Abstract
BAG3, a member of the Hsc70 binding co-chaperone BAG-family proteins, has critical roles in regulating actin organization, cell adhesion, cell motility and tumor metastasis. The PDZ domain containing Guanine Nucleotide Exchange Factor 2 (PDZGEF2) was cloned as a BAG3 interacting protein. PDZGEF2 induces activation of Rap1 and increases integrin mediated cell adhesion. The PPDY motif at the C-terminus of PDZGEF2 binds to the WW domain of BAG3 in vitro and in vivo. BAG3 deletion mutant lacking the WW domain lose its cell adhesion and motility activity. Gene knockdown of PDZGEF2 leads to the loss of cell adhesion on fibronectin-coated plates while BAG3 overexpression increases cell adhesion in Cos7 cells, but not in PDZGEF2 gene knockdown cells indicating that PDZGEF2 is a critical partner for BAG3 in regulating cell adhesion.
Keywords: Hsp70, molecular chaperone, Rap1, Guanine nucleotide exchange factor, WW domain, cell adhesion
Introduction
The BAG3 protein contains a WW domain near its N-terminus, a proline-rich region (multiple PXXP motifs) and a BAG domain. The WW domain represents a protein interaction module that binds proteins carrying a proline-rich motif having the sequence PPXY [1,2]. Some WW domains recognize their peptide ligands in a phosphorylation-dependent manner. These domains have been identified in several signal transduction proteins that interact with plasma-membrane receptor complexes or with components of the submembranous cytoskeleton (reviewed in [2,3]. BAG3 also contains several SH3-ligand motifs of the sequence PXXP [4], indicating that BAG3 could bind specific SH3-containing proteins that would allow BAG3 to play a role in signal transduction pathways. BAG3 associates with actin at the leading edge of migrating cells and controls cell motility [5]. In many human tumor cell lines, especially adenocarcinomas, the BAG3 protein is highly expressed. Reduction of BAG3 levels by gene knockdown inhibits the invasive and metastatic activity of an epithelial cancer cell line (ALVA31) in vivo, indicating that BAG3 may contribute to the invasive or metastatic phenotype of cancers [5].
During cell movement, the actin cytoskeleton is tightly regulated in a spatial and temporal manner. Adhesion is mediated by integrin-family proteins attached to the extracellular matrix. Integrins are rapidly recycled via internalization, endocytosis, trafficking and exocytosis while adhesion molecules are internalized into the cell and transported to the leading-edge for reuse in attachment. Recent studies indicated that clathrin-mediated endocytosis of integrins has a dominant role at the leading edge of cells and polarized cells move in response to elevated recycled/internalization at the leading edge. Thus, tightly controlled signaling mechanisms of the actin cytoskeleton, endocytosis and adhesion molecules cooperate to regulate cell movement (reviewed in [6,7,8]).
To date, many small GTPase proteins are reported to be involved in cell movement. Recently, the PDZ domain containing guanine nucleotide exchange factors, PDZGEF1 (also known as RA-GEF1, nRapGEP and CNrasGEF) and PDZGEF2 were cloned and characterized [9]. Both PDZGEF1 and PDZGEF2 proteins have similar biochemical activity and activate Rap1 and Rap2 small GTPases [9]. The RA domain in PDZGEF1 interacts with Rap1, but not with Ras [10] while its PY domain binds the WW domain of Nedd4 [11]. PDZGEF2 increases integrin-mediated cell adhesion and also has roles in maturation of the adhesion junction [12,13].
Here we show that the co-chaperone BAG3 directly interacts with PDZGEF2 to regulate integrin-mediated cell adhesion.
Materials & Methods
Two-hybrid Screens
Yeast two-hybrid library screening of a human Jurkat cell cDNA library was performed as described previously [14]. Mating tests were then performed using RFY206 yeast strain transformed with pGilda, pGilda BAG3 ΔBAG, or pGilda BAG3 full length. The pJG4–5 cDNAs were recovered using KC8 Escherichia coli strain that is auxotrophic for Trp [14].
Cell Culture and antibody
Low passage Cos7 cells were cultured in DMEM medium with 10% Fetal Calf Serum (FCS) with penicillin/streptomycin. Transfections were performed with FuGENE (Roche, Indianapolis). Anti-PDZGEF2 polyclonal antibody generated against GST fusion protein of PDZGEF2 clone F11 and affinity purified. Anti-GFP, -Rap1, Myc (SantaCruiz Biotech, SantaCuiz). Anti-Actin (Calbiochem, La Jolla). Anti-Flag, -alpha tubulin (Sigma, Saint Louis).
Gene silencing using shRNA expression vector
Gene silencing of bag3 was accomplished using the shRNA expression vector pLTRH1puro as described previously [15]. For PDZGEF2 knockdown, we generated pLTRH1 puro vector with oligonucleotides (shGEF2#1 CUGCAUGAAUUGUAACUGAA, shGEF2#2 CAGGAAGAAGGGACAAACAAA) as previously reported by Dube et al [12].
Generation of plasmid encoding cDNA of BAG3 and PDZGEF2 deletion mutants
Deletion mutants were made using two-step PCR-mediated mutagenesis as described previously [14]. Full-length BAG3 cDNA was used as a template to create a Flag tagged version of full-length bag3 and deletion WW (Deletion nucleotide 52–159 from the first ATG, amino acid 17–53) [14]. PDZGEF2 cDNA was amplified using RNA extracted from HEK293 cell, followed by RT-PCR (GenBank ID: NM_001164386). Deletion mutants were generated using two-step PCR (ΔPPGY: deletion amino acid 1516–1519, ΔPPDY: 1537–1540).
Cell Motility and adhesion assays
Cos7 cells were transfected with pEGFP-BAG3, pEGFP-BAG3ΔWW, pcDNA3-FAK, pEGFP vector with or without PDZGEF2 knockdown and used for motility and adhesion assays as previously described [5]. A paired Student’s t-test was used to analyze differences between two groups, and p vales of <0.5, <0.05 or <0.01 were considered significant.
Results
Cloning of PDZGEF2 as an interacting partner of BAG3
To identify proteins that interact with BAG3, we carried out yeast two-hybrid screens of human Jurkat T cell cDNA libraries of 2×106 clones, using the full-length BAG3 protein as bait. After performing several tests to confirm the specificity of these interactions, we found several candidate BAG3-binding proteins: i) PDZGEF2, a guanine nucleotide exchange factor that contains a PDZ domain and reportedly activates the Ras-family member Rap1 [9], ii) cDNA clones with sequences for uncharacterized SH3 or praline-rich proteins, and iii) overlapping clones of Hsc70. Three different fragments were cloned as overlapping clones of PDZGEF2 (RapGEF6), containing amino acid 1326–1609 (clone A7), 1444–1609 (5 clones including F11 clone) and 1506–1609 (2 clones) as shown in Figure 1A.
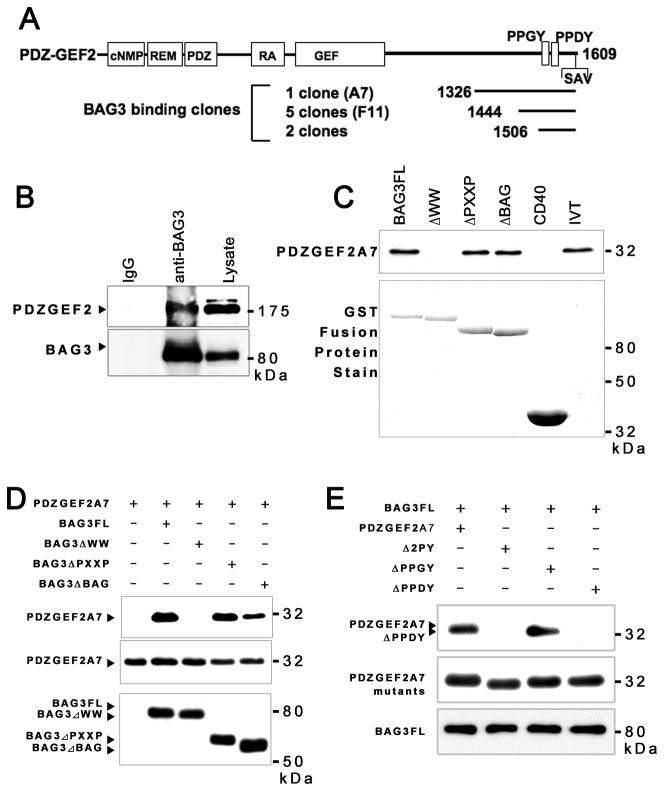
Figure 1. Analysis of PDZGEF2 interaction with BAG3.
(A) Schematic structure of PDZGEF2 domains. The cNMP-binding homology, REM, PDZ, RA, GEF, PPXY domains are indicated by open boxes. The regions corresponding to the eight BAG3 binding cDNA clones obtained by two-hybrid screens are indicated by 3 lines located at the right upper corner. (B) Endogenous BAG3 and PDZGEF2 interact in cells. Endogenous BAG3 was immunoprecipitated by AR100 (polyclonal anti-BAG3 antibody), followed by immunoblot using anti-PDZGEF2 antibody. (C) GST-fusion proteins encoding BAG3, BAG3 deletion mutants (ΔWW, ΔPXXP, ΔBAG), CD40 cytosolic domain (negative control) were used for in vitro binding assay with 35S-L-methionine labeled PDZGEF2A7. As a control, an equivalent amount of input in vitro translated PDZGEF2A7 (IVT) was loaded directly onto the gels. (D) Immunoprecipitation assay using Myc-tagged PDZGEF2A7 and Flag-tagged BAG3 (FL, ΔWW, ΔPXXP, ΔBAG). Immune-complexes were precipitated by Flag antibody, followed by western blot detected by myc for PDZGEF2 (upper panel). Cell lysate were directly applied for immunoblotting, followed by detection with Myc antibody (middle panel) and Flag for BAG3 constructs. (E) Deletion of two PPXY motifs in PDZGEF2 indicating specific interaction with BAG3 by PPDY. Myc tagged PPGY deletion (ΔPPGY), PPDY deletion (ΔPPDY), or both (Δ2PY) or control were made in C-terminal fragments of PDZGEF2 (A7 fragment), followed by immunoprecipitation assay using BAG3 co-transfection in 293 cells. Immune-complex was prepared by Flag antibody and immunoblotted by Myc antibody (upper panel).
The BAG3-binding protein PDZGEF2 is a close homologue of PDZGEF1 and has similar peptide sequences at the N-terminus, but has limited similarity at C-terminus, the site of BAG3 interaction. The clone of PDZGEF2 (A7) having the longest sequence has only 39% identity and 52% similarity with the PDZGEF1. In contrast, the N-terminal domain has 72%–85% identity and 82–93% similarity, suggesting a functional similarity but different binding affinity with BAG3. In the homologous region, PDZGEF1 and PDZGEF2 contain (from N-terminus to C-terminus) a cyclic nucleotide monophosphate (cNMP)-binding homology domain, Ras-exchange domain (REM), a PDZ domain, a Ras/Rap association (RA) domain, a putative GDP/GTP Exchange Factor (GEF) domain, a PY domain (PPXY motif, which is known as WW domain interacting motif), and a C-terminal PDZ-ligand motif (S/TXV) motif [9]. The highest PDZGEF1 expression levels detected by Northern blot analysis were localized to brain, heart, placental, skeletal muscle, kidney and pancreatic tissue [9]. In contrast, PDZGEF2 is more broadly expressed among tissues.
In our screens, all BAG3-interacting clones carried the C-terminal PPXY motif. For in vitro binding assays, the A7 and F11 clones were subcloned and used for further analysis. The shortest interacting clone had only 103 nucleotides and was not used for subsequent assays.
PDZGEF2 binds BAG3 in vitro and in cells
PDZGEF2 binding to BAG3 was confirmed by in vivo and in vitro binding assays using endogenous protein, an overexpression system in 293T cells and GST-fusion proteins in combination with in vitro- translated 35S methionine labeled PDZGEF2.
To detect endogenous PDZGEF2, we generated an anti-PDZGEF2 polyclonal antibody (F11) against a recombinant PDZGEF2-F11 clone corresponding to amino acids 1444–1609 as described above. Rabbit anti-PDZGEF2 antibody was purified by an affinity purification method described previously [5]. Using the prostate cancer cell line ALVA31, immunoprecipitation experiments were performed using anti-BAG3 and F11 antibodies. ALVA31 cells were used for initial report describing regulation of motility and adhesion by BAG3 [5]. Immune complexes were isolated with an anti-BAG3 antibody or F11 antibody, followed by immunodection using anti-F11 or anti-BAG3 antibodies. In Figure 1B, immunoprecipitation of BAG3 co-purifies with PDZGEF2 (anti-BAG3). In the third lane, cell lysates were loaded directly to detect endogenous expression of BAG3. IgG1 was also used for immunoprecipitation as negative control (IgG). Using the anti-PDZGEF2 antibody and anti-BAG3 polyclonal antibody, we detected an endogenous protein-protein complex in ALVA31 cells.
To determine the BAG3 interaction domain of PDZGEF2, we constructed BAG3 deletion mutants lacking either the WW (ΔWW), PXXP (ΔPXXP), or BAG domains (ΔBAG) as described previously [5]. The interaction of BAG3 with PDZGEF2 C-terminal fragments (PDZGEF2A7) was demonstrated by an in vitro binding assay using GST fusion protein of the full-length and BAG3 deletion mutants. The in vitro-translated PDZGEF2 protein fragments represented the same cDNA clones (A7 clone) as those pulled out by two-hybrid screening, and encompassed the C-terminal portions of PDZGEF2 protein (A7 clone). From these results, we concluded that the WW domain of BAG3 and C-terminal portion of PDZGEF2 is sufficient for binding in vitro (Figure 1C).
Domain dependent interactions were also confirmed by co-immunoprecipitation assays using lysates from human HEK293T cells transfected with plasmids encoding Myc epitope-tagged versions PDZGEF2. Flag-tagged versions of full-length BAG3 and the three deletion mutants were recovered in Myc-PDZGEF2A7 (1326–1609) immune-complexes, as determined by immunoblotting using anti-myc antibody (Figure 1D).
The cDNA clones obtained in the 2-hybrid screens using BAG3 as bait encode the C-terminus of PDZGEF2, which includes the two PPXY motifs (PPGY, starting at amino acid 1516, PPDY, starting at amino acid 1537). We generated deletion mutants of PPGY (ΔPPGY), PPDY (ΔPPDY) and both (Δ2PY) in the C-terminal fragment of A7 clone of PDZGEF2 with a myc tag. Overexpression of Flag-tagged BAG3 and Myc-tagged wild type PDZGEF2A7 fragment or mutants were used for immunoprecipitation assays (Flag antibody), followed by immunodetection by myc antibody to detect co-precipitated C-terminal fragments of PDZGEF2. As shown in Figure 1E, ΔPPDY and Δ2PY lost binding activity with BAG3, suggesting PPDY, but not the PPGY motif specifically interacting with BAG3. These data provide supporting evidence that PDZGEF2 may be a physiologically relevant target of BAG3.
WW domain of BAG3 is critical for regulating cell motility and adhesion
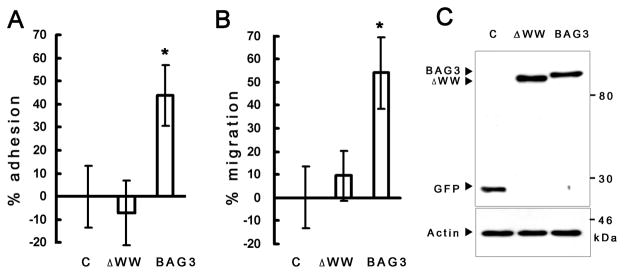
We observed that overexpression of BAG3 in Cos7 cells increased cell adhesion and cell motility in transient motility assays conducted under serum-depleted conditions [5]. Figure 2A shows a comparison of the motility of Cos7 cells transfected with pEGFP BAG3 ΔWW, pEGFP wild type BAG3 or pEGFP plasmids. Cell motility assays were performed using a transwell system. Deletion of the BAG3 WW domain caused a significant reduction in cell motility. Since our cDNA cloning data and interaction assay indicated that the WW domain interacts with PDZGEF2 (RapGEF6) and Rap1 is known as positive regulator of integrin-mediated cell adhesion [13,16], we performed adhesion assays using fibronectin-coated plates. Figure 2B shows that BAG3ΔWW significantly reduces adhesion activity on fibronectin coated plates, suggesting that the WW domain of BAG3 is important for integrin-mediated cell adhesion. Figure 2C indicates the stable expression of pEGFP-BAG3 and pEGFPBAG3ΔWW in this assay.
Figure 2. WW domain of BAG3 is critical for cell motility and adhesion.
(A) Cells transfected with BAG3ΔWW deletion have impaired integrin-mediated adhesion. Cos7 cells were transfected by BAG3 wild type (BAG3), WW domain deleted (ΔWW) and pEGFP empty vector, followed by adhesion assay (*p<0.01, mean ± SD, n=6). (B) Impaired cell motility found in Cos7 cells with BAG3ΔWW (*p<0.01, mean ± SD, n=6). (C) BAG3 wild type (pEGFP-BAG3) and pEGFP-BAG3ΔWW were transfected into 293T cells. Western blotting with GFP antibody indicated stable expression.
PDZGEF2 induces Rap1 activation and increases integrin mediated adhesion
Full-length cDNA of PDZGEF2 was cloned by RT-PCR, using total RNA extracted from HEK293T cells. The small GTP-binding protein Rap1 is known to be a downstream target of PDZGEF2. To detect Rap1 activation by PDZGEF2, we compared the activity of Rap1 in control Cos7 vs. cells overexpressing PDZGEF2. Cell lysates were prepared, followed by recovery of the GTP-bound form of Rap1 by adsorption to immobilized GST-RalGDS fusion protein. For positive and negative controls, we used lysates incubated with non-hydrolyzable GTP (GTPγS) and GDP, respectively. Greater amounts of active Rap1 were recovered from lysates of PDZGEF2 overexpressing cells compared to control Cos7 cells (Figure 3A). In contrast, GTPγS-treated lysates demonstrated comparable total levels of Rap1 in both control and PDZGEF2 overexpressing cells (not shown). This result suggests that PDZGEF2 regulates Rap1 activity. Since Rap1 is reported to be a positive regulator of integrin-mediated adhesion, we compared the rate of attachment of PDZGEF2 overexpressing cells to fibronectin-coated (integrin-dependent ligand) plastic plates. In short-term adhesion assays (20 min), more PDZGEF2 over-expressing Cos7 cells adhered to fibronectin-coated plates than control transfected Cos7 cells (Figure 3B). Next, we used a transwell assay to address whether PDZGEF2 increases cell motility. Since PDZGEF2 interacts with the WW domain of BAG3 and BAG3ΔWW expressing cells have reduced motility, we examined whether PDZGEF2 overexpression increases cell motility in a transwell assay. As shown in Figure 3C, PDZGEF2 and control-transfected Cos7 cells displayed no difference in cell motility.
Figure 3. PDZGEF2 activates Rap1 and regulates cell adhesion.
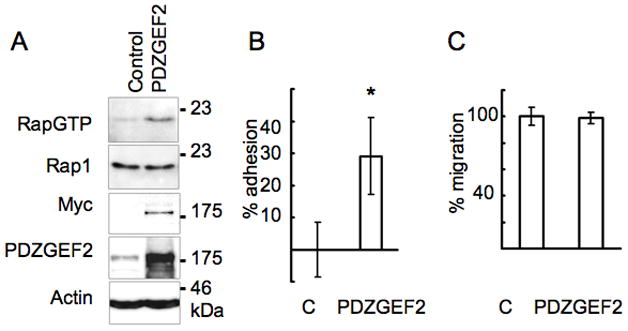
(A) Rap1 activation was detected in PDZGEF2-transfected COS7 cells using GST-RalGDS pull down (1st panel). Total Rap1 expression (2nd panel). The overexpression of MycPDZGEF2 was detected by Myc antibody (3rd panel). Anti-PDZGEF2 antibody F11 detects endogenous PDZGEF2 in control transfected cells as well as overexpressed myc PDZGEF2 (4th panel). Actin as a loading control (5th panel). (B) PDZGEF2-transfected Cos 7 cell increase adhesion on fibronectin-coated coated plates. Data represent percentage change in the number of adherent cells relative to control-transfected cells, 20 minutes after plating (*p<0.01, mean ± SD, n=6). (C) Cell motility assay. Cos7 cells were transfected with plasmids encoding pcDNA3 mycPDZGEF2 or with empty control pcDNA3 plasmid, followed by serum starvation and transwell assay (8 μm pore size) for 4 hours. Data represent mean ± SD (n = 6).
BAG3 lose its cell adhesion activity in Cos7 cells with PDZGEF2 gene knockdown
We examined whether inhibition of PDZGEF2 function influences BAG3 function in cell adhesion. We generated PDZGEF2 shRNA expression vector using two different oligomers, shGEF2#1 and shGEF2#2, as described in the Materials and Methods section. Cos7 cells were infected with retrovirus containing PDZGEF2 shRNA and stable lines were selected using puromycin-containing medium (1μg/ml). Expression of PDZGEF2 protein were confirmed by western blot, which indicated that shGEF2#2 knocked down PDZGEF2 expression more effectively than shGEF2#1, achieving suppression of PDZGEF2 expression by ~80% and ~50%, respectively. Using shGEF2#2 transfectants, pcDNA3mycbag3 full-length (pcDNA3BAG3) and control plasmids (pcDNA3) were transiently transfected (Figure 4A), followed by adhesion assays on fibronectin coated plates. As shown in Figure 4B, BAG3 overexpression does not increase cell adhesion in PDZGEF2 knockdown cells, indicating that PDZGEF2 interaction is sufficient for adhesion activity controlled by BAG3. Exogenously overexpressed myc-tagged bag3 was also confirmed by immunoblot with Myc antibody (not shown).
Figure 4. BAG3 lose its cell adhesion activity in PDZGEF2 gene knockdown cells.
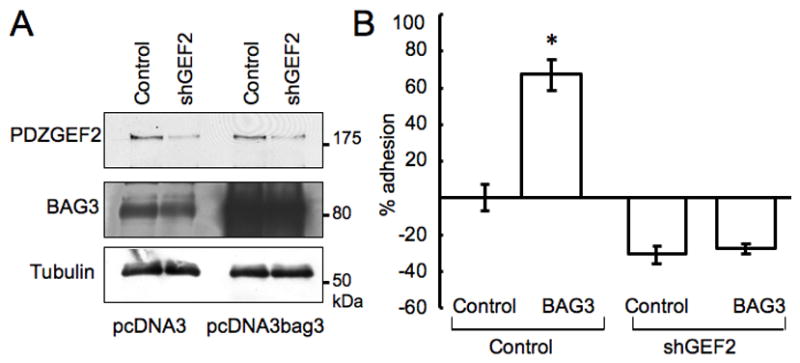
(A) PDZGEF2 knockdown cells (shGEF2) or control retrovirus infected cells were used for overexpression with pcDNA3 myc-bag3 construct (pcDNA3bag3) or pcDNA3 vector alone, followed by immunodetection with an anti-PDZGEF2 antibody (upper panel) and anti-BAG3 antibody (middle panel). Tubulin served as a loading control (lower panel). (B) PDZGEF2 gene knockdown or control infected cells were transfected with full length expression vector of BAG3, followed by an adhesion assay using crystal violet. Data represent mean± SE (n=6, *p<0.01).
Discussion
In this report, we demonstrate that BAG3 direct interacts with the guanine exchange factor of Rap1, PDZGEF2. The BAG3 WW domain interacts with the PPXY motif of PDZGEF2, a known motif of WW domain interaction. There are two PPXY sequences in PDZGEF2. Deletion mutant of either of the PPXY sequences indicated that only the second PPXY motif (PPDY) is critical for the BAG3 interaction. BAG3 WW domain deletion mutants were made and used for cell motility or adhesion assays. These suggested the importance of the WW domain in regulating motility and adhesion. The PPXY motif is also found in PDZGEF1, which is a close homologue of PDZGEF2. Although the C-terminal region of PDZGEF2 and PDZGEF1 has only low similarity, PDZGEF1 and PDZGEF2 have two PPXY motifs, (PPGY and PPDY), suggesting that PDZGEF1 may also interact with BAG3. The PPXY domain of PDZGEF1 binds to the WW domain of the E3 ubiquitin ligase Nedd4, which targets it for ubiquitin proteasomal degradation [17]. Both the PPGY and PPDY motifs of PDZGEF1 interact with Nedd4 ubiquitin ligase, but BAG3 may only interact with the PPDY motif. It will be of interest to investigate the Nedd4 ubiquitin ligase interaction with PDZGEF2 and whether BAG3 can compete with Nedd4 for interaction. The BAG3 WW domain also interacts with adenovirus penton base protein. The adenovirus penton base is viral capsid protein responsible for viral internalization and direct interaction of BAG3 with this protein increases nuclear migration, suggesting that BAG3 may contribute to the adenovirus lifecycle by [18].
Both of PDZGEF1 and PDZGEF2 have the C-terminal S/TXV motif, which is known as a PDZ ligand motif. Two PDZ proteins (S-CAM and MAGI-1) have been reported to associate with PDZGEF1 via this motif [19,20]. It will be interesting to find whether PDZGEF2 partners with PDZ ligand motifs in the BAG3 complex to better understand the function of BAG3 in regulating PDZGEF2 activity.
Previously we showed that BAG3 overexpression increased, and shRNA-mediated BAG3 protein reduction decreased cell adhesion on fibronectin coated plates. In this report, our results suggest that BAG3 is a regulator of PDZGEF2 and this interaction may potentially connect the activation of Rap1. Since PDZGEF2 increase cell adhesion, but not cell motility, further investigation is necessary to elucidate the molecular mechanisms of BAG3 regulation of cell motility. BAG3 interact with PLCγ and deletion of PXXP domain alter FAK phosphorylation [21,22]. It is possible that multiple SH3 proteins may link BAG3 with cell movement processes. Further analysis of physiologically relevant proteins that interact with the PXXP motifs of BAG3 may reveal the importance of SH3 proteins in BAG3-mediated regulation of cell motility.
A close homologue of PDZGEF2, PDZGEF1, is functionally similar in that both regulate GDP/GTP exchange on Rap1 and Rap2 [9], which are small GTPases known to regulate cytoskeletal assembly and cell migration (reviewed in [23]). The RA domain of PDZGEF1, which permits interaction with Rap1 but not Ras, is required for GEF activity and the PDZGEF1-Rap1 interaction stimulates Rap1 function via positive feedback [10]. The RA domain of PDZGEF2 interacts with M-Ras, which activates and translocates PDZGEF2 to the plasma membrane [24]. Gao et al. originally cloned a C-terminal truncated isoform as full-length PDZGEF2 and showed Rap1 activation with this isoform [24], which should not interact with BAG3. Although we obtained only a single cDNA fragment by RT-PCR, there are six potential alternatively spliced forms of PDZGEF2 found in the genome database that result in different C-terminal sequences. Isoforms #1 (GenBank ID: NM_001164386) and #2 (NM_016340.5) contain PPXY motifs, but other isoforms lack the C-terminal peptides, including the PPXY motif. Thus, we expect isoforms #3 (NM_001164387.1), #4 (NM_001164388.1), #5 (NM_001164389.1) and #6 (NM_001164390.1) would be functionally independent from BAG3 association or could work as dominant negative inhibitors of the BAG3/PDZGEF2 complex. According to sequence data of splicing variants, our shRNA knockdown of PDZGEF2 should be ineffective for isoform 6. Isoforms #2 and #5 have an 8 amino acid deletion in the GEF domain, although whether this affects their GEF activity is unknown. Our antibody detects one major band at a molecular weight of about 180kDa. Overexpression of full-length constructs produces the same molecular weight band. Thus, we expect isoform #1 (1609 amino acid) transcript to be the major product in our system, but it will be very important to examine expression pattern of each isoform in different cell types or tissues, especially whether the PPXY motif is present.
Recently, PDZGEF2 was found to be a partner protein of junctional adhesion molecule-A (JAM-A), which interacts with Afadin. PDZGEF2 regulates cell-cell adhesion to integrin medicated cell-ECM regulation by interacting with JAM-A [25]. Rap1 signaling is reported to be activated by E-cadherin internalization and increase intergrin mediated cell-ECM adhesion, suggesting regulation of signals from cell-cell adhesion to cell-ECM adhesion by Rap1. Maturation of the adhesion junction is also regulated by PDZGEF2 [12]. This evidence suggests that PDZGEF2 regulates both cell-cell and cell-ECM adhesion. The physiological activators of PDZGEF2 remain unclear, but our investigation may help reveal the role of BAG3 in the physiological function of PDZGEF2 in regulating cell adhesion and motility.
Conclusion
The WW domain of BAG3 interacts with the PPDY motif of PDZGEF2 (RapGEF6). Deletion of the BAG3 WW domain causes a significant reduction in cell motility and adhesion, while BAG3 over-expression does not increase cell adhesion in PDZGEF2 knockdown cells. These results indicate that PDZGEF2 is a critical partner of BAG3 in regulating cell adhesion.
Research Highlights.
PDZGEF2 was cloned as a BAG3 interacting protein.
The PPDY motif of PDZGEF2 binds to the WW domain of BAG3.
BAG3 deletion WW mutant loses cell adhesion and motility.
PDZGEF2 is a critical partner of BAG3 for regulating cell adhesion.
Acknowledgments
We thank the NIH for their generous support (CA107793, AR052925).
Nonstandard abbreviations used
- BAG
Bcl-2-associated athanogene
- PDZGEF2
PDZ domain containing guanine nucleotide exchange factor 2
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Einbond A, Sudol M. Towards prediction of cognate complexes between the WW domain and proline-rich ligands. FEBS Lett. 1996;384:1–8. doi: 10.1016/0014-5793(96)00263-3. [DOI] [PubMed] [Google Scholar]
- 2.Sudol M, Chen HI, Bougeret C, et al. Characterization of a novel protein-binding module--the WW domain. FEBS Lett. 1996;365:198–202. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 3.Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 4.Feller SM, Ren R, Hanafusa H, et al. SH2 and SH3 domains as molecular adhesives: the interactions of Crk and Abl. Trends Biochem Sci. 1994;19:453–458. doi: 10.1016/0968-0004(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 5.Iwasaki M, Homma S, Hishiya A, et al. BAG3 regulates motility and adhesion of epithelial cancer cells. Cancer Res. 2007;67:10252–10259. doi: 10.1158/0008-5472.CAN-07-0618. [DOI] [PubMed] [Google Scholar]
- 6.Juliano RL, Reddig P, Alahari S, et al. Integrin regulation of cell signalling and motility. Biochem Soc Trans. 2004;32:443–446. doi: 10.1042/BST0320443. [DOI] [PubMed] [Google Scholar]
- 7.Klemke RL, Cai S, Giannini AL, et al. Regulation of cell motility by mitogen-activated protein kinase. J Cell Biol. 1997;137:481–492. doi: 10.1083/jcb.137.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Small JV, Stradal T, Vignal E, et al. The lamellipodium: where motility begins. Trends Cell Biol. 2002;12:112–120. doi: 10.1016/s0962-8924(01)02237-1. [DOI] [PubMed] [Google Scholar]
- 9.Kuiperij HB, de Rooij J, Rehmann H, et al. Characterisation of PDZ-GEFs, a family of guanine nucleotide exchange factors specific for Rap1 and Rap2. Biochim Biophys Acta. 2003;1593:141–149. doi: 10.1016/s0167-4889(02)00365-8. [DOI] [PubMed] [Google Scholar]
- 10.Liao Y, Kariya K, Hu CD, et al. RA-GEF, a novel Rap1A guanine nucleotide exchange factor containing a Ras/Rap1A-associating domain, is conserved between nematode and humans. J Biol Chem. 1999;274:37815–37820. doi: 10.1074/jbc.274.53.37815. [DOI] [PubMed] [Google Scholar]
- 11.Pham N, Cheglakov I, Koch CA, et al. The guanine nucleotide exchange factor CNrasGEF activates ras in response to cAMP and cGMP. Curr Biol. 2000;10:555–558. doi: 10.1016/s0960-9822(00)00473-5. [DOI] [PubMed] [Google Scholar]
- 12.Dube N, Kooistra MR, Pannekoek WJ, et al. The RapGEF PDZ-GEF2 is required for maturation of cell-cell junctions. Cell Signal. 2008;20:1608–1615. doi: 10.1016/j.cellsig.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Yoshikawa Y, Satoh T, Tamura T, et al. The M-Ras-RA-GEF-2-Rap1 pathway mediates tumor necrosis factor-alpha dependent regulation of integrin activation in splenocytes. Mol Biol Cell. 2007;18:2949–2959. doi: 10.1091/mbc.E07-03-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274:781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 15.Homma S, Iwasaki M, Shelton GD, et al. BAG3 deficiency results in fulminant myopathy and early lethality. Am J Pathol. 2006;169:761–773. doi: 10.2353/ajpath.2006.060250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katagiri K, Maeda A, Shimonaka M, et al. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat Immunol. 2003;4:741–748. doi: 10.1038/ni950. [DOI] [PubMed] [Google Scholar]
- 17.Pham N, Rotin D. Nedd4 regulates ubiquitination and stability of the guanine-nucleotide exchange factor CNrasGEF. J Biol Chem. 2001;276:46995–47003. doi: 10.1074/jbc.M108373200. [DOI] [PubMed] [Google Scholar]
- 18.Gout E, Gutkowska M, Takayama S, et al. Co-chaperone BAG3 and adenovirus penton base protein partnership. Journal of Cellular Biochemistry. 2010 doi: 10.1002/jcb.22756. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mino A, Ohtsuka T, Inoue E, et al. Membrane-associated guanylate kinase with inverted orientation (MAGI)-1/brain angiogenesis inhibitor 1-associated protein (BAP1) as a scaffolding molecule for Rap small G protein GDP/GTP exchange protein at tight junctions. Genes Cells. 2000;5:1009–1016. doi: 10.1046/j.1365-2443.2000.00385.x. [DOI] [PubMed] [Google Scholar]
- 20.Ohtsuka T, Hata Y, Ide N, et al. nRap GEP: a novel neural GDP/GTP exchange protein for rap1 small G protein that interacts with synaptic scaffolding molecule (S-SCAM) Biochem Biophys Res Commun. 1999;265:38–44. doi: 10.1006/bbrc.1999.1619. [DOI] [PubMed] [Google Scholar]
- 21.Doong H, Price J, Kim YS, et al. CAIR-1/BAG-3 forms an EGF-regulated ternary complex with phospholipase C-gamma and Hsp70/Hsc70 [In Process Citation] Oncogene. 2000;19:4385–4395. doi: 10.1038/sj.onc.1203797. [DOI] [PubMed] [Google Scholar]
- 22.Kassis JN, Guancial EA, Doong H, et al. CAIR-1/BAG-3 modulates cell adhesion and migration by downregulating activity of focal adhesion proteins. Exp Cell Res. 2006;312:2962–2971. doi: 10.1016/j.yexcr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 23.Bos JL. Linking Rap to cell adhesion. Curr Opin Cell Biol. 2005;17:123–128. doi: 10.1016/j.ceb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Gao T, Newton AC. The turn motif is a phosphorylation switch that regulates the binding of Hsp70 to protein kinase C. J Biol Chem. 2002;277:31585–31592. doi: 10.1074/jbc.M204335200. [DOI] [PubMed] [Google Scholar]
- 25.Severson EA, Lee WY, Capaldo CT, et al. Junctional adhesion molecule A interacts with Afadin and PDZ-GEF2 to activate Rap1A, regulate beta1 integrin levels, and enhance cell migration. Mol Biol Cell. 2009;20:1916–1925. doi: 10.1091/mbc.E08-10-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]