Summary
Acetylation of lysine residues at the H3 N-terminus is proposed to have a role in replication-coupled (RC) nucleosome assembly, a process critical for the inheritance of epigenetic information and maintenance of genome stability. However, the role of H3 N-terminal lysine acetylation and the corresponding lysine acetyltransferase (KAT) in RC nucleosome assembly are not known. Here we show that Gcn5, a KAT with a well-studied role in gene transcription, functions in parallel with Rtt109, the H3 lysine 56 KAT, to promote RC nucleosome assembly. Cells lacking both Gcn5 and Rtt109 are highly sensitive to DNA damaging agents. Moreover, cells lacking GCN5 or expressing an H3 mutant with mutations at the H3 N-terminus result in compromised deposition of new H3 onto replicating DNA and a reduction in the binding of H3 with CAF-1, a histone chaperone involved in RC nucleosome assembly. These results demonstrate that Gcn5 regulates RC nucleosome assembly, in part, through promoting the association of H3 with CAF-1 via H3 acetylation.
Introduction
The nucleosome, the fundamental unit of chromatin, is comprised of 147 base pairs of DNA wrapped around a histone octamer of H2A, H2B, H3, and H4. During S phase of the cell cycle, parental nucleosomes are disassembled to facilitate access to DNA for the replication machinery. Replicated DNA must then be immediately reassembled into nucleosomes using parental histones as well as newly-synthesized histones in a process referred to as DNA replication-coupled nucleosome assembly. This process plays an important role in the inheritance of epigenetic states and the maintenance of genome integrity (Groth et al., 2007b; Morrison and Shen, 2009). While it is not well understood how parental histones are reassembled into nucleosomes following DNA replication, assembly of newly-synthesized histones into nucleosomes requires histone chaperones such as chromatin assembly factor 1 (CAF-1) (Stillman, 1986), Asf1 and Rtt106. These three proteins bind histone H3-H4 and function coordinately in nucleosome assembly during S phase of the cell cycle (Groth et al., 2007b; Li et al., 2008).
Newly-synthesized histone H3-H4 is acetylated by lysine acetyltransferases (KAT) before being assembled into nucleosomes (Roth et al., 2001). Histone H4 is acetylated at lysine residues 5 and 12 (K5, K12) by Hat1 (Ai and Parthun, 2004; Kleff et al., 1995), an acetylation pattern that is conserved from yeast to humans (Sobel et al., 1995). Patterns of acetylation on newly-synthesized H3 are not as conserved among species. In HeLa cells, acetylation of newly-synthesized histone H3.1 is barely detectable, while new H3 is diacetylated at K9 and K14 in Tetrahymena and K14 and K23 in Drosophila (Benson et al., 2006; Sobel et al., 1995). In yeast cells, newly-synthesized H3 is acetylated at lysine 56 (H3K56Ac) (Masumoto et al., 2005). We and others have shown that this modification is important for nucleosome assembly during DNA replication and DNA repair (Chen et al., 2008; Li et al., 2008). A recent study indicates that the function of this modification in nucleosome assembly appears to be conserved in mammalian cells (Das et al., 2009). In yeast cells, H3K56Ac is catalyzed by Rtt109 (Kat11) (Collins et al., 2007; Driscoll et al., 2007; Han et al., 2007a) and is dependent upon the histone chaperone Asf1 (Recht et al., 2006). We have shown that the binding of H3 with Rtt106 is barely detectable in cells lacking H3K56Ac, whereas the association of H3 with CAF-1 is reduced in cells lacking this modification (Li et al., 2008), suggesting that other modifications on H3 may also regulate the binding of H3 with CAF-1.
In addition to H3K56Ac, new H3 is predominantly acetylated at K9, followed by acetylation of K27 (Kuo et al., 1996). However, the yeast KAT that is responsible for acetylation of these lysine residues of newly-synthesized H3 is not well defined. Genetic evidence indicates that the N-terminus of H3, in particular the acetylation of five lysine residues (K9, K14, K18, K23, and K27), is important for nucleosome assembly (Li et al., 2008; Ma et al., 1998; Qin and Parthun, 2002). However, it is not known which KAT acetylates these five lysine residues and regulates nucleosome assembly.
Gcn5 is the catalytic subunit of three KAT complexes including SAGA, SLIK, and ADA. All of these Gcn5-containing complexes regulate transcription. In vitro, recombinant Gcn5 acetylates predominantly K14 of free H3 and shows little or no activity against nucleosomal H3 (Kuo et al., 1996). On the other hand, the SAGA and ADA complexes acetylate both free and nucleosomal H3. While ADA preferentially acetylates K14 and K18 of nucleosomal H3, SAGA acetylates K14 and K18 and to a lesser degree, K23 and K9 (Grant et al., 1999). Thus, the activity and specificity of Gcn5 is regulated by its associated proteins.
Cells lacking Gcn5 are sensitive to DNA damaging agents, suggesting that Gcn5, in addition to its role in gene transcription, may have a role in DNA replication and DNA repair (Choy and Kron, 2002; Tamburini and Tyler, 2005). However, how Gcn5 is involved in DNA replication or DNA repair is not well understood. Here we show that gcn5Δ rtt109Δ double mutant cells are highly sensitive to DNA damaging agents due to the loss of enzymatic activities of both enzymes. Moreover, GCN5 genetically interacts with genes known to be involved in DNA replication, the DNA damage response, as well as nucleosome assembly. Furthermore, cells lacking GCN5 or expressing an H3 mutant containing mutations at five lysine residues of the N-terminus of histone H3 exhibit compromised deposition of new H3 onto replicating DNA and reduced binding of H3 to CAF-1. Together, these results indicate that Gcn5 promotes nucleosome assembly, in part, by acetylating lysine residues at the H3 N-terminus which contributes to the binding of H3 with CAF-1.
Results
Gcn5 and Rtt109 function in parallel in cell growth and response to DNA damaging agents
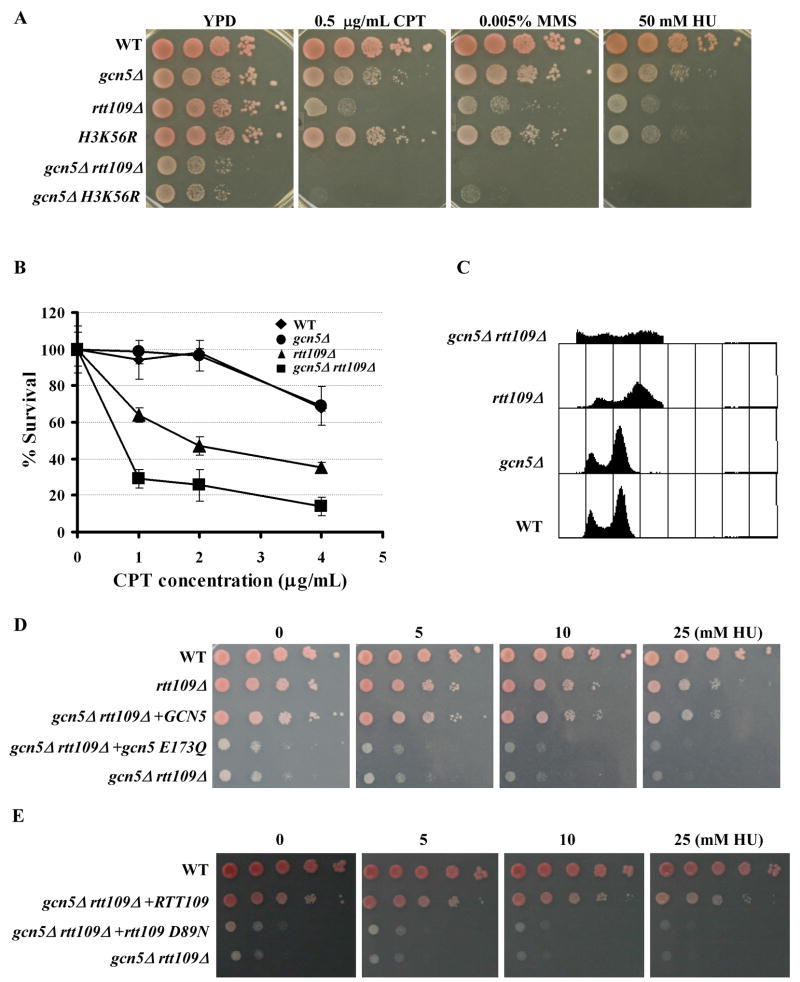
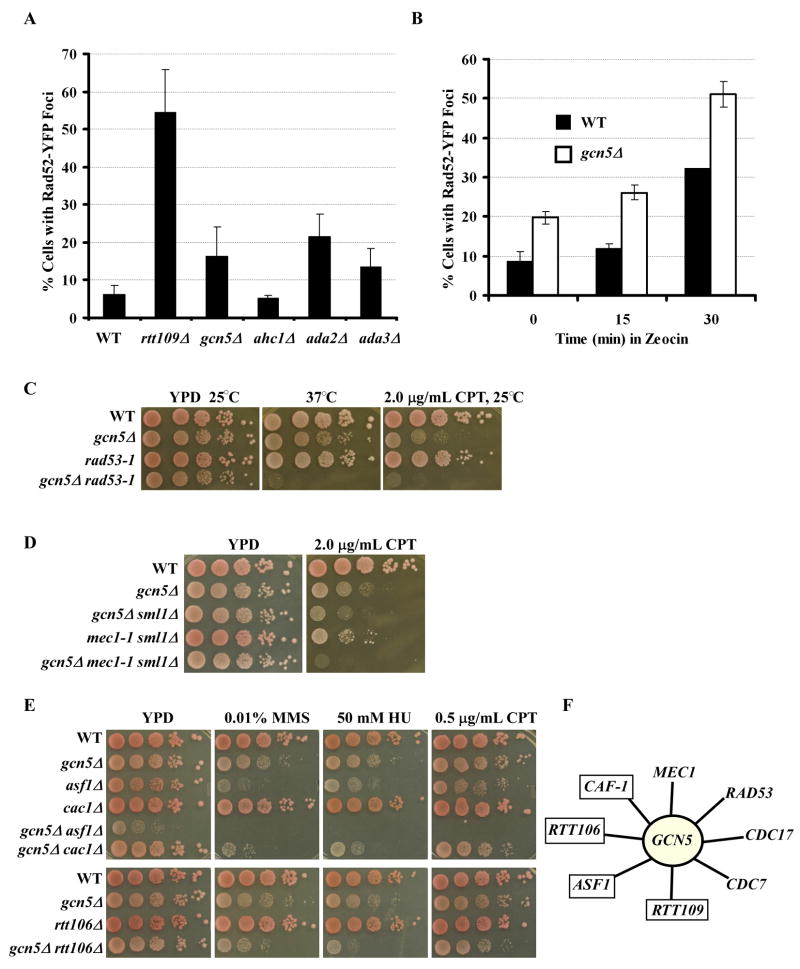
Acetylation of N-terminal lysine residues of H3 is likely to function in parallel with H3K56Ac to resist DNA damage (Li et al., 2008). To find the lysine acetyltransferase (s) (KAT) that functions with H3K56Ac to promote genome stability, we tested the cell growth and DNA damage sensitivity of double mutant cells lacking ASF1, the H3K56Ac regulator (Han et al., 2007a), and one of the non-essential KATs in the budding yeast, S. cerevisiae. We found that mutations in GCN5 and ELP3, but not mutations in other H3 KATs such as SAS3, resulted in severe growth defects and a high degree of DNA damage sensitivity when combined with the asf1Δ mutation (Table S1). The phenotypes observed in gcn5Δ asf1Δ double mutant cells are consistent with recently published reports showing that gcn5Δ mutant cells exhibit a synthetic sick phenotype with the rtt109Δ mutant (Fillingham et al., 2008; Lin et al., 2008). We, therefore, tested whether Gcn5 was the KAT that functioned with H3K56Ac to resist DNA damaging agents. First, we asked whether the gcn5Δ mutant exhibited synthetic growth defects and sensitivity towards DNA damaging agents when combined with the rtt109Δ mutation or an H3 lysine 56 mutant with lysine 56 mutated to arginine (H3K56R). The gcn5Δ mutant cells were sensitive to the three DNA damaging agents camptothecin (CPT), hydroxyurea (HU), and methyl methane sulfonate (MMS) but to a lesser degree than rtt109Δ mutant cells (Figure 1A–B and Supplemental Figure 1). However, gcn5Δ rtt109Δ cells were more sensitive to DNA damaging agents than gcn5Δ or rtt109Δ single mutants alone (Figure 1B), and these double mutant cells exhibited an abnormal cell cycle profile with an increased amount of debris, an indication of cell death (Figure 1C). Furthermore, gcn5Δ H3K56R double mutant cells grew slowly and exhibited a similar pattern of DNA damage sensitivity to that of gcn5Δ rtt109Δ cells (Figure 1A and Supplemental Figure 1). Lastly, expression of wild-type Gcn5 or Rtt109 from a plasmid in gcn5Δ rtt109Δ double mutant cells rescued the synthetic growth defects and DNA damage sensitivity of the double mutant cells whereas expression of the Gcn5 catalytic dead mutant, gcn5 E173Q (Marmorstein, 2004), or Rtt109 catalytic dead mutant, rtt109 D89N (Han et al., 2007a), had no effect (Figure 1D and E). These results suggest that Gcn5 functions in parallel with Rtt109 in growth and response to DNA damaging agents and that the slow growth and DNA damage sensitivity of gcn5Δ rtt109Δ cells are due to loss of the enzymatic activity of both Rtt109 and Gcn5.
Figure 1.
Gcn5 and H3 lysine 56 acetylation function in parallel in growth and response to DNA damaging agents. Data presented in Table S1 indicate the presence of synthetic genetic interactions between the HAT, GCN5, and ASF1, a regulator of H3K56Ac. (A) The gcn5Δ mutant exhibits synthetic phenotypes with rtt109Δ and H3K56R mutants. Ten fold serial dilutions of wild-type (WT) or mutant yeast cells with relevant genotype indicated were spotted onto normal growth media, YPD, or media containing the indicated concentration of the DNA damaging agents hydroxyurea (HU), methyl methane sulfonate (MMS), or camptothecin (CPT). Full data presented in Figure S1. (B) The gcn5Δ rtt109Δ double mutant cells are more sensitive to CPT than either single mutant. Yeast cells were treated with the indicated concentration of CPT for 2 hours, and the percentage of surviving cells was reported. (C) FACS analysis of the DNA content of unsynchronized yeast cells. (D–E) The catalytic activity of Gcn5 (D) and Rtt109 (E) is required for cell growth and sensitivity towards DNA damaging agents. Mutant cells transformed with either plasmid for wild-type GCN5, gcn5 E173Q, or empty vector (gcn5Δ rtt109Δ) were spotted onto media containing different concentrations of HU (D). Similar experiments were also performed for wild-type RTT109 or the rtt109 D89N (E).
Both Gcn5 and Rtt109 acetylate histone H3 lysine 27
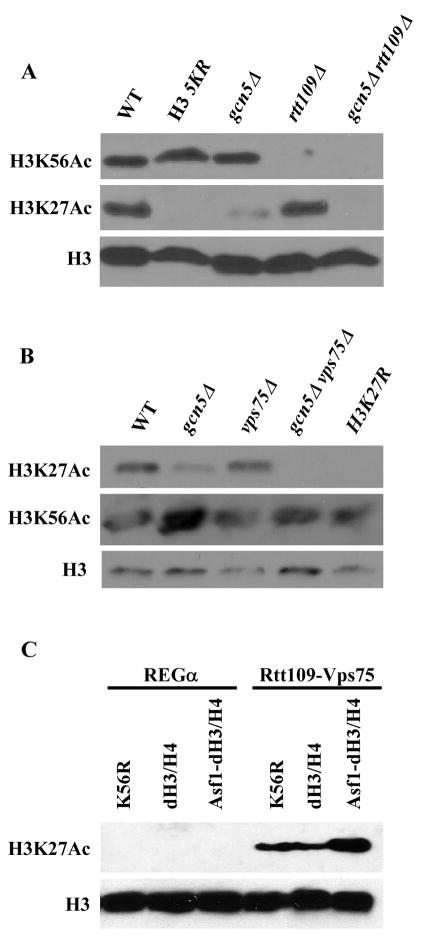
It is known that both Gcn5 and Rtt109 can acetylate lysine 9 and 23 of histone H3 (Berndsen et al., 2008; Fillingham et al., 2008). Recently, it was reported that Gcn5 is involved in H3K56Ac in human cells (Tjeertes et al., 2009). Because acetylation of lysine residues 9, 27 and 56 is present on newly-synthesized H3 (Kuo et al., 1996; Masumoto et al., 2005), we first asked whether Gcn5 is required for H3K56Ac in yeast cells. As reported (Han et al., 2007a), deletion of RTT109 abolished H3K56Ac, whereas deleting GCN5 had no detectable effect on H3K56Ac (Fig. 2A). Next, we analyzed how the gcn5Δ and rtt109Δ mutations affected acetylation of histone H3 lysine 27 (H3K27Ac). H3K27Ac was significantly reduced in gcn5Δ mutant cells compared to wild-type cells, whereas deletion of RTT109 had no apparent effect on the level of H3K27Ac. Importantly, H3K27Ac was not detected in gcn5Δ rtt109Δ mutant cells, suggesting that both Gcn5 and Rtt109 are required for H3K27Ac in vivo.
Figure 2.
Both Gcn5 and Rtt109 acetylate H3 lysine 27 (H3K27Ac). (A) Cells lacking both GCN5 and RTT109 result in a significant loss of H3K27Ac. The H3 5KR mutant contains mutations at five lysine residues of the N-terminus of H3 (9, 14, 18, 23 and 27). (B) Vps75 is essential for H3K27Ac in the absence of Gcn5. Western blot analysis of whole cell extracts showing loss of H3K27Ac in gcn5Δ vps75Δ cells. H3K27R: lysine 27 of H3 was mutated to arginine. (C) The Rtt109-Vps75 complex acetylates H3K27 in vitro. HAT assays were performed using recombinant histone H3-H4 in the presence or absence of the histone chaperone Asf1. Western blot was performed using antibodies against H3 or H3K27Ac.
Vps75 is a component of the Rtt109-Vps75 histone acetyltransferase complex (Han et al., 2007c). Therefore, we asked whether Vps75 was also required for H3K27Ac in yeast cells. Cells lacking VPS75 had no apparent effect on H3K27 acetylation compared to wild-type cells (Figure 2B). However, H3K27Ac was not detected in gcn5Δ vps75Δ cells, whereas H3K56Ac was not affected in gcn5Δ vps75Δ mutant cells compared to wild-type cells (Figure 2B). These results indicate that both Gcn5 and the Rtt109-Vps75 complex are involved in H3K27 acetylation in yeast cells.
Lastly, we tested whether the Rtt109-Vps75 complex acetylated H3K27 in vitro and whether Asf1 enhanced the activity of the Rtt109-Vps75 complex towards H3K27. Recombinant Rtt109-Vps75 complex was incubated with recombinant H3-H4 and co-factor, acetyl-CoA, in the presence or absence of Asf1. The reaction mixtures were analyzed by Western blot using antibodies recognizing H3K27Ac. As shown in Fig. 2C, the Rtt109-Vps75 complex acetylated H3K27 in vitro, and Asf1 stimulated the activity of the Rtt109-Vps75 complex towards H3K27Ac (Figure 2C). Interestingly, replacement of H3K56 with arginine (H3K56R) had no apparent effect on the ability of the Rtt109-Vps75 complex to acetylate H3K27, suggesting that the acetylation of H3K56 by the Rtt109-Vps75 complex is not a prerequisite for this complex to acetylate H3K27. Thus, the Rtt109-Vps75 complex acetylates both H3K27 and H3K56 in vitro and in vivo.
A Gcn5-containing complex works together with Rtt109 in a common process
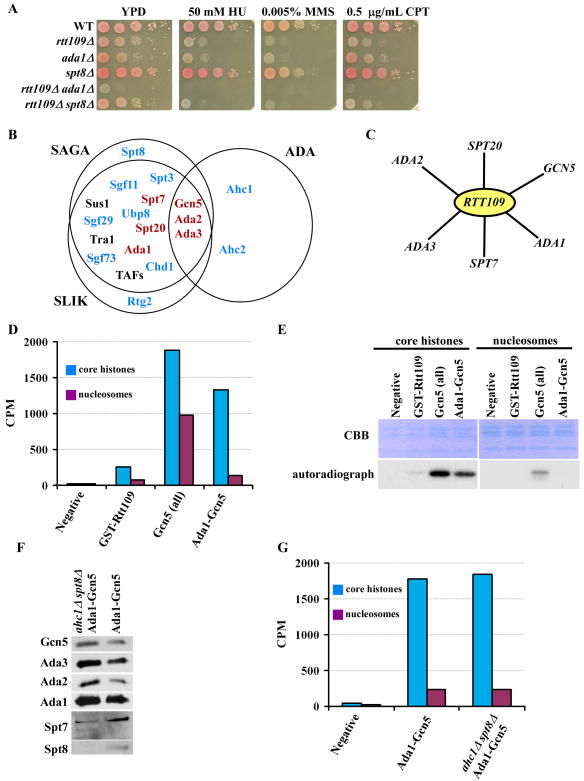
Gcn5 is the catalytic subunit of three KAT complexes, SAGA, ADA and SLIK, with well-known roles in gene transcription. To identify which known Gcn5-containing complex functions in parallel with Rtt109, we deleted all of the non-essential subunits of the SAGA, SLIK and ADA complexes in rtt109Δ mutant cells and compared growth and DNA damage sensitivity of each of the double mutants with that of gcn5Δ rtt109Δ cells (Figure 3A, Supplemental Figure 2). We found that deletion of five subunits, ADA1, ADA2, ADA3, SPT7, and SPT20, in rtt109Δ mutant cells phenocopied gcn5Δ rtt109Δ cells. In contrast, deletion of the unique components of the SAGA (SPT8), SLIK (RTG2) or ADA (AHC1 and AHC2) complexes, as well as genes shared among the Gcn5-containing complexes, SGF29, CHD1, UBP8, SGF73, SGF11, and SPT3, had no apparent effect on the growth and DNA damage sensitivity of rtt109Δ mutant cells (Figure 3A and Supplemental Figure 2). These results demonstrate that Ada1, Ada2, Ada3, Spt7, and Spt20 function with Gcn5 to maintain cell growth and resist DNA damage in the absence of Rtt109.
Figure 3.
A Gcn5-containing complex functions in parallel with Rtt109 in a common process. (A) Mutations in ADA1 but not SPT8 phenocopy the gcn5Δ mutant in the absence of Rtt109. (B) Venn diagram depicting the subunits of the SAGA, SLIK, and ADA complex. Genes in black represent those that were not tested, those in blue are genes that did not show synergistic phenotypes, and in red, are genes that exhibited synthetic phenotypes when combined with deletion of RTT109. Complete genetic data is in Figure S2. (C) Web depicting genetic interactions with RTT109. (D and E) The proteins that co-purified with Ada1 and Gcn5 (Ada1-Gcn5) have HAT activities different from those purified with Gcn5-TAP (Gcn5-all). Core histones and mono-nucleosomes were used as substrates for the HAT assays and incorporated 3H-CoA acetate was quantified (D). The HAT assay samples were also resolved on SDS-PAGE and revealed by Coomassie Brilliant Blue staining (CBB) (E, top) or by fluorography (E, bottom). (F) Western blot analysis of indicated components of the Ada1-Gcn5 protein complex purified from wild-type (Ada1-Gcn5, Ada1-TAP Gcn5-Flag) cells or cells lacking Ahc1 and Spt8 (ahc1Δ spt8Δ). (G) The HAT activity of complexes purified from wild-type (Ada1-Gcn5) cells and ahc1Δ spt8Δ cells.
Cells lacking Ada1, Spt7 and Spt20 exhibit sensitivity towards DNA damaging agents (Supplemental Figure 2) as well as defects in transcription (Sterner et al., 1999), suggesting that Ada1, Spt7 and Spt20 function together in a complex. Ada2, Ada3 and Gcn5 are known to form the core subunits of the SAGA, ADA and SLIK complexes (Balasubramanian et al., 2002; Candau and Berger, 1996; Grant et al., 1997). Therefore, in an attempt to characterize the complex containing Gcn5, Ada1, Ada2, Ada3, Spt7 and Spt20, tagged Ada1 (Ada1-TAP) was first purified from yeast cells, and the resulting proteins were further purified using Gcn5 tagged with the Flag epitope (see Experimental Procedures) (Ada1-Gcn5). As a comparison, we also purified Gcn5 from yeast cells using tandem affinity purification (Gcn5-all). The Gcn5-containing complex purified using the Ada1-Gcn5 procedure acetylates predominantly H3 of core histones, with limited activity against nucleosomal H3. On the other hand, the Gcn5-all complex acetylated both nucleosomal H3 as well as free H3 (Figure 3D and E). Because the SAGA and ADA complexes acetylated nucleosomal H3 (Grant et al., 1997), the Ada1-Gcn5 complex we purified may be distinct from the SAGA and ADA complex. To test this idea further, we followed the same Ada1-Gcn5 purification procedure and purified a complex from cells lacking both Ahc1 (the structural component of the ADA complex (Eberharter et al., 1999) and Spt8 (the unique subunit of SAGA). Deletion of AHC1 and SPT8 did not affect co-purification of Ada2, Ada3 and Spt7 with Ada1 and Gcn5 (Figure 3F) or the activity profile of the Ada1-Gcn5 complex (Figure 3G). Based on these genetic and biochemical studies, we suggest that a Gcn5-containing complex acetylates the N-terminal lysine residues of newly-synthesized histone H3.
Cells lacking Gcn5 or expressing the H3 5KR mutant exhibit defects in cell cycle progression
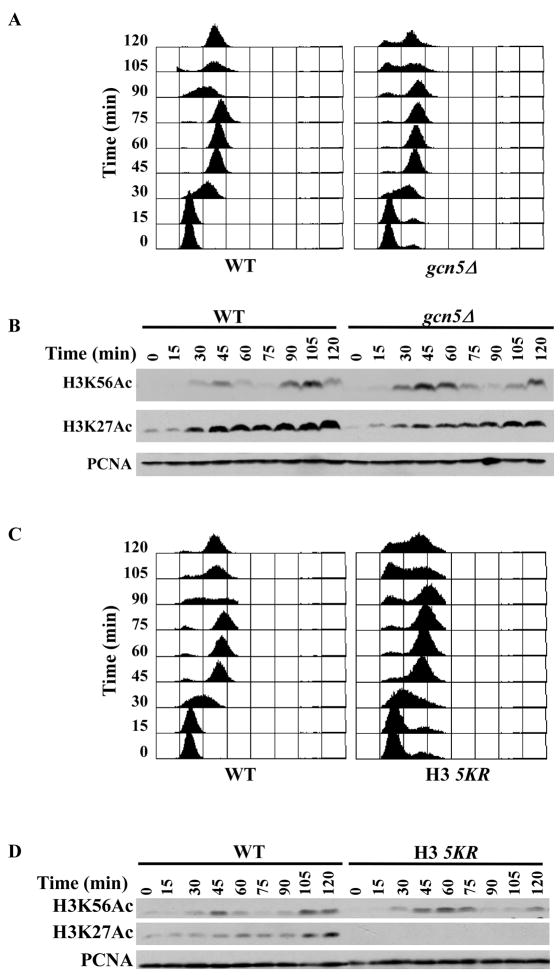
The complex integrity of SAGA, SLIK and ADA, as well as their role in transcription, is mediated partly by their unique subunits (Eberharter et al., 1999). The fact that loss of the unique components of these complexes had no apparent effect on the cell growth and DNA damage sensitivity of rtt109Δ mutant cells suggests that the DNA damage sensitivity observed in gcn5Δ rtt109Δ (as well as ada1Δ rtt109Δ, ada2Δ rtt109Δ and ada3Δ rtt109Δ, spt7Δ rtt109Δ, and spt20 rtt109Δ) cells is not likely due to the loss of function of any of the three previously characterized Gcn5-containing complexes in gene transcription. Instead, we suggest that Gcn5 may function in parallel with Rtt109 to maintain genome stability in a manner that is independent of the transcriptional function of the ADA, SAGA and SLIK complexes. To test this idea further, we first asked whether the gcn5Δ mutant affected cell cycle progression. Because Gcn5 acetylates the H3 N-terminus, we also analyzed the cell cycle progression of cells expressing the H3 mutant containing mutations at five lysine residues (H3 5KR: K9R, K14R, K18R, K23R and K27R) of the H3 N-terminus. Wild-type, gcn5Δ and H3 5KR cells were arrested at G1 using α-factor and then released into fresh media to allow cell cycle progression. Every 15 minutes, aliquots of cells were removed for analysis of DNA content by FACS and H3K56Ac by Western blot. As shown in Figure 4A and C, wild-type, gcn5Δ and H3 5KR mutant cells entered S phase with similar kinetics. However, both gcn5Δ and H3 5KR mutant cells entered the next cell cycle 15 minutes later than wild-type cells. This result is consistent with previous observations that gcn5Δ mutant cells accumulate at G2/M phase of the cell cycle (Zhang et al., 1998). Like wild-type cells, H3K56Ac started to peak at 45 minutes in gcn5Δ and H3 5KR mutant cells. However, the decline of this modification was 15 minutes later in the gcn5Δ and H3 5KR mutant cells compared to wild-type cells (Figure 4B and D). Together, these results suggest that gcn5Δ and H3 5KR mutant cells exhibit abnormal cell cycle progression and deregulation of H3K56Ac during the cell cycle.
Figure 4.
The gcn5Δ and H3 5KR mutants affect cell cycle progression and cell cycle dynamics of H3K56Ac. (A-B) The gcn5Δ cells exhibit a prolonged G2/M phase and persistence of high levels of H3K56Ac compared to wild-type cells. Every 15 minutes after release from G1, aliquots of cells were removed for analysis of DNA content (A) or analysis of H3K56Ac and H3K27Ac by Western blot (B). (C-D) Cells expressing the H3 5KR mutant showed a prolonged G2/M phase and persistence of a high level of H3K56Ac compared to wild-type cells. The experiments were performed as described in A and B.
Gcn5 is required for the maintenance of genome stability
Next, we asked whether the gcn5Δ mutant had a higher level of spontaneous chromosome breaks than wild-type cells using the Rad52 foci assay (Lisby et al., 2001). Rad52, a mediator of DNA double strand break repair, is localized throughout the nucleus in the absence of chromosome breaks. In the presence of chromosome breaks, Rad52 molecules accumulate and form foci at chromosome break sites. As reported, about 10% of wild-type cells formed Rad52 foci, and all these cells were in S/G2/M phase of the cell cycle (Figure 5A) (Lisby et al., 2001). Moreover, significantly more rtt109Δ mutant cells displayed Rad52 foci than wild-type cells (Driscoll et al., 2007; Han et al., 2007b). About two-fold (20% vs 10%) more gcn5Δ mutant cells formed Rad52 foci than wild-type cells under normal growth conditions (Figure 5A). Thus, like rtt109Δ mutant cells, gcn5Δ mutant cells exhibit spontaneous chromosome breaks during normal growth conditions that may contribute to abnormal H3K56Ac and cell cycle progression.
Figure 5.
GCN5 is involved in DNA replication-coupled nucleosome assembly. (A) Cells lacking GCN5, ADA2 or ADA3 exhibited a higher level of Rad52 foci compared to wild-type cells. The percentage of cells with Rad52-YFP foci was determined as described in Experimental Procedures. Data represent the mean percentage ± SEM. Mock experiments were also performed in which the genotype of each strain was unknown to the person who performed the experiment. (B) The gcn5Δ mutant cells exhibited higher levels of chromosome breaks than wild-type cells when challenged with a DNA damaging agent. Wild-type or gcn5Δ cells expressing Rad52-YFP were treated with zeocin for 0, 15, or 30 minutes and the percentage of cells containing Rad52 foci was determined. (C–D) The gcn5Δ mutation exhibited a synthetic genetic interaction with mutations in the checkpoint kinases RAD53 (C, rad53-1) and MEC1 (D, mec1-1). (E) The gcn5Δ mutant exhibited slow growth and increased DNA damage sensitivity with mutations in ASF1, CAC1, and RTT106. (F) A summary of genetic interactions observed for GCN5. Those in blue boxes indicate genes that are known to be involved in nucleosome assembly. Complete genetic data is presented in Figure S3.
Like gcn5Δ mutant cells, significantly more ada2Δ and ada3Δ cells formed Rad52 foci than wild-type cells. In contrast, the ahc1Δ mutant, which did not exaggerate growth defects or DNA damage sensitivity of rtt109Δ mutant cells, did not exhibit a significant change in cells with Rad52 foci compared to wild-type cells (Figure 5A). Lastly, we observed that significantly more gcn5Δ mutant cells had Rad52 foci than wild-type cells when both cells were challenged with zeocin, a drug that induces double strand breaks (Figure 5B). These results suggest that Gcn5 functions to maintain genome stability during normal S phase as well as when under insult from DNA damaging agents. Consistent with this idea, gcn5Δ mutant cells exhibited synthetic growth defects with mutations in the checkpoint kinase RAD53 as well as exaggerated sensitivity towards the DNA damaging agent CPT when combined with mutations in RAD53 and MEC1 (another checkpoint kinase) (Figure 5C–D and Supplemental Figure 3A–C). These results provide additional support for the idea that Gcn5 is required to maintain genome stability and that the synthetic phenotypes of gcn5Δ rtt109Δ cells are likely due to the compromised ability of these double mutant cells to maintain genome stability.
GCN5 genetically interacts with genes involved in DNA replication and replication-coupled nucleosome assembly
RTT109 genetically interacts with genes involved in DNA replication (Collins et al., 2007; Han et al., 2007a). Therefore, we tested whether GCN5 genetically interacted with CDC7 and CDC17 using the temperature sensitive (ts) mutants of CDC7 and CDC17, cdc7-1 and cdc17-1, respectively. Cdc7 is a protein kinase required for initiation of DNA replication, and Cdc17 is the catalytic subunit of the DNA polymerase α –primase complex. The gcn5Δ cdc7-1 mutant grew more slowly at permissive and semi-permissive temperatures compared to either single mutant alone. The same was true for the gcn5Δ cdc17-1 double mutant at the semi-permissive temperature (33°C) (Supplemental Figure 3D). The genetic interactions among GCN5, CDC7 and CDC17 suggest that Gcn5, like Rtt109, has a role in DNA replication.
Recently, we have shown that CAF-1 and Rtt106 are two effectors that bind H3K56Ac and promote nucleosome assembly during S phase (Li et al., 2008). Therefore, we examined whether GCN5 genetically interacted with CAC1 (the large subunit of the histone chaperone CAF-1) and RTT106 (Figure 5E and supplemental Figure 3E). The gcn5Δ cac1Δ and gcn5Δ rtt106Δ cells were more sensitive to at least two DNA damaging agents compared to the cac1Δ, rtt106Δ or gcn5Δ single mutants. Moreover, when compared to gcn5Δ asf1Δ double mutant cells, gcn5Δ cac1Δ and gcn5Δ rtt106Δ cells exhibited lesser growth defects and were less sensitive to DNA damaging agents (Figure 5E and supplemental Figure 3E). This web of genetic interactions (Figure 5F) between GCN5 and genes involved in DNA replication, replication-coupled nucleosome assembly and the DNA damage response suggests that Gcn5, like Rtt109, plays a role in DNA replication and/or replication-coupled nucleosome assembly.
Gcn5 and acetylation of five lysine residues at the H3 N-terminus promote efficient nucleosome assembly
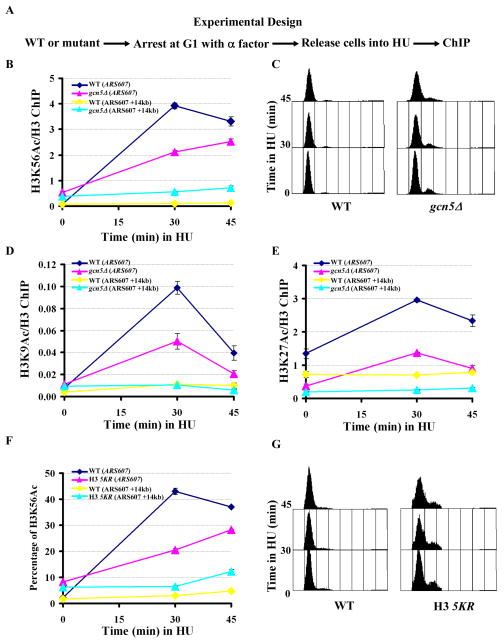
To determine whether Gcn5 is needed for efficient nucleosome assembly during S phase, we used the chromatin immunoprecipitation assay (ChIP) to analyze how the gcn5Δ mutation affected the deposition of newly-synthesized H3, marked by H3K56Ac (Masumoto et al., 2005), onto replicating DNA. Briefly, wild-type and gcn5Δ mutant cells were synchronized at G1 using α-factor and then released into fresh media containing HU for different periods of time. ChIP assays were performed using antibodies recognizing H3K56Ac or H3 (Figure 6A). The ChIP DNA was analyzed using primers amplifying the early replication origin ARS607 and a DNA fragment 14 Kb from ARS607 (ARS607+14kb). While HU has no effect on the firing of early replication origins such as ARS607, it prevents firing of late replication origins and impedes progression of the DNA replication fork (Tercero and Diffley, 2001). Therefore, the replication fork originating from ARS607 cannot reach ARS607+14 Kb in the presence of HU. Significantly more H3K56Ac was detected at replicating DNA (ARS607) than at ARS607+14 kb in wild-type cells, confirming earlier results indicating that replication fork passage is required for deposition of H3K56Ac onto replicating DNA (Li et al., 2008). Importantly, H3K56Ac was significantly reduced at replicating DNA (ARS607) in gcn5Δ mutant cells compared to wild-type cells (Figure 6B and C). Because the overall level of H3K56Ac was not affected in gcn5Δ mutant cells (Figure 4 and Supplemental Figure 4A), the reduction of H3K56Ac at replicating DNA was not likely due to reduction of the overall level of H3K56Ac in gcn5Δ mutant cells. Lastly, compared to extracts prepared from wild-type cells, cell extracts prepared from gcn5Δ mutant cells exhibited a compromised ability to assemble plasmid DNA into nucleosomes (Supplemental Figure 4B). These results provide direct evidence supporting the idea that Gcn5 is required for efficient deposition of H3 onto replicating DNA.
Figure 6.
Gcn5 and five lysine residues at the N-terminus of H3 are required for efficient deposition of newly-synthesized H3 onto replicating DNA. (A) A schematic diagram showing ChIP experimental design. Wild-type or mutant cells (gcn5Δ or H3 5KR) were arrested in G1 using α-factor and then released into HU. At different time points, samples were removed for FACS analysis and ChIP assays using antibodies against H3 or H3K56Ac. (B) The deposition of H3K56Ac onto replicating DNA is compromised in gcn5Δ mutant cells. The ChIP DNA was analyzed using primers that amplify the replication origin ARS607 or a fragment 14 kb away from ARS607 (ARS607+14 kb). The ratio of H3K56Ac ChIP signal over that of H3 was calculated. (C) FACS analysis of the DNA content of wild-type and gcn5Δ mutant cells used in B. (D–E) H3K9Ac (D) and H3K27Ac (E) were detected at replicating DNA. The experiment was performed as described in A and B except that antibodies against H3K9Ac or H3K27Ac were used. (F) Deposition of H3K56Ac was compromised in cells expressing the H3 5KR mutant cells. The experiment was performed as described in A and B except that the percentage of DNA that was precipitated by antibodies against H3K56Ac was calculated by analyzing both ChIP DNA and DNA from whole cells using real-time PCR. (G) FACS analysis of the DNA content of yeast strains used in experiments described in F. Each ChIP experiment was performed independently at least twice. The data represent mean ± SEM of three real-time PCR samples from one experiment. See also Figure S4.
Next we asked whether H3 acetylated at lysine 9 (H3K9Ac) or lysine 27 could be detected on replicating DNA during S phase of the cell cycle using the ChIP assay described above. The antibodies against H3K9Ac and H3K27Ac were specific because more DNA was precipitated from wild-type cells than from H3 mutant cells where lysine 9 and lysine 27 were mutated to arginine (Supplemental Figure 4C). In wild-type cells, the level of H3K9Ac and H3K27Ac at replicating DNA (ARS607) increased when cells entered S phase, whereas the level of H3K9Ac and H3K27Ac at ARS607+14kb did not change significantly (Figure 6D and E). These results confirm that H3K9Ac and H3K27Ac are present on newly-synthesized H3 and demonstrate that replication fork passage is required for the deposition of these two modified forms of H3 onto replicating DNA.
Compared to wild-type cells, deposition of H3K9Ac was significantly compromised in gcn5Δ mutant cells (Figure 6D). Because the level of H3K27Ac was low in G1 (Time 0) in gcn5Δ mutant cells compared to wild-type cells, it was not clear whether deposition of H3K27Ac was also compromised in gcn5Δ mutant cells. We also detected Gcn5 at early replication origins (ARS305 and ARS607) at G1 and at replicating DNA during early S phase. Moreover, the level of Gcn5 at these two origins did not change significantly when cells were released from G1 to early S phase (Supplemental Figure 4D–E). Together, these results are consistent with the idea that Gcn5 acetylates lysine residues of the N-termini of H3 molecules prior to their assembly into nucleosomes.
Lastly, we determined whether cells expressing the H3 5KR mutant affected the deposition of H3K56Ac onto replicating DNA. Compared to wild-type cells, the deposition of H3K56Ac was significantly reduced in cells expressing the H3 5KR mutant. The reduction of H3K56Ac on replicating DNA was not due to a reduced level of H3K56Ac in H3 5KR mutant cells (Supplemental Figure 4A and Figure 4). Thus, acetylation of some or all five lysine residues at the H3 N-terminus, possibly by Gcn5 and Rtt109, is important for the deposition of H3K56Ac onto replicating DNA.
Gcn5 and acetylation of five lysine residues at the H3 N-terminus are important for efficient association of H3 with CAF-1
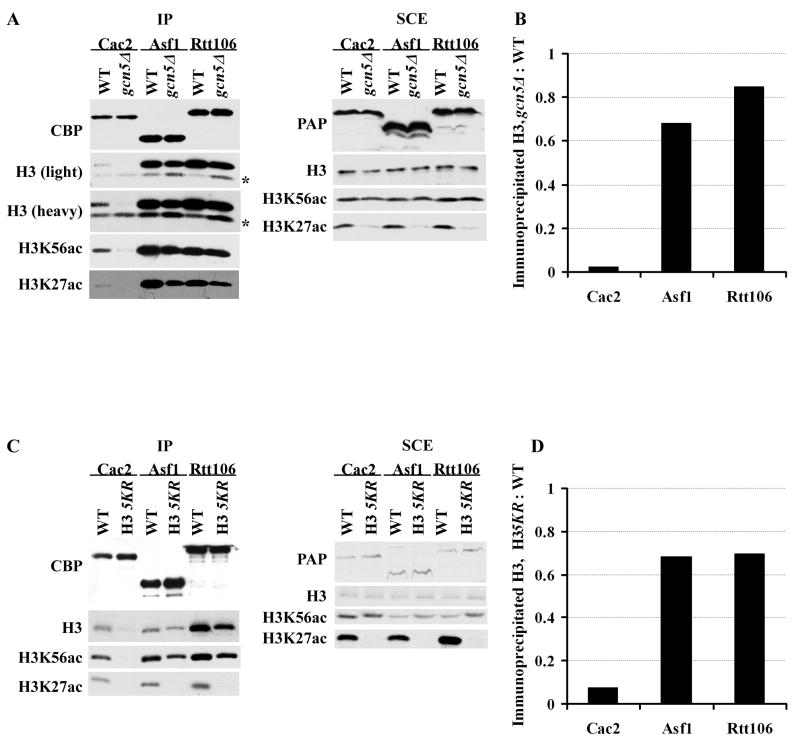
Deposition of newly-synthesized H3, marked by H3K56Ac, requires the histone chaperones CAF-1 and Rtt106 (Li et al., 2008). To understand how Gcn5 and acetylation of lysine residues at the H3 N-terminus impact H3K56Ac deposition, we tested whether Gcn5 and acetylation of five lysine residues at the H3 N-terminus are required for efficient binding of H3 with CAF-1, Asf1 and Rtt106. First, Cac2-, Asf1- or Rtt106-TAP was purified from wild-type and gcn5Δ mutant cells, and co-purified H3 molecules were detected by Western blot using antibodies against H3. As shown in Fig. 7A–B, the association of H3 with Asf1 and Rtt106 was slightly reduced in gcn5Δ mutant cells compared to wild-type cells, whereas the binding of H3 with CAF-1 was reduced dramatically in gcn5Δ mutant cells compared to wild-type cells. The reduction of H3 binding with CAF-1 was not likely due to the reduced level of Cac2, H3 or H3K56Ac in gcn5Δ mutant cells (Fig. 7A, right panel). Thus, Gcn5 is required for the efficient binding of H3 with CAF-1.
Figure 7.
Gcn5 and acetylation of five lysine residues of the H3 N-terminus regulate the binding of H3 with CAF-1. (A–B) The binding of H3 with CAF-1 is significantly reduced in gcn5Δ mutant cells. (A) Cac2-, Asf1- and Rtt106-TAP were purified from wild-type and gcn5Δ mutant cells, and co-purified proteins as well as proteins in soluble cell extracts (SCE) were detected by Western blot using antibodies against H3, H3K56ac, H3K27ac, calmodulin binding peptide (CBP) and IgG (PAP). * indicates non-specific band. (B) Quantification of the co-purified H3. The H3 intensity shown in A was quantified, and the ratio of the H3 that co-purified from each protein from the gcn5Δ cells over wild type cells was reported. (C–D) The association of H3 with CAF-1 is significantly reduced in cells expressing the H3 5KR mutant cells. The experiment was performed as described in A and quantification in D was performed as described in B.
Using the same approach, we also observed that mutation of five lysine residues at the H3 N-terminus also resulted in a significant reduction in the binding of H3 with CAF-1 and a modest effect on the association of H3 with Asf1 and Rtt106 (Figure 7C and D). These results are consistent with our previous studies indicating that in cells lacking H3K56Ac, the binding of H3 with Rtt106 is reduced to an undetectable level by Western blot, and the association of H3 with CAF-1 is significantly reduced (Li et al., 2008). Together, these results indicate that acetylation of five lysine residues at the H3 N-terminus, possibly by Gcn5 and Rtt109, is important for the binding of H3 with CAF-1.
Our results appear to contradict a previously published report showing that deletion of the H3 N-terminus has no apparent effect on human CAF-1’s ability to assemble the H3-H4 mutant onto replicating DNA using the in vitro SV40 DNA replication-coupled nucleosome assembly assay (Shibahara et al., 2000). There are two potential explanations for this discrepancy. First, it is possible that human CAF-1 and yeast CAF-1 are regulated differently by acetylation of H3 N-termini. In this regard, we notice that unlike the acetylation pattern on newly-synthesized H4, acetylation patterns on newly-synthesized H3 appear to diverge from yeast to human (Benson et al., 2006; Sobel et al., 1995). Alternatively, the in vivo assays used in this study are more sensitive towards the detection of the contribution of the H3-N-terminus to CAF-1-mediated nucleosome assembly than the in vitro assay used previously. Future studies are needed to address these two possibilities. Nonetheless, our studies are consistent with yeast genetic studies suggesting that the N-terminus of yeast H3 has a role in nucleosome assembly (Li et al., 2008; Ma et al., 1998).
Discussion
Here we show that Gcn5 functions in parallel with Rtt109 to promote DNA replication-coupled nucleosome assembly. Cells lacking GCN5 exhibit synthetic growth defects and sensitivity to DNA damaging agents in combination with mutations that perturb H3K56 acetylation (rtt109Δ, asf1Δ, and H3K56R), DNA replication (CDC7 and CDC17), the DNA damage response (RAD53 and MEC1), and two histone chaperones (CAC1 and RTT106) that are involved in DNA replication-coupled nucleosome assembly. This web of genetic interactions suggests that Gcn5 has a role in DNA replication independent of its role in gene transcription. Moreover, we have shown that Gcn5 and five lysine residues of the H3 N-terminus are required for efficient deposition of new H3 onto replicating DNA and for efficient binding of H3 with CAF-1. Together, these results demonstrate that Gcn5 promotes DNA replication-coupled nucleosome assembly, partly through acetylation of lysine residues at the N terminus of new H3 molecules prior to their deposition onto replicating DNA.
A role for Gcn5 in DNA replication-coupled nucleosome assembly
In yeast and mammalian cells, Gcn5-containing complexes are known for their roles in transcriptional activation and elongation (Sterner et al., 1999). Therefore, it is possible that the gcn5Δ mutant phenotype displayed in the absence of Rtt109 may be an indirect effect of mis-regulated transcription of genes involved in nucleosome assembly following DNA replication. However, two lines of evidence argue against this idea. First, gene expression profiling using microarrays indicates that the expression level of genes critical for DNA replication-coupled nucleosome assembly such as PCNA, CAF-1 and Rtt106 are not affected to a detectable degree in gcn5Δ cells (Lee et al., 2000). Second, loss of ten genes, including SPT8 and UBP8, known to function with Gcn5 in transcriptional regulation did not display the same phenotype as the gcn5Δ mutant when combined with the rtt109Δ mutation. Because genes whose transcription are regulated by Gcn5 overlap with or depend on these ten genes (Grant et al., 1997; Grant et al., 1999; Lee et al., 2000; Sterner et al., 1999), it is not likely that the defects in nucleosome assembly observed in gcn5Δ mutant cells, as well as the synthetic defect in response to DNA damaging agents of gcn5Δ rtt109Δ mutant cells, are solely due to consequences of impaired gene transcription. Because Rtt109 promotes nucleosome assembly following DNA replication and DNA repair (Chen et al., 2008; Li et al., 2008), we propose that Gcn5 and Rtt109 function in parallel to promote efficient DNA replication-coupled nucleosome assembly. We provide the following data to further support this conclusion. First, like other genes involved in replication-coupled nucleosome assembly, Gcn5 is required for maintaining genome stability. Second, GCN5 genetically interacts in a negative way with genes involved in DNA replication (CDC7 and CDC17), the DNA damage response (RAD53 and MEC1) as well as three histone chaperones involved in DNA replication-coupled nucleosome assembly (CAF-1, ASF1 and RTT106). More importantly, deposition of newly-synthesized H3, marked by H3K56Ac, onto replicating DNA is compromised in gcn5Δ mutant cells, and cell extracts prepared from gcn5Δ mutant cells are less efficient at assembling DNA into nucleosomes. Together, these results reveal a role for Gcn5 in replication-coupled nucleosome assembly.
How is Gcn5 involved in nucleosome assembly? We presented several lines of evidence supporting a model in which Gcn5 promotes nucleosome assembly in part by acetylating lysine residues at the H3 N-terminus that contribute to the association of H3 with CAF-1. CAF-1 is the primary histone chaperone that promotes replication-coupled nucleosome assembly in yeast and mammalian cells (Stillman, 1986). First, cells lacking Gcn5 exhibit similar defects in cell cycle progression as those expressing the H3 5KR mutant that contains mutations at five lysine residues of the H3 N-terminus. Second, gcn5Δ and H3 5KR mutant cells exhibit similar defects in the deposition of new H3 onto replicating DNA and a similar reduction of H3 binding to CAF-1. Thus, we propose that Gcn5 acetylates the N-terminus of H3, promoting replication-coupled nucleosome assembly, in part by increasing the binding of H3 with CAF-1.
However, cells lacking Gcn5 or expressing the H3 5KR mutant are more sensitive to DNA damaging agents than cells lacking Cac1, the large subunit of CAF-1. Therefore, the DNA damage sensitivity of gcn5Δ or H3 5KR mutant cells is not solely due to the reduced binding of H3 with CAF-1. In budding yeast, Gcn5 is also involved in DNA repair by unknown mechanisms (Tamburini and Tyler, 2005). It has been shown that Gcn5 is recruited to the break site, and acetylation of the H3 N-terminus increases following generation of chromosome breaks (Tamburini and Tyler, 2005). Thus, analogous to H3K56Ac that promotes nucleosome assembly following DNA replication as well as DNA repair (Chen et al., 2008; Li et al., 2008), Gcn5 may also promote nucleosome assembly following repair of damaged DNA. Thus, the DNA damage sensitivity observed in gcn5Δ or H3 5KR mutant cells may be a manifestation of the combined defects in both DNA replication and DNA repair of these mutant cells.
In mammalian cells, it has been reported recently that Gcn5 and p300/CBP are the mammalian H3K56Ac KATs (Das et al., 2009; Tjeertes et al., 2009). Moreover, H3K56Ac is important to maintain genome stability (Yuan et al., 2009) and promote replication-coupled nucleosome assembly in mammalian cells (Das et al., 2009). Therefore, it would be interesting to determine whether Gcn5 also has a role in replication-coupled nucleosome assembly in mammalian cells, and if so, whether Gcn5 functions in this process through acetylation of lysine residues at the H3 N-terminus and/or H3K56.
Gcn5 functions with other proteins to promote nucleosome assembly
Gcn5 is a component of three distinct complexes, ADA, SAGA and SLIK. Other components regulate the integrity and activity of the Gcn5-containing complexes and modulate Gcn5’s role in gene transcription. For instance, Ahc1, one of the unique components of the ADA complex, is required for the structural integrity of the ADA complex (Eberharter et al., 1999). Ada2 and Ada3, two components of the SAGA complex, target Gcn5 to acetylate nucleosomal histones (Grant et al., 1997). Ada1, Spt7 and Spt20 are required for the structural integrity of the SAGA complex (Sterner et al., 1999). We have utilized a genetic approach to identify five genes, ADA1, ADA2, ADA3, SPT7 and SPT20, that when mutated in rtt109Δ mutant cells, displayed a similar cell growth and DNA damage sensitivity phenotype as deletion of GCN5. These genetic interactions are consistent with recent genome-wide studies indicating that RTT109 exhibits synthetic growth defects with mutation of the core subunits of the SAGA complex, including ADA1, ADA2, ADA3, and GCN5 (Lin et al., 2008). Moreover, we have shown that cells lacking ADA1, ADA2, ADA3, SPT7, or SPT20 alone were also sensitive to DNA damaging agents. These results suggest that these five gene-products function with Gcn5 to promote nucleosome assembly. Supporting this idea, we purified a complex using Ada1 and Gcn5 as bait, and this complex acetylates H3 in core histones more efficiently than nucleosomal H3. While it remains to be determined how these five gene products function with Gcn5 to promote replication-coupled nucleosome assembly, it is possible that these five gene products regulate the ability of Gcn5 to acetylate newly-synthesized H3 for efficient deposition by CAF-1.
Involvement of multiple acetylation sites and KATs in replication-coupled nucleosome assembly
Acetylation at multiple sites on H3 and H4 has been implicated in replication-coupled nucleosome assembly. These include acetylation of lysine residues 5, 8, and 12 of newly-synthesized H4, acetylation of the five lysine residues at the H3 N-terminus and H3K56Ac. Because H3K56Ac promotes nucleosome assembly by increasing the binding affinity of H3 with CAF-1 and Rtt106 (Li et al., 2008), and acetylation of lysine residues of the H3 N-terminus contributes to the binding of H3 with CAF-1, it is likely that acetylation of lysine residues on H4 also regulate the binding of histones with histone chaperones involved in replication-coupled nucleosome assembly. A recent study shows that acetylation of both K5 and K8 of H4 cooperate to bind one binding pocket of a single bromodomain of Brdt (Moriniere et al., 2009). Therefore, it is tempting to speculate that acetylation of multiple sites on H3 and H4 may coordinate the binding of H3-H4 with CAF-1 and other histone chaperones.
In addition to Gcn5, several KATs including Rtt109, Elp3 (Table 1, (Li et al., 2009)) and Hat1 have implicated roles in replication-coupled nucleosome assembly. This provides a possible explanation for why assembly of new H3-H4 is essential for cell viability (Kim et al., 1988), yet none of these KATs are. Moreover, recent studies suggest that histone acetyltransferases may participate in distinct steps of replication-coupled nucleosome assembly. In addition to acetylating new histone H3-H4 to promote nucleosome assembly, some of these lysine acetyltransferases may acetylate parental nucleosomes. Following DNA replication, parental nucleosomal histones must be transferred to newly-synthesized DNA. It is proposed that human Asf1 can disrupt parental nucleosomes and promote the transfer of parental H3-H4 to DNA behind replication forks (Groth et al., 2007a). How this process occurs is still an enigma. Analogous to the role of acetylation in the promotion of nucleosome remodeling during gene transcription, it is possible that some of these KATs acetylate parental nucleosomes for nucleosome disassembly by Asf1 or remodeling by chromatin remodeling complexes. In this regard, we found that Gcn5 can be detected at replication origins in both G1 and early S phase. Thus, it is possible that in addition to its role in acetylating new H3, Gcn5 may regulate acetylation of parental nucleosomes, facilitating their disassembly. Future studies are needed to address how these lysine acetyltransferases coordinate to promote efficient nucleosome assembly and to determine the molecular mechanisms by which Gcn5 functions in replication and repair.
Experimental Procedures
Yeast strains and Plasmids
All yeast strains were derived from W303-1 or BY4741 and are listed in the supplemental yeast strain table. Strains were made using standard yeast procedures and media. All plasmids were constructed using standard molecular biology procedures and were confirmed by sequencing.
DNA damage sensitivity assays
Freshly grown cells were taken and diluted to 0.6 (unless otherwise indicated) at OD600. Then 10-fold serial dilutions of each yeast stain were performed. Five microliters of yeast cells at different dilutions were then dotted on YPD media or YPD media containing different concentrations of drugs, camptothecin (CPT), methyl methane sulfonate (MMS) and hydroxyurea (HU). To select plasmids, SCM dropout media containing different concentrations of DNA damaging agents was used. Images were taken after incubation for 2–5 days. Unless indicated, plates were incubated at 30°C.
Detection of Rad52-YFP foci
Cells expressing Rad52 tagged with yellow fluorescent protein (YFP) at its C-terminus were grown to exponential phase at 25 °C, harvested and fixed in PBS with 4% formaldehyde. To test the effect of the DNA damaging agent zeocin, cells were treated with 100 μg/ml zeocin for varying amounts of times and then fixed. After washing and sonication, cells were placed on a glass slide with 1% agarose. A single DIC image and 11 YFP images obtained at 0.3-μm intervals along the z-axis were captured for each frame using a Zeiss LSM10 confocal microscope. Rad52-YFP foci were counted by inspecting images of all focal planes. Percentages of cells in S/G2/M phase containing YFP foci were calculated. For each strain, about 200–1000 cells were scored for Rad52-YFP foci.
Gcn5 complex purification
Gcn5 alone was purified using the TAP purification procedures as described (Li et al., 2008). We also designed a two-step strategy to enrich Ada1-Gcn5 containing complex. First, we made a yeast strain (ada1-TAP, gcn5Δ. GCN5-Flag), purified Ada1 first using the IgG sepharose. After elution of proteins using the TEV protease, proteins were further purified using antibodies against the Flag epitope and eluted with Flag peptides (see Supplemental Experimental Procedures for details). Eluted proteins were used for HAT assays and analyzed by Western blot.
Chromatin immunoprecipitation (ChIP) Assays
To determine the forms of histones deposited onto replication forks, ChIP assays were performed as previously described (Huang et al., 2007; Li et al., 2008). Following collection of ChIP DNA, quantitative PCR was performed to quantify ChIP DNA as well as whole cell DNA using primer pairs amplifying replication origins or fragments of downstream of the replication origins as indicated in Figure legends. The data were presented either as percentage of ChIP DNA over while cell DNA or ratio of ChIP signals from samples using antibodies against a modified form of histones over those using antibodies against histone H3.
Western blot analysis of modified forms of histone H3 and whole histone H3 was described in detail in the Supplemental Experimental Procedures. The HAT assays were performed as described (Han et al., 2007a).
Supplementary Material
Acknowledgments
We would like to thank Drs. Alain Verreault, Shelly Berger, Michael Weinreich, Jessica Tyler and Steve Kron for reagents and Dr. Alabert for the protocol to visualize Rad52 foci. This work is supported by grants from the NIH (GM719729 and GM 81838). RB is supported by the Mayo Clinic Sydney Luckman Family Predoctoral Fellowship, and JH is supported by Kendall-Mayo postdoctoral fellowship. We would also like to thank Brian Davies, Qing Li and Michael Guy for their critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ai X, Parthun MR. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol Cell. 2004;14:195–205. doi: 10.1016/s1097-2765(04)00184-4. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- Benson LJ, Gu Y, Yakovleva T, Tong K, Barrows C, Strack CL, Cook RG, Mizzen CA, Annunziato AT. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J Biol Chem. 2006;281:9287–9296. doi: 10.1074/jbc.M512956200. [DOI] [PubMed] [Google Scholar]
- Berndsen CE, Tsubota T, Lindner SE, Lee S, Holton JM, Kaufman PD, Keck JL, Denu JM. Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75. Nat Struct Mol Biol. 2008;15:948–956. doi: 10.1038/nsmb.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candau R, Berger SL. Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo. J Biol Chem. 1996;271:5237–5245. doi: 10.1074/jbc.271.9.5237. [DOI] [PubMed] [Google Scholar]
- Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Kron SJ. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol Cell Biol. 2002;22:8215–8225. doi: 10.1128/MCB.22.23.8215-8225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Sterner DE, Schieltz D, Hassan A, Yates JR, 3rd, Berger SL, Workman JL. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol. 2008;28:4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grant PA, Eberharter A, John S, Cook RG, Turner BM, Workman JL. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007a;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007b;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007a;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem. 2007b;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- Han J, Zhou H, Li Z, Xu RM, Zhang Z. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J Biol Chem. 2007c;282:14158–14164. doi: 10.1074/jbc.M700611200. [DOI] [PubMed] [Google Scholar]
- Huang S, Zhou H, Tarara J, Zhang Z. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. Embo J. 2007;26:2274–2283. doi: 10.1038/sj.emboj.7601670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim UJ, Han M, Kayne P, Grunstein M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 1988;7:2211–2219. doi: 10.1002/j.1460-2075.1988.tb03060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- Li Q, Fazly AM, Zhou H, Huang S, Zhang Z, Stillman B. The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet. 2009;5:e1000684. doi: 10.1371/journal.pgen.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Qi Y, Lu JY, Pan X, Yuan DS, Zhao Y, Bader JS, Boeke JD. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 2008;22:2062–2074. doi: 10.1101/gad.1679508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci U S A. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XJ, Wu J, Altheim BA, Schultz MC, Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc Natl Acad Sci U S A. 1998;95:6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R. Structural and chemical basis of histone acetylation. Novartis Found Symp. 2004;259:78–98. discussion 98–101, 163–109. [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Shen X. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol. 2009;10:373–384. doi: 10.1038/nrm2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Parthun MR. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol Cell Biol. 2002;22:8353–8365. doi: 10.1128/MCB.22.23.8353-8365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci U S A. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Shibahara K, Verreault A, Stillman B. The N-terminal domains of histones H3 and H4 are not necessary for chromatin assembly factor-1- mediated nucleosome assembly onto replicated DNA in vitro. Proc Natl Acad Sci U S A. 2000;97:7766–7771. doi: 10.1073/pnas.97.14.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009 doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8:1747–1753. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.