Abstract
Nuclear receptors (NRs) are ligand-regulated transcription factors that recruit coregulators and other transcription factors to gene promoters to effect regulation of tissue-specific transcriptomes. The prodigious rate at which the NR signaling field has generated high content gene expression and, more recently, genome-wide location analysis datasets has not been matched by a committed effort to archiving this information for routine access by bench and clinical scientists. As a first step towards this goal, we searched the MEDLINE database for studies, which referenced either expression microarray and/or genome-wide location analysis datasets in which a NR or NR ligand was an experimental variable. A total of 1122 studies encompassing 325 unique organs, tissues, primary cells, and cell lines, 35 NRs, and 91 NR ligands were retrieved and annotated. The data were incorporated into a new section of the Nuclear Receptor Signaling Atlas Molecule Pages, Transcriptomics and Cistromics, for which we designed an intuitive, freely accessible user interface to browse the studies. Each study links to an abstract, the MEDLINE record, and, where available, Gene Expression Omnibus and ArrayExpress records. The resource will be updated on a regular basis to provide a current and comprehensive entrez into the sum of transcriptomic and cistromic research in this field.
The Nuclear Receptor Signaling Atlas (NURSA) has archived expression array and genome-wide location analysis studies for many receptors and ligands on its website, www.nursa.org.
Nuclear receptors (NRs) comprise a large superfamily of evolutionarily conserved ligand-activated transcription factors that regulate target genes controlling essential biological processes, such as development, reproduction, and metabolism (1). Included in this family are receptors for endocrine steroids (corticosteroids, progesterone, androgens, and estrogens), fat-soluble vitamins A and D, thyroid hormone, fatty acids, and numerous dietary and environmental endocrine-disrupting chemicals (EDCs). Additional members of this family identified by sequence similarity are referred to as orphan receptors, because their ligands remain unknown. NR coregulators are a large, disparate class of molecules that are required for the efficient function of NRs. Because of their specific functional interactions with small molecules, NRs encompass one of the most successful group of targets for drugs for the treatment of a broad range of therapeutic indices, including obesity, diabetes, cancer, and a variety of cardiovascular, metabolic, environmental, senescent, and reproductive pathologies.
Since the advent of high content expression array technology in the early part of the last decade, the NR field has generated a large volume of data that collectively describe the cognate transcriptomes of a wide variety of NR ligands and receptors in many different tissues and cell lines. In addition, chromatin immunoprecipitation (ChIP) using antibodies for specific NRs, paired either with array technology (ChIP-chip) or, more recently, massively parallel sequencing (ChIP-seq), has given rise to an increasing number of genome-wide location analysis (GWLA) studies describing tissue/cell line-specific and ligand-specific NR cistromes (2,3). The sheer volume and scale of these studies is daunting, and a variety of factors have combined to obstruct their accessibility and usability by scientists in the field. Article abstracts are necessarily terse and often equivocally worded where experimental details are concerned. Moreover, the rudimentary integration between MEDLINE and public dataset repositories, such as Gene Expression Omnibus and the European Bioinformatics Institute’s ArrayExpress, typically requires a separate search to locate the raw data. One of the mandates of the Nuclear Receptor Signaling Atlas (NURSA) Bioinformatics Resource is to facilitate access of the NR community to existing public datasets. Accordingly, we have embarked upon an effort to catalog these disparate transcriptomic and cistromics studies in a single central repository, namely the NURSA Molecule Pages. Our initial study functions both as a detailed portrait of the spectrum of tissues, in which programs of gene expression directed by NRs and their ligands function, and as a convenient, freely accessible portal for both bench and clinical scientists into this complex body of data.
Results and Discussion
As part of our curation efforts for NURSA, we maintain lists of common names and synonyms for approximately 50 mammalian NRs, over 250 ligands (including physiological hormones, pharmaceuticals, selective receptor modulators, EDCs, and dietary ligands) (see Fig. 1), and over 300 coregulators (NURSA, 2009, available at www.nursa.org). Articles employing expression microarray-based or GWLA-based technology to characterize NR, NR ligand, or coregulator-dependent gene expression were identified through a systematic search of all fields within the MEDLINE database from July 2000 through May 2010. The initial Perl script-based search of PubMed yielded over 7000 PubMed identifier records which were then manually curated to remove false positives, leaving 1122 true positive expression array datasets corresponding to a total of 325 unique tissues and cell lines, 35 NRs and 91 NR ligands. Platforms ranged from early limited-coverage and two-color expression arrays to those with more comprehensive coverage and, more recently, include tiling genomic arrays for GWLA and microRNA platforms to evaluate NR regulation of microRNAs. The species, tissue/cell RNA source, NR, and NR ligand annotations to which each expression microarray or GWLA dataset was mapped are shown in the Supplemental data (published on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org) along with their corresponding PubMed identifiers.
Figure 1.
Nuclear receptor ligands used to generate expression microarray datasets described in this study. The Physiological category includes endogenously synthesized bioactive ligands, their precursors and metabolites. The Clinical category includes approved therapeutic ligands. EDCs are compounds that have therapeutic or industrial uses but which in the context of nuclear receptor signaling are classified as disrupting chemicals. The Experimental category includes ligands used in basic research as well as therapeutic candidates not yet approved for use in the clinic. Dietary ligands include those ingested as part of the diet. For receptor subfamilies, ligands are pan-agonists unless otherwise indicated by superscript symbols representing the specific isoforms. 15PJG2, 15-Deoxy-δ-12,14-prostaglandin J2; 5α-diol, 5α-androstane 3α,17β-diol; CDCA, chenodeoxycholic acid; DDT, 1-chloro-4-[2,2,2-trichloro-1-(4-chlorophenyl)ethyl]benzene; DHEA, dihydroepiandrosterone; DOCO, docosahexanoic acid; EICO, eicosapentaenoic acid; PCN, pregnenolone carbonitrile. Only ligands currently curated by NURSA were included in the Figure. Datasets for all ligands have been compiled and annotated and are accessible in the Transcriptomics and Cistromics section of the individual Ligand Pages on the NURSA website at www.nursa.org.
The vast majority of the expression array datasets identified involved treatment of a system with a NR ligand. We applied an empirical approach to the molecular annotation of these datasets, such that only the ligand was annotated and no necessary inference was made as to the involvement of a specific receptor. To illustrate the significance of this, an expression profiling study involving the effect of 17β-estradiol (17βE2) on MCF-7 cells was annotated only for 17βE2 and not estrogen receptor (ER)α and ERβ, because this might be taken to imply the direct involvement of these receptors in the regulation of any differentially expressed genes. Given the evidence supporting the role of pregenomic 17βE2 signaling, indirectly regulated genes, and alternative pathways of activation of ERs (4), an unqualified assumption of complete overlap between 17βE2 and ER target genes is invalid. That said, it is widely recognized that ligands and their cognate NRs represent a functional continuum, and for this reason, the gene expression study listings for ligands and their cognate NRs are extensively cross referenced in the web resource user interface.
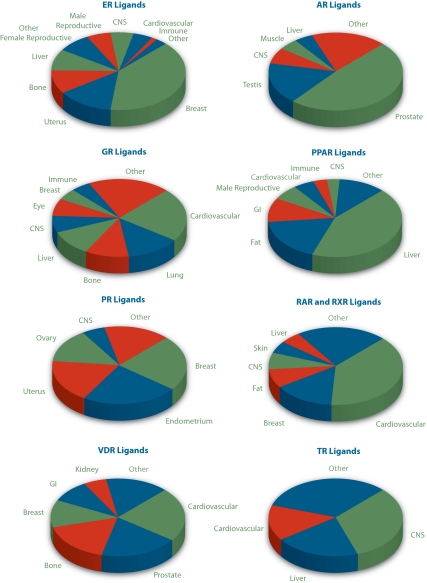
Our study reiterates the large transcriptional compass of NR signaling in physiology and disease and depicts overlapping but distinct functional tissue profiles of individual NR signaling pathways (Fig. 2 and Supplemental data). For the purposes of displaying the data, the individual RNA sources listed in Supplemental data were collapsed into the organ categories shown in the figures. Steroid receptor members of the superfamily (ERs, AR, and PR) and their ligands have significant roles in reproductive tissues; metabolic organs, such as fat, liver, and cardiovascular tissues, figure prominently in the transcriptional profiles of PPARs, glucocorticoid receptor (GR), TR and orphan receptors; the importance of ERs, VDR, and GR in skeletal function can be readily appreciated; and studies involving the central nervous system are well represented in nearly all NR signaling pathways. Conversely, some NR signaling pathways were conspicuous by their absence. The dearth, for example, of published expression array studies involving MR ligands (five in total) was particularly surprising given the critical renal and cardiovascular roles of this NR pathway in regulating blood pressure and fluid balance via the renin-angiotensin system.
Figure 2.
Distinct NR signaling pathways direct tissue-specific programs of gene expression. Data were generated as described in the text. For raw data, please see Supplemental data. Note that due to space constraints, some ligand-receptor groups referred to in Figure 1 are omitted from this figure. These include those for which studies were present in too few numbers (MR and LXR ligands) or which were carried out almost exclusively in a single tissue (PXR, FXR, and CAR ligands in the liver). These studies have, however, been annotated and can be found, along with all the other studies, in the Transcriptomics and Cistromics section of the appropriate Receptor and Ligand Pages on the NURSA website. GI, Gastrointestinal; CNS, central nervous system.
The number of studies devoted to documenting the transcriptomic and, more recently, cistromic properties of orphan NRs in metabolic contexts is striking (Fig. 3A). In a panel of 47 expression array and GWLA experiments involving perturbation of an orphan NR, nearly 75% were performed in tissues or cell lines from metabolically important organs, including fat, liver, and pancreas. In the context of translation, the functional tissue profiles of individual NRs reflect their prominence as druggable targets in a wide range of pathologies and the extent to which both their agonist and antagonist biologies are being leveraged in these therapeutic strategies (Fig. 2 and Supplemental data). For example, synthetic analogs of the RAR agonist all-trans retinoic acid are used in the treatment of acne, acute promyelocytic leukemia, and breast cancer. Moreover, a host of GR agonists is either under development or actively used in the treatment of asthma, macular edema, and other inflammatory conditions, as are VDR agonists in the treatment of osteoporosis and kidney disease and PPAR agonists in hyperlipidemia and type II diabetes. Conversely, the treatment of diseases such as breast cancer and prostate cancer is benefiting from antagonism of signaling pathways mediated by ERα (tamoxifen and raloxifene) and AR (flutamide), respectively.
Figure 3.
A, Orphan NRs are key regulators of gene expression in metabolic organs. Tissues with critical roles in the regulation of energy metabolism [liver, central nervous system, cardiovascular, fat, pancreas, gastrointestinal (GI) tract, and other metabolic tissues, such as kidney, adrenal gland, and bone] constitute nearly 75% of orphan NR-based expression array datasets. Data were compiled from 47 expression array and GWLA datasets in which the function of an orphan NR was experimentally perturbed. Receptors studied were DAX1, SHP, RORα, RORγ, HNF4α, COUPTF-I, ERRα, ERRβ, ERRγ, NUR77, NURR1, and SF-1. Raw data are available in Supplemental data. Datasets for all orphan NRs have been compiled and annotated and are accessible in the Transcriptomics and Cistromics section of the individual Receptor Pages on the NURSA website. B, EDCs are transcriptionally active in female and male reproductive tissues. For details, please see text. Datasets for all EDCs have been compiled and annotated and are accessible in the Transcriptomics and Cistromics section of the individual Receptor and Ligand Pages on the NURSA website. CNS, Central nervous system.
This study also conveys a sense of the importance of selective receptor modulation, which seeks to dial out the undesirable effects of therapeutic NR ligands while retaining their beneficial actions. For example, bone emerged as one of the major settings for the study of GR agonists, reflecting the fact that glucocorticoid treatment for the management of autoimmune and inflammatory diseases is associated with decreased bone formation and increased risk of fracture. Finally, our analysis mirrors the increasing interest in the role of EDCs as disrupting agents in human physiology and as contributing agents in the etiology of cancer and metabolic disorders. Indeed, the gravity with which these compounds are viewed can be appreciated by the fact that The Endocrine Society recently issued a scientific statement on the subject (5). We identified 96 studies involving EDCs since our analysis began in July 2000, which collectively catalog the transcriptional signatures of these molecules, principally in female (breast and uterus) and male (prostate and testis) reproductive tissues but also in metabolic organs (liver and cardiovascular) (Fig. 3B).
The annotated studies are accessible on the NURSA website (www.nursa.org) in a new module in the NURSA Molecule Pages, entitled Transcriptomics and Cistromics. The studies are organized initially into top level headings corresponding to the three species annotated (Homo sapiens, Mus musculus, and Rattus norveggicus), then lower level headings that correspond to the cell line or the organism tissue in question. Cultured cell lines are annotated according to their tissue of origin, (e.g. mammary gland, mammary carcinoma cells, and MCF-7), such that these are logically listed alongside studies with normal tissue or primary cells. Although the archiving of gene expression studies in Gene Expression Omnibus and ArrayExpress regrettably remains far from routine (6), direct links to these records are provided where available, thereby negating the need for time-consuming searches of these databases. The Transcriptomics and Cistromics section of the site will be updated by the NURSA Bioinformatics Resource on a regular quarterly basis as resources permit and we welcome suggestions from members of the community for additions to the database. We anticipate our resource providing the community with a reference tool through which to access both current and legacy datasets to inform and facilitate their research in this area.
Materials and Methods
Article screening strategy
A Perl based regular expression was used in conjunction with National Center for Biotechnology Information’s eUtils (7) to implement the search (scripts available upon request). Specifically, the eUtils eSearch and eFetch were combined in a three-component regular expression to identify and download abstracts from target articles. The first component consisted of terms used to identify expression microarray or GWLA technology. The second component consisted of terms used to eliminate single nucleotide polymorphism genome-wide association studies. The third component consisted of terms specific to the NRs, NR ligands, and coregulators currently curated by the NURSA Bioinformatics Resource.
Supplementary Material
Acknowledgments
We acknowledge helpful comments from Austin Cooney.
Footnotes
This work was supported by National Institute of Diabetes, Digestive and Kidney Disease Nuclear Receptor Signaling Atlas Grant U19 DK62434, with supplementary support from the National Institute of Environmental Health Sciences and the National Heart, Lung and Blood Institute.
Disclosure Summary: The authors have nothing to disclose.
First Published Online August 4, 2010
Abbreviations: AR, Androgen receptor; CAR, constitutive androstane receptor; CDCA, chenodeoxycholic acid; ChIP, chromatin immunoprecipitation; 17βE2, 17β-estradiol; COUP-TF1, chicken ovalbumin upstream promoter factor-1; DAX1, dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1; DDT, 1-chloro-4-[2,2,2-trichloro-1-(4-chlorophenyl)ethyl]benzene; DHEA, dihydroepiandrosterone; 5α-diol, 5α-androstane 3α,17β-diol; DOCO, docosahexanoic acid; EDC, endocrine-disrupting chemical; EICO, eicosapentaenoic acid; ER, estrogen receptor; ERR, estrogen-related receptor; FXR, farnesoid X receptor; GR, glucocorticoid receptor; GWLA genome-wide location analysis; HNF, hepatocyte nuclear factor; LXR, liver X receptor; MR, mineralocorticoid receptor; NR, nuclear receptor; NURR1, Nur-related protein 1; NURSA, Nuclear Receptor Signaling Atlas; PCN, pregnenolone carbonitrile; 15PJG2, 15-deoxy-δ-12,14-prostaglandin J2; PPAR, peroxisome proliferator-activated receptor; PXR, pregnane X receptor; RAR, retinoic acid receptor; RXR, retinoid X receptor; SF-1, steroidogenic factor-1; SHP, short heterodimeric partner; ROR, RAR-related orphan receptor; TR, thyroid hormone receptor; VDR, vitamin D receptor.
References
- Zhang Z, Burch PE, Cooney AJ, Lanz RB, Pereira FA, Wu J, Gibbs RA, Weinstock G, Wheeler DA 2004 Genomic analysis of the nuclear receptor family: new insights into structure, regulation, and evolution from the rat genome. Genome Res 14:580–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung E, Kraus WL 2010 Genomic analyses of hormone signaling and gene regulation. Annu Rev Physiol 72:191–218 [DOI] [PubMed] [Google Scholar]
- Lupien M, Brown M 2009 Cistromics of hormone-dependent cancer. Endocr Relat Cancer 16:381–389 [DOI] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Ström A, Treuter E, Warner M, Gustafsson JA 2007 Estrogen receptors: how do they signal and what are their targets. Physiol Rev 87:905–931 [DOI] [PubMed] [Google Scholar]
- Diamanti-Kandarakis E, Bourguignon JP, Giudice LC, Hauser R, Prins GS, Soto AM, Zoeller RT, Gore AC 2009 Endocrine-disrupting chemicals: an Endocrine Society scientific statement. Endocr Rev 30:293–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner SA, Steffen DL, Stoeckert Jr CJ, McKenna NJ 2008 Much room for improvement in deposition rates of expression microarray datasets. Nat Methods 5:991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Mizrachi I, Ostell J, Panchenko A, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang Y, John Wilbur W, Yaschenko E, Ye J 2010 Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 38:D5–D16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.