, 2865–2868. [DOI] [PubMed] [Google Scholar]; f Nickols N. G.; Jacobs C. S.; Farkas M. E.; Dervan P. B. Nucleic Acids. Res. 2007, 35, 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Dose C.; Farkas M. E.; Chenoweth D. M.; Dervan P. B. J. Am. Chem. Soc. 2008, 130, 6859–6866. [DOI] [PMC free article] [PubMed] [Google Scholar]; h Hsu C. F.; Dervan P. B. Bioorg. Med. Chem. Lett. 2008, 18, 5851–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Suto R. K.; Edayathumangalam R. S.; White C. L.; Melander C.; Gottesfeld J. M.; Dervan P. B.; Luger K. J. Mol. Biol. 2003, 326, 371–380. [DOI] [PubMed] [Google Scholar]; b Edayathumangalam R. S.; Weyermann P.; Gottesfeld J. M.; Dervan P. B.; Luger K. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 6864–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Gottesfeld J. M.; Melander C.; Suto R. K.; Raviol H.; Luger K.; Dervan P. B. J. Mol. Biol. 2001, 309, 615–629. [DOI] [PubMed] [Google Scholar]
- a Olenyuk B. Z.; Zhang G. J.; Klco G. J.; Nickols N. G.; Kaelin W. G.; Dervan P. B. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 16768–16773. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kageyama Y.; Sugiyama H.; Ayame H.; Iwai A.; Fujii Y.; Huang L. E.; Kizaka-Kondoh S.; Hiraoka M.; Kihara K. Acta Oncol. 2006, 45, 317–324. [DOI] [PubMed] [Google Scholar]; c Nickols N. G.; Jacobs C. S.; Farkas M. E.; Dervan P. B. ACS Chem. Biol. 2007, 2, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Nickols N. G.; Dervan P. B. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 10418–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Matsuda H.; Fukuda N.; Ueno T.; Tahira Y.; Ayame H.; Zhang W.; Bando T.; Sugiyama H.; Saito S.; Matsumoto K.; Mugishima H.; Serie K. J. Am. Soc. Nephrol. 2006, 17, 422–432. [DOI] [PubMed] [Google Scholar]; f Yao E. H.; Fukuda N.; Ueno T.; Matsuda H.; Matsumoto K.; Nagase H.; Matsumoto Y.; Takasaka A.; Serie K.; Sugiyama H.; Sawamura T. Hypertension 2008, 52, 86–92. [DOI] [PubMed] [Google Scholar]; g Muzikar K. A.; Nickols N. G.; Dervan P. B. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 16598–16603. [DOI] [PMC free article] [PubMed] [Google Scholar]; h Gearhart M. D.; Dickinson L.; Ehley J.; Melander C.; Dervan P. B.; Wright P. E.; Gottesfeld J. M. Biochemistry 2005, 44, 4196–4203. [DOI] [PubMed] [Google Scholar]
- a Chenoweth D. M.; Harki D. A.; Phillips J. W.; Dose C.; Dervan P. B. J. Am. Chem. Soc. 2009, 131, 7182–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Harki D. A.; Satyamurthy N.; Stout D. B.; Phelps M. E.; Dervan P. B. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 13039–13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- In vitro ADMET (absorption, distribution, metabolism, excretion, and toxicity) studies of hairpin and cyclic polyamides coupled with in vivo PET (positron emission tomography) studies have revealed favorable drug-like properties and excellent metabolic stability for both hairpin and cyclic polyamides.18a,18b
- a de Clairac R. P. L.; Geierstanger B. H.; Mrksich M.; Dervan P. B.; Wemmer D. E. J. Am. Chem. Soc. 1997, 119, 7909–7916. [Google Scholar]; b Hawkins C. A.; de Clairac R. P. L.; Dominey R. N.; Baird E. E.; White S.; Dervan P. B.; Wemmer D. E. J. Am. Chem. Soc. 2000, 122, 5235–5243. [Google Scholar]; c Hawkins C. A.; Baird E. E.; Dervan P. B.; Wemmer D. E. J. Am. Chem. Soc. 2002, 124, 12689–12696. [DOI] [PubMed] [Google Scholar]; d Zhang Q.; Dwyer T. J.; Tsui V.; Case D. A.; Cho J.; Dervan P. B.; Wemmer P. B. J. Am. Chem. Soc. 2004, 126, 7958–7966. [DOI] [PubMed] [Google Scholar]
- Chenoweth D. M.; Dervan P. B. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 13175–13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CI = 5-iodocytosine.
- a Auffinger P.; Hays F. A.; Westhof E.; Ho P. S. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 16789–16794. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Voth A. R.; Hays F. A.; Ho P. S. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 6188–6193. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Voth A. R.; Khuu P.; Oishi K.; Ho P. S. Nature Chem. 2009, 1, 74–79. [DOI] [PubMed] [Google Scholar]; d Metrangolo P.; Neukirch H.; Pilati T.; Resnati G. Acc. Chem. Res. 2005, 38, 386–395. [DOI] [PubMed] [Google Scholar]; e Sarwar M. G.; Dragisic B.; Salsberg L. J.; Gouliaras C.; Taylor M. S. J. Am. Chem. Soc. 2010, 132, 1646–1653. [DOI] [PubMed] [Google Scholar]
- Kielkopf C. L.; Ding S.; Kuhn P.; Rees D. C. J. Mol. Biol. 2000, 296, 787–801. [DOI] [PubMed] [Google Scholar]
- Shaffer P. L.; Jivan A.; Dollins D. E.; Claessens F.; Gewirth D. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 4758–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Luisi B. F.; Xu W. X.; Otwinowski Z.; Freedman L. P.; Yamamoto K. R.; Sigler P. B. Nature 1991, 352, 497–505. [DOI] [PubMed] [Google Scholar]; b Meijsing S. H.; Pufall M. A.; So A. Y.; Bates D. L.; Chen L.; Yamamoto K. R. Science 2009, 324, 407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R.; West S. M.; Sosinsky A.; Liu P.; Mann R. S.; Honig B. Nature 2009, 461, 1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A. G. W. Newsl. Protein Crystallogr. 1992, 26. [Google Scholar]
- Acta Crystallogr. D 1994, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Sheldrick G. M. Acta Crystallogr. A 2008, 64, 112–122. [DOI] [PubMed] [Google Scholar]
- Schneider T. R.; Sheldrick G. M. Acta. Crystallogr. D 2002, 58, 1772–1779. [DOI] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. Acta Crystallogr. D 2004, 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Murshudov G. N.; Vagin A. A.; Dodson E. J. Acta. Crystallogr. D 1997, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Lavery R.; Sklenar H. Biochim. Theor. CNRS URA 1997, 77. [Google Scholar]
- a Lu X.-J.; Olson W. K. Nucleic Acids. Res. 2003, 31, 5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Lu X.-J.; Olson W. K. Nat. Protoc. 2008, 3, 1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; et al. J. Comput. Chem. 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- a Schmidt M. W.; et al. J. Comput. Chem. 1993, 14, 1347–1363. [Google Scholar]; b Binkley J. S.; Pople J. A.; Hehre W. J. J. Am. Chem. Soc. 1980, 102, 939–947. [Google Scholar]; c Pietro W. J.; et al. J. Am. Chem. Soc. 1982, 104, 5039–5048. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.