Abstract
GnRH, the central regulator of reproductive function, is produced by only approximately 800 highly specialized hypothalamic neurons. Previous studies identified a minimal promoter [GnRH minimal promoter (GnRH-P)] (−173/+1) and a neuron-specific enhancer [GnRH-enhancer (E)1] (−1863/−1571) as regulatory regions in the rat gene that confer this stringent specificity of GnRH expression to differentiated GnRH neurons. In transgenic mice, these two elements target only GnRH neurons but fail to drive expression in the entire population, suggesting the existence of additional regulatory regions. Here, we define two novel, highly conserved, upstream enhancers in the GnRH gene termed GnRH-E2 (−3135/−2631) and GnRH-E3 (−4199/−3895) that increase neuron-specific GnRH expression through interactions with GnRH-E1 and GnRH-P. GnRH-E2 and GnRH-E3 regulate GnRH expression through similar mechanisms via Oct-1, Msx1, and Dlx2, which bind both GnRH-E2 and the GnRH-E3 critical region at −3952/−3895. Overexpression of Dlx2 increases transcription through GnRH-E2 and GnRH-E3. Remarkably, these novel elements are contained within the 3′ untranslated region of the neighboring upstream gene, yet are marked endogenously by histone modification signatures consistent with those of enhancers. Thus, GnRH-E2 and GnRH-E3 are novel regulatory elements that, together with GnRH-E1 and GnRH-P, confer the specificity of GnRH expression to differentiated and mature GnRH neurons.
Two novel enhancers of GnRH expression are located in the 3’UTR of the neighboring upstream gene and specify hypothalamic expression through homeodomain transcription factors.
Neuroendocrine regulation of physiologic processes, including stress responses, metabolism, and reproduction, is mediated by peptide hormones produced by specialized neurosecretory cells in the hypothalamus. The targeting of hypothalamic neuropeptide hormone genes to individual populations of neurons is specified during development as the cells adopt unique cell fates defined by specific patterns of gene expression. The mature differentiated phenotype and function of these neurosecretory cells is ultimately determined by the cell-specific expression of the gene encoding the neuropeptide hormone itself (1).
GnRH is the central regulator of reproductive function at the peak of the hypothalamic-pituitary-gonadal axis, responding to cues and afferent inputs from the central nervous system to control gonadotropin production from the anterior pituitary. GnRH is a decapeptide hormone produced by a small population of approximately 800-1000 highly specialized, terminally differentiated neurons that are dispersed throughout the basal forebrain and medial preoptic area in the anterior hypothalamus and project to the median eminence (2,3). During mouse development, GnRH neurons originate in the olfactory placode and migrate through the nasal septum and cribriform plate to the forebrain, then form a scattered population in the hypothalamus. The differentiated, mature phenotype of the GnRH neuron is defined by the expression of the GnRH gene (4,5,6,7,8,9), because there is a gradual increase in the expression of GnRH as these neurons migrate and acquire their terminally differentiated phenotype (10). Accurate expression of GnRH and proper development of the GnRH neuron are critical for normal reproductive function, because dysregulation leads to reproductive diseases, such as idiopathic hypogonadotropic hypogonadism, Kallmann syndrome, and/or infertility. The precise molecular mechanisms involved in the pathogenesis of these reproductive disorders are unclear and complex due to phenotypic and genetic heterogeneity (11).
GnRH production is tightly regulated at the level of transcription (4,5,6,7,8,9). Neuronal specification of GnRH transcription is controlled by coordinate regulation by combinations of narrowly and more broadly expressed transcription factors acting on GnRH regulatory elements. Evolutionarily conserved regulatory regions of the rat GnRH gene have been characterized using the GT1-7 immortalized hypothalamic neuronal cell line, a model of the mature, terminally differentiated GnRH neuron (12). The 173-bp minimal promoter [GnRH minimal promoter (GnRH-P)] (13) is controlled by the action of several homeodomain transcription factors, including Oct-1 (14), Pbx and Prep (15), Msx and Dlx (16), Nkx2.1 (17), and Otx2 (18). The proximal enhancer [here termed GnRH-enhancer (E)1] (4), which lies between 1.5 and 1.8 kb upstream of the transcription start site (TSS), enhances GnRH expression only in GT1-7 cells and is therefore a neuron-specific enhancer (4). GnRH enhancer (GnRH-E) interacts and synergizes with GnRH-P (19) and binds many transcription factors including Oct-1 (20), Pbx and Prep (15), Msx and Dlx (16), Nkx2.1 (17), and GATA-4 (21). Cell-specific expression of GnRH is thought to be mediated by interactions of a ubiquitous transcription factor like Oct-1, with additional transcription factors exhibiting more tissue-specific patterns of expression. For example, Oct-1 has been shown to interact with Pbx, Prep, and Meis (15), and with Dlx2 (22), to increase neuron-specific GnRH expression and also with Msx1 and Grg/TLE coregulators to repress GnRH expression (22).
A transgene driven by the combination of the GnRH-E1 and GnRH-P (GnRH-E1/GnRH-P), upstream of a β-galactosidase reporter gene, targets expression exclusively to GnRH neurons but to only 50–60% of the total population in adult mice (9). There is an increase in the number of GnRH neurons targeted in transgenic mice by reporter genes driven by regions that include sequences further upstream. Transgenic mice that contain approximately 5 kb of sequence upstream of the GnRH TSS target expression to nearly the entire population of GnRH neurons (6,8,23,24,25). These results suggest the presence of additional discrete regulatory elements likely involved in targeting GnRH expression to the entire population of GnRH neurons. Such elements could include the region between −2.6 and −2.9 kb, previously termed the upstream enhancer (26), but transgenic mouse studies suggest the involvement of regions even further upstream.
In this study, we define two novel, evolutionarily conserved, regulatory regions of the GnRH gene between 3 and 5 kb upstream of the TSS that function as enhancers of GnRH expression. Although located in the 3′ untranslated region (3′ UTR) of the neighboring upstream gene, these regulatory regions are marked in the genome by histone modification signatures of enhancers, bind homeodomain proteins, and confer specificity of GnRH transcription to mature, differentiated GnRH neurons.
Results
Evolutionarily conserved sequences 3–5 kb upstream of the GnRH gene contain novel GnRH regulatory elements within the 3′ UTR of the neighboring upstream gene, Kctd9
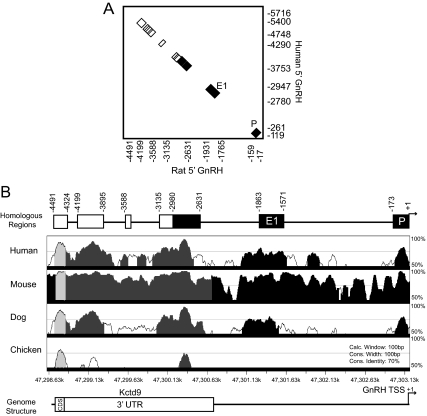
Regions of the genome that are evolutionarily conserved likely serve important biological functions (27). The complete sequencing of the rat genome (28) allowed for analysis of previously unavailable sequences beyond 3 kb upstream of the GnRH gene. To identify putative GnRH regulatory elements, comparative genomics was applied to GnRH gene sequences across mammalian species, an effective strategy used in previous studies (13,26). Approximately 5 kb of rat GnRH gene sequence upstream of the TSS was used in a pairwise Basic Local Alignment Search Tool (BLAST) alignment with the corresponding human sequences. The rat and human BLAST alignment revealed seven regions of homology defined as significantly greater than 75% identity (Fig. 1A). Four highly conserved sequence blocks were identified between −5 and −3 kb. These regions of significant homology were located at −4491/−4324 bp, −4199/−3895 bp, −3588/−3560 bp, and −3135/−2980 bp in the rat sequence. The proximal three regions corresponded to the previously characterized GnRH regulatory elements (4,9,13,26).
Figure 1.
Novel evolutionarily conserved regions between 3 and 5 kb upstream of the GnRH gene are located in the 3′ UTR of the neighboring upstream gene Kctd9. A, Pairwise BLAST analysis of the rat GnRH gene sequence 5 kb upstream of the TSS (x-axis) with the human GnRH gene sequence approximately 6 kb upstream of the TSS (y-axis) is shown. Previously characterized regions are colored in black. P, Promoter; E1, enhancer-1. Four novel highly conserved sequence blocks between −3 and −5 kb of the rat sequence are colored in white. Locations of boundaries of conserved elements for rat and human are denoted by numbers on the x- and y-axes, respectively, relative to the TSS. B, VISTA Browser multispecies alignment of rat upstream GnRH gene sequence as the base genome with the corresponding human, mouse, dog, and chicken upstream GnRH gene sequences. The level of sequence conservation (y-axis) is plotted against the coordinates of the rat chromosomal location (x-axis). Shaded areas indicate regions of significant homology above 70% that are conserved in noncoding sequence (black), the 3′ UTR of Kctd9 (dark gray), and the coding sequence of Kctd9 (light gray), respectively. Schematic diagrams of the rat GnRH gene sequence denoting relative locations of corresponding regions identified by BLAST (A), as well as of the genomic structure of this region are above and below the VISTA plot, respectively. CDS, Coding sequence.
Additional pairwise BLAST analyses were performed with the rat GnRH 5-kb upstream sequence against the corresponding genomic regions of mouse, dog, and cow (Table 1). The percent homology for the total −5-kb region was the highest between rat and mouse (97% conservation) and ranged from approximately 30 to 50% for rat vs. cow, dog, or human. The novel upstream regions have equivalent, if not higher, levels of identity (76–95%) compared with the previously characterized elements, which displayed approximately 80 to 95% homology between rat and mouse or human and approximately 50–80% homology between rat and dog or cow (Table 1).
Table 1.
Homology of conserved GnRH upstream elements among mammalian species
| Rat location | −5 kb | −4491/−4324 | −4199/−3895 | −3588/−3560 | −3135/−2631 | −1863/−1571 | −173/+1 |
|---|---|---|---|---|---|---|---|
| Rat vs. mouse (%) | 97 | 95 | 93 | 90 | 89 | 88 | 94 |
| Rat vs. human (%) | 46 | 85 | 82 | 96 | 82 | 77 | 80 |
| Rat vs. dog (%) | 37 | 85 | 81 | 77 | 76 | 62 | 67 |
| Rat vs. cow (%) | 28 | 82 | 77 | 80 | 76 | 53 | 71 |
Further comparative genomic analysis was performed using the VISTA Browser multiple sequence alignment (29) with the −5-kb rat GnRH sequence as the base genome for comparison with the corresponding human, mouse, dog, and chicken sequences. Comparison of the rat and mouse sequences by VISTA showed that nearly the entire 5-kb upstream region was significantly conserved. The alignment between the rat, human, and dog upstream GnRH sequences identified regions of significant homology that corresponded to six of the seven regions revealed by BLAST, including the novel regions at −4491/−4324 bp, −4199/−3895 bp, and −3135/−2980 bp (Fig. 1B). This conservation of sequences upstream of the GnRH gene appears to be limited only to mammalian species, as the VISTA Browser (Fig. 1B), as well as University of California, Santa Cruz Genome Browser (data not shown), analyses do not show significant conservation with lower vertebrates, except for a small region of homology in chicken from −4491 to −4324 bp and at roughly −2600 bp.
Surprisingly, the VISTA comparison also revealed that the conserved regions corresponding to −4199/−3895 bp and −3135/−2631 bp were contained within the 3′ UTR of the upstream neighboring gene, potassium channel tetramerization domain containing 9 (Kctd9) (Fig. 1B). This gene and its protein product have not been studied but were predicted based on an open reading frame, and mRNAs were found in GenBank for rat, mouse, human, and many other species. The termination of the predicted Kctd9 open reading frame is at −4366 bp upstream of the GnRH TSS, and therefore, the region from −4491 to −4324 bp is most likely conserved due to its function as protein coding for the Kctd9 gene. The polyadenylation signal for Kctd9 is located at −2660 bp, near the 3′ end of region −3135/−2631, and therefore, conserved regions −4199/−3895 and −3135/−2631 are contained entirely within the Kctd9 3′ UTR.
Because the novel evolutionarily conserved regions between 3–5 kb upstream of the GnRH gene were present in the 3′ UTR of the neighboring gene Kctd9, these elements could possibly be involved in the regulation of Kctd9 expression in addition to GnRH expression. GnRH expression is strongest and primarily restricted to mature, differentiated GnRH neurons (10,12). The GT1-7 cell line, a representative model of the mature, terminally differentiated GnRH neuron, was used to study GnRH transcription. These cells secrete high levels of GnRH in a pulsatile fashion and have ultrastructural characteristics of neurosecretory cells (12). To further study neuronal specification of GnRH transcription, expression in GT1-7 cells was compared with the GN11 cell line, a representative model of immature, migratory GnRH neurons that secrete very low levels of GnRH (30), and also the NIH3T3 mouse fibroblast cell line as a nonneuronal cell line control.
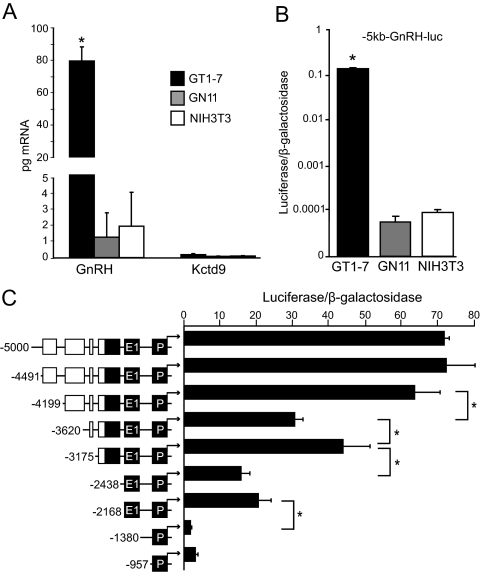
To determine the role of the upstream sequence in GnRH gene regulation, the cell-specific expression patterns of the endogenous GnRH and Kctd9 mRNAs were compared with that of the −5-kb rat GnRH upstream sequence cloned upstream of a luciferase reporter (−5-kb-GnRH-luc). GnRH and Kctd9 mRNA expression levels were compared by reverse transcription of mRNA isolated from GT1-7, GN11, and NIH3T3 cells, followed by quantitative real-time PCR (qPCR). Quantities of GnRH and Kctd9 mRNA were normalized to mRNA levels of cyclophilin B, which showed similar expression levels among the cell lines. GnRH transcript was approximately 40- to 60-fold higher in GT1-7 cells compared with GN11 or NIH3T3 cells (Fig. 2A). The quantities of Kctd9 mRNA, although higher than a control lacking reverse transcriptase (data not shown), were extremely low in all three cell lines and not differentially expressed. These observations are supported by Kctd9 in situ hybridization analysis in the Allen Mouse Brain Atlas (31) that show Kctd9 expressed at most weakly in the adult hypothalamus. The −5-kb-GnRH-luc reporter was expressed over 1500-fold more strongly in GT1-7 cells than in GN11 or NIH3T3 cells, indicating that this sequence conferred specificity almost exclusively to mature GnRH neurons (Fig. 2B) and mimicked the differential, cell-specific expression pattern of GnRH mRNA (Fig. 2, A and B). Because Kctd9 mRNA was present at such low levels in these three cell lines compared with GnRH, the sequence 5 kb upstream of the GnRH gene, which contained conserved regions within the 3′ UTR of the upstream gene Kctd9, is much more likely to be involved in the regulation and specification of GnRH transcription to differentiated GnRH neurons than Kctd9 transcriptional regulation.
Figure 2.
The 5 kb of GnRH upstream sequence displays cell specificity similar to GnRH but not Kctd9 mRNA. A, RT-qPCR analysis of GnRH and Kctd9 mRNA from GT1-7, GN11, and NIH3T3 cells. Data are presented as picograms of mRNA of each gene normalized to cyclophilin B and indicate mean ± sd. Statistical analysis was performed using two-way ANOVA followed by post hoc least squares mean differences Tukey-Kramer HSD. *, Significantly different from GnRH mRNA levels in GN11 and NIH3T3 cells and Kctd9 mRNA levels in all three cell lines (P < 0.05). B, Transient transfection of the −5-kb GnRH gene sequence upstream of a luciferase reporter gene (−5 kb-GnRH-luc) into GT1-7, GN11, and NIH3T3 cells. Luciferase values were normalized to a cotransfected β-galactosidase reporter to control for transfection efficiency, and data were normalized to RSVe/RSVp-luc transfected in parallel. Data represent mean ± sd plotted on a logarithmic scale. Statistical analysis was performed using one-way ANOVA followed by post hoc Tukey Kramer HSD. *, Significantly different from GN11 and NIH3T3 cells (P < 0.05). C, Truncation analysis of −5 kb of the GnRH gene sequence in GT1-7 cells. Transient transfections were performed in GT1-7 cells using 5′ truncations of −5-kb-GnRH-luc reporter plasmids as indicated by schematic diagrams. Locations of truncations are denoted as base-pair positions upstream of the GnRH TSS to the left of schematic diagrams. Previously characterized GnRH regulatory elements are colored in black, and novel conserved sequence blocks are colored in white. Data represent luciferase/β-galactosidase values normalized to empty pGL3 vector and are shown as mean ± sd. *, Significant difference between constructs by one-way ANOVA and post hoc Tukey-Kramer HSD (P < 0.05). P, Promoter; E1, enhancer-1.
Truncation analysis was performed to determine critical upstream regions involved in the regulation of GnRH expression (Fig. 2C). Luciferase reporter plasmids driven by 5′ truncations of −5-kb-GnRH-luc were transiently transfected into GT1-7 cells to assay for reporter activity. Deletion of −4491/−4199 did not alter GnRH expression, confirming the likely role of conserved region −4491/−4324 as protein coding sequence for Kctd9. Two deletions in the previously unavailable −5-kb upstream GnRH sequence resulted in significant decreases in reporter gene expression: −4199 to −3620 bp and also −3175 to −2438 bp. Deletion of −3620/−3175, which contained conserved region −3588/−3560, resulted in a significant increase in expression, suggesting the presence of a putative silencer element. The importance of the characterized GnRH-E1 (4) was confirmed by the significant decrease in expression upon deletion of −2168 to −1380 bp. These results indicate the presence of novel putative regulatory elements between −5 and −3 kb of the GnRH gene.
Regions identified by truncation analysis as important in positive GnRH gene regulation overlapped with evolutionarily conserved regions identified by BLAST. Critical region −4199/−3620 corresponded to conserved region −4199/−3895, region −3175/−2438 included conserved region −3135/−2980 and the previously characterized region −2980/−2631, and critical region −2168/−1380 contained GnRH-E1. This study will further investigate the novel positive GnRH regulatory elements contained within the Kctd9 3′ UTR at −4199/−3895 and −3135/−2980.
Two novel upstream conserved regions enhance neuron-specific expression of GnRH regulatory elements
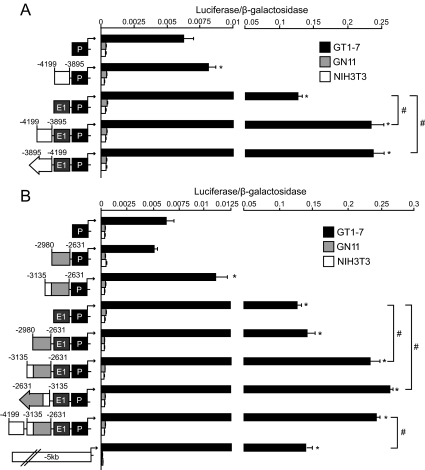
To determine whether the conserved sequence between −4199/−3895 could function as an enhancer of GnRH expression, luciferase reporter plasmids containing −4199/−3895 bp upstream of combinations of characterized GnRH regulatory elements were transiently transfected into GT1-7, GN11, and NIH3T3 cells to assay for reporter gene expression (Fig. 3A). The −4199/−3895 region significantly enhanced expression of GnRH-P and more dramatically enhanced GnRH-E1/GnRH-P, a result also observed with −4199/−3895 in the reverse orientation, confirming a characteristic enhancer function for this element (Fig. 3A). No combination of GnRH regulatory elements enhanced expression over GnRH-P in GN11 or NIH3T3 cells. These results showed that the −4199/−3895 region functions as an enhancer and increases specification of GnRH expression to differentiated GnRH neurons.
Figure 3.
Conserved regions −4199/−3895 and −3135/−2980 enhance neuron-specific GnRH expression. Transient transfections were performed in GT1-7 (black), GN11 (gray), and NIH3T3 cells (white) using luciferase reporter plasmids containing the −4199/−3895 (A) and the −3135/−2980 (B) regions upstream of previously characterized GnRH regulatory elements as indicated by schematic diagrams. Data represent luciferase/β-galactosidase values normalized to RSVe/RSVp-luc and are shown as mean ± sd. *, Significant difference compared with GnRH-P alone; and #, significant difference between constructs by one-way ANOVA and post hoc Tukey-Kramer HSD (P < 0.05). P, Promoter; E1, enhancer-1.
Although the −2980/−2631 region has previously been shown to increase cell-specificity of GnRH expression (26), this element individually does not significantly increase overall expression of GnRH in GT1-7 cells. To determine the role of the adjacent conserved −3135/−2980 region in regulation of GnRH expression, luciferase reporter plasmids driven by −3135/−2980 upstream of combinations of previously characterized GnRH regulatory regions were transiently transfected into GT1-7, GN11, and NIH3T3 cells to assay for reporter gene activity. Neither the novel −3135/−2980 region (Supplemental Fig. 1A, published on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org) nor the −2980/−2631 region (Fig. 3B) alone significantly enhanced expression in GT1-7 cells. However, reporters containing a contiguous sequence with these regions combined (−3135/−2631) significantly increased expression of GnRH-P, as well as of GnRH-E1/GnRH-P in either forward or reverse orientation (Fig. 3B). Similar results were observed upstream of the heterologous Rous sarcoma virus promoter (RSVp) (data not shown). These increases in expression were observed only in GT1-7 cells, because combinations of enhancer elements did not increase expression over GnRH-P in GN11 or NIH3T3 cells, indicating that this region enhances neuron-specificity of GnRH expression. These results define −3135/−2631 as a classical enhancer of GnRH expression.
Interestingly, −4199/−3895 did not further enhance expression of −3135/−2631-GnRH-E1/GnRH-P, suggesting that these two novel regions may serve similar functions. In addition, expression of the reporter construct containing all four GnRH regulatory elements was significantly higher than −5-kb-GnRH-luc, which generated expression levels similar to GnRH-E1/GnRH-P (Fig. 3B). These two observations suggest that the three GnRH enhancers together with GnRH-P likely comprise the positive regulatory elements required for maximal GnRH expression. The reporters containing the defined GnRH regulatory elements were expressed roughly 20- to 800-fold higher in GT1-7 cells than in GN11 or NIH3T3 cells (Fig. 3), indicating that these regulatory regions were sufficient to confer specificity almost exclusively to mature GnRH neuronal cells. The −3135/−2631 enhancer region will hereby be termed GnRH-E2 and −4199/−3895 will be termed GnRH-E3.
Neuron-specific GnRH expression mediated by GnRH-E2 and GnRH-E3 requires a contribution from GnRH-E1
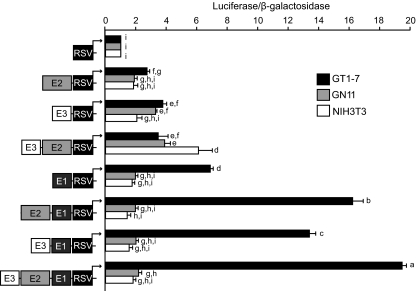
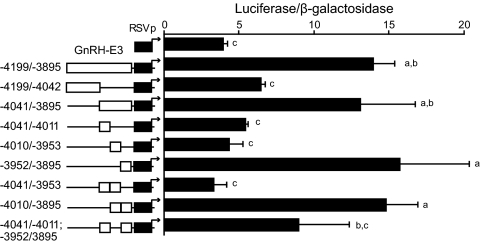
GnRH-P has been shown to have a critical role in specifying transcription to differentiated GnRH neurons (Fig. 3) (9). To examine the roles of each of the three GnRH enhancer regions in mediating neuron-specific GnRH expression in the absence of the GnRH promoter, the activity of each enhancer was evaluated upstream of the heterologous RSVp in GT1-7, GN11, and NIH3T3 cells (Fig. 4). In GT1-7 cells, GnRH-E2 and GnRH-E3 each significantly enhanced expression over RSVp alone to similar levels, confirming characteristic enhancer function, although their activities were not as strong as GnRH-E1. GnRH-E2 and GnRH-E3 in combination did not enhance activity further, providing additional evidence for functional similarity. GnRH-E2 and GnRH-E3 individually, and also in combination, enhanced expression of GnRH-E1/RSVp. Overall, in GT1-7 cells, GnRH-E2 and GnRH-E3 enhanced expression independently, as well as in the presence of GnRH-E1.
Figure 4.
Neuron-specific GnRH gene expression mediated by GnRH-E2 and GnRH-E3 requires a contribution from GnRH-E1. Transient transfections were performed in GT1-7 (black), GN11 (gray), and NIH3T3 cells (white) using heterologous RSVp-luciferase reporter plasmids containing GnRH-E2 and/or GnRH-E3 in the absence or presence of GnRH-E1, as indicated by schematic diagrams. Data represent luciferase/β-galactosidase values normalized to RSVp and are shown as mean ± sd. Levels not assigned the same letter are significantly different by two-way ANOVA followed by post hoc least squares mean differences Tukey-Kramer HSD (P < 0.05). E, Enhancer.
In GN11 cells, in the absence of GnRH-E1, GnRH-E3 alone or combined with GnRH-E2 significantly enhanced expression of RSVp to similar levels as GT1-7 cells, whereas GnRH-E1 alone did not significantly increase expression. Contrary to GT1-7 cells, however, the observed increases caused by GnRH-E3, with or without GnRH-E2, were muted in the presence of GnRH-E1. Similarly, in NIH3T3 cells, the combination of GnRH-E2 and GnRH-E3 upstream of RSVp resulted in a significant enhancement of expression that was, in fact, higher than in GT1-7 or GN11 cells but was abolished upon the addition of GnRH-E1. No other combination of enhancer elements caused significant increases in expression in NIH3T3 cells.
In summary, GnRH-E2 and GnRH-E3, in the absence of GnRH-E1, activated expression to similar levels in all three cell lines. Upon addition of GnRH-E1, GnRH-E2 and GnRH-E3 increased expression further in GT1-7 cells, but expression in GN11 or NIH3T3 cells was blunted. Therefore, GnRH-E2 and GnRH-E3 likely act as general enhancers that interact with GnRH-E1 to enhance and direct GnRH expression to differentiated GnRH neurons.
Critical region of GnRH-E3 maps to −3952/−3895
To determine a region of GnRH-E3 critical for enhancer activity, GnRH-E3, upstream of RSVp, was first divided in half, and luciferase activity was tested in GT1-7 cells (Fig. 5). The proximal half of GnRH-E3, −4041/−3895, was sufficient to activate expression to levels equivalent to the full-length GnRH-E3, whereas the distal half, −4199/−4042, did not significantly increase expression. The proximal half of GnRH-E3 was divided into three parts to more precisely map a critical region (Fig. 5). The proximal third, −3952/−3985, was both necessary and sufficient to activate expression to the level of full-length GnRH-E3. These results define the proximal −3952/−3895 section as the critical subregion of GnRH-E3.
Figure 5.
The critical region of GnRH-E3 maps to −3952/−3895. Transient transfections were performed in GT1-7 cells using luciferase reporter plasmids containing portions of GnRH-E3 on RSVp, as indicated by schematic diagrams. Positions of subregions studied are left of schematic diagrams. Data represent luciferase/β-galactosidase values normalized to empty pGL3 vector and are shown as mean ± sd. Levels not assigned the same letter are significantly different by one-way ANOVA followed by post hoc Tukey-Kramer HSD (P < 0.05).
A similar approach was used to identify critical subregions of the novel −3135/−2980 section of GnRH-E2. This approximately 150-bp region was divided into thirds in the presence of −2980/−2631 (because the full GnRH-E2 was required for enhancer function), and various subregion combinations were tested upstream of GnRH-P and RSVp in luciferase reporter assays in GT1-7 cells. Every tested subregion combination enhanced expression to similar, if not slightly higher levels, compared with intact GnRH-E2 (Supplemental Fig. 1, B and C), indicating redundancy within the novel region of GnRH-E2. The sequences in this novel region were not repetitive and showed unique in silico potential transcription factor binding profiles by Matinspector (32) and TFSearch (33), suggesting that GnRH-E2 may require a threshold complement of transcription factors provided by any portion of −3135/−2980 to activate expression.
Critical region of GnRH-E3 is regulated by Oct-1, Msx1, and Dlx2
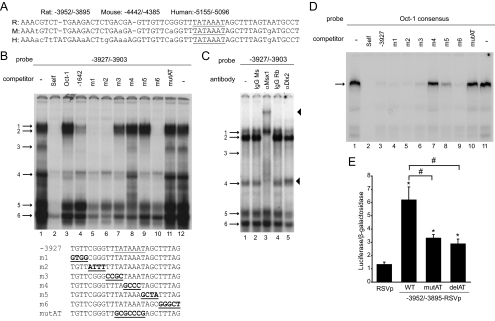
Alignment of the rat GnRH-E3 proximal critical region −3952/−3895 sequence with the corresponding human and mouse sequence revealed that the proximal portion, −3931/−3895, showed nearly 100% conservation (Fig. 6A). Contained within this sequence is a 100% conserved AT-rich region at −3917/−3911 (TATAAAT), which was similar to the Oct-1 consensus site (ATGCAAAT), and also AT-rich sequences (i.e. TCAATTAAAT and CAATTT) reported to bind Q50 homeodomain proteins in the regulation of GnRH-P, GnRH-E1, and portions of GnRH-E2 (14,16,20,26,34,35). Because of its high conservation, this AT-rich region could be important for GnRH-E3 function.
Figure 6.
The critical region of GnRH-E3 is regulated by Oct-1, Ms-1, and Dlx2. A, Rat, mouse, and human sequences corresponding to the GnRH-E3 critical region aligned by VISTA. Bases conserved in all three species are in capital letters. AT-rich region −3917/−3911 is underlined. B, EMSA was performed using GT1-7 nuclear extracts using a radiolabeled oligonucleotide probe containing −3927/−3903 of GnRH-E3 (lanes 1 and 12). Competition assays were performed with unlabeled oligonucleotides: −3927/−3903 (Self) (lane 2), Oct-1 consensus (Oct-1) (lane 3), Q50 homeodomain binding region in −1642/−1623 in GnRH-E1 (−1642) (lane 4), 4-bp scanning mutants of −3927/−3903 (m1–m6) (lanes 5–10), and mutation of AT-rich region −3917/−3911 (mutAT) (lane 11). Protein complexes are indicated by numbered arrows. Sequences of wild-type (WT) and mutant −3927/−3903 EMSA probes for competition assays are below the gel. The AT-rich −3917/−3911 region is underlined, and mutations are bold and underlined. C, EMSA with GT1-7 extracts using a radiolabeled probe containing −3927/−3903. Supershift assays were performed with mouse (Ms) and rabbit (Rb) IgG controls (lanes 2 and 4, respectively) and with antibodies recognizing Msx1 and Dlx2 (lanes 3 and 5, respectively). Protein complexes are indicated by numbered arrows, supershifted complexes are indicated by arrowheads. D, EMSA with GT1-7 nuclear extracts using a radiolabeled probe containing the Oct-1 consensus sequence. The Oct-1 complex is indicated by an arrow. Competition assays were performed with: Oct-1 consensus (Self) (lane 2), −3927/−3903 (−3927) (lane 3), scanning mutants m1–m6 in −3927/−3903 (lanes 4–9), and mutAT in −3927/−3903 (lane 10). E, Transient transfections were performed in GT1-7 cells using luciferase reporter plasmids containing the GnRH-E3 critical region (−3952/−3895) on RSVp containing a mutated (mutAT) or deleted (delAT)AT-rich region −3917/−3911. Data represent luciferase/β-galactosidase values normalized to empty vector and are shown as mean ± sd. *, Significantly different from RSVp; and #, significantly different from WT by one-way ANOVA and post hoc Tukey-Kramer HSD (P < 0.05).
EMSA using GT1-7 nuclear extracts was used to elucidate transcription factors that bind this region. EMSA using a radiolabeled probe corresponding to −3927/−3903 resulted in six specific protein complexes competed by self-unlabeled oligonucleotide, numbered 1–6 (Fig. 6B, lanes 1 and 2). Some of these complexes were competed by either or both the Oct-1 consensus probe or a probe containing GnRH-E1 sequences (−1642/−1623) shown previously to bind Q50 homeodomain proteins (16), suggesting that complexes containing Oct-1 and homeodomain proteins bind GnRH-E3 (Fig. 6B, lanes 3 and 4).
Complexes 1 and 2 likely contain Q50 homeodomain proteins, because these complexes were competed only by the −1642/−1623 probe. Competition assays with unlabeled oligonucleotide probes containing scanning mutations (m) (m1–m6) of region −3927/−3903, as well as a mutation of 7 bp of the AT-rich sequence at −3917/−3911 (mutAT), showed that complexes 1 and 2 were not competed by m3–m5 or the mutAT probes, indicating that these complexes bound the AT-rich region (Fig. 6B, lanes 5–11). Supershift assays using antibodies to Msx1 and Dlx2, Q50 homeodomain factors previously shown to be involved in GnRH gene regulation (16,22), showed that complex 1 formation was not altered by either Msx1 or Dlx2 antibody (Fig. 6C, lanes 3 and 5) and likely contained an unknown homeodomain protein. However, incubation with the Msx1 antibody resulted in a supershifted complex (upper arrowhead), as well as inhibition of complex 2 formation (Fig. 6C, lane 3), suggesting Msx1 is a component of complex 2.
Complex 3 was competed by an unlabeled oligonucleotide containing either the Oct-1 consensus or −1642/−1623 sequences but was not competed by scanning mutant probes m4 and mutAT (Fig. 6B, lanes 3, 4, 8, and 11), suggesting that Oct-1 could bind to this AT-rich region. Because no currently available commercial antibodies are suitable for EMSA supershift assays in our hands, Oct-1 binding to the −3927/−3903 region of GnRH-E3 was studied by EMSA using GT1-7 nuclear extracts with radiolabeled Oct-1 consensus probe (14), which resulted in one predominant protein complex corresponding to Oct-1 (Fig. 6D), consistent with previous studies (19). This Oct-1 complex was strongly competed by self oligonucleotide and also by the −3927/−3903 probe (Fig. 6D, lanes 1–3). The m4, m5, and mutAT probes either weakly or did not compete binding to this Oct-1 complex (Fig. 6D, lanes 7, 8, and 10). This binding pattern was similar to complex 3 on the −3927/−3903 probe (Fig. 6B) and suggested that a complex likely containing Oct-1 mapped to the AT-rich −3917/−3911 region.
Complex 4 was competed by both Oct-1 consensus and −1642/−1623 probes but was not competed by the m4 and mutAT mutant probes, indicating that the middle bases of the AT-rich region were critical for binding of this complex (Fig. 6B, lanes 3, 4, 8, and 11). Complex 4 formation was inhibited by Msx1 antibody, indicating its presence in this complex (Fig. 6C, lane 3). Complexes 5 and 6 both bound the AT-rich region (Fig. 6B, lane 11), but neighboring bases were also important, because complex 5 was not completely competed by m2–m5, and complex 6 was inefficiently competed by probes m3–m6 (Fig. 6B, lanes 5–12). Incubation with the Dlx2 antibody caused a supershifted complex to form (lower arrowhead) and also reduced the formation of complexes 4 and 6 (Fig. 6C, lane 5), which were also competed by the −1642/−1623 probe (Fig. 6B, lane 4), confirming that complexes 4 and 6 contained Dlx2. The Dlx2 antibody also reduced the formation of complex 5 (Fig. 6C, lane 5), which was competed by the Oct-1 consensus but not the −1642/−1623 probe (Fig. 6B, lanes 3 and 4). Thus, Dlx2 was also present in complex 5 and may bind alone or as part of a complex that preferentially binds AT-rich sequences present in the Oct-1 consensus.
Taken together, EMSA analyses demonstrated Msx1, Dlx2, and likely Oct-1 binding to the conserved AT-rich region −3917/−3911 in GnRH-E3. To determine whether this binding site was involved in transcriptional activity of GnRH-E3, these 7 bp were either mutated (mutAT) or deleted in the context of the −3952/−3895-RSVp plasmid, and reporter activity was measured in GT1-7 cells. Either mutation or deletion of this site resulted in a significant reduction in expression of nearly 50% (Fig. 6E), indicating that the Oct-1 and Q50 homeodomain binding sites in GnRH-E3 are important for enhancer activity.
GnRH-E2 binds Msx1 and Dlx2 in addition to Oct-1
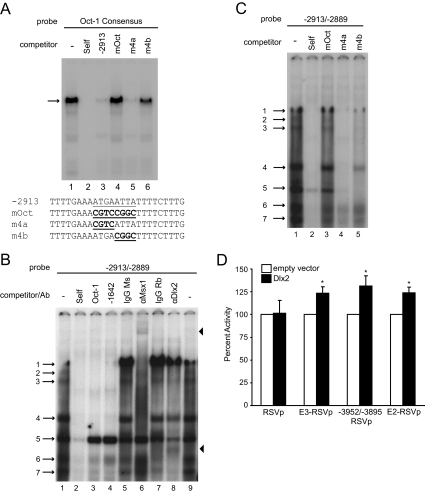
GnRH-E2 and GnRH-E3 do not further enhance expression when combined (Fig. 3B) and therefore may function by similar mechanisms. This may be the result of common transcription factors binding to these enhancer elements. The −2980/−2631 portion of GnRH-E2 contains numerous sites critical for GnRH gene expression that previously have been shown to bind Oct-1 (26). Many consensus ATTA sites for Msx1 and Dlx2 surround these Oct-1 binding sequences (22) and could, similar to GnRH-E3, bind these factors in addition to Oct-1. One putative GnRH-E2 location for Msx1 and Dlx2 binding is −2905/−2898, where the Oct-1 binding sequence at this location is ATGAATTA. The presence of an ATTA element suggests that Msx1 and Dlx2 could bind this region. This Oct-1 binding site was further characterized by performing EMSA with radiolabeled Oct-1 consensus probe. The Oct-1 complex was competed efficiently by both self and −2913/−2889 probe, which contained the −2905/−2898 Oct-1 binding site (Fig. 7A, lanes 1–3). Competition assays with a probe containing an 8-bp mutation of the Oct-1 binding site (mOct), as well as 4-bp mutations corresponding to the two halves of this site (m4a and m4b), showed that the mOct and m4b probes were unable to compete for Oct-1 complex binding (Fig. 7A, lanes 4–6), providing further evidence of the importance of this −2905/−2898 GnRH-E2 site for Oct-1 binding.
Figure 7.
Regulation by GnRH-E2 is mediated by Oct-1 and Q50 homeodomain proteins. A, EMSA with GT1-7 nuclear extracts using a radiolabeled probe containing the Oct-1 consensus sequence. The Oct-1 complex is indicated by an arrow. Competition assays were performed with: Oct-1 consensus (Self) (lane 2), −2913/−2889 of GnRH-E2 (−2913) (lane 3), and mutations of the Oct-1 binding site in −2913/−2889 (mOct, m4a, and m4b) (lanes 4–6). Sequences of mutant probes for competition assays are listed below the gel. The Oct-1 binding site is underlined, and mutations are bold and underlined. B, EMSA with GT1-7 nuclear extracts using a radiolabeled oligonucleotide probe containing −2913/−2889. Competition assays were performed with unlabeled oligonucleotides corresponding to: −2913/−2889 (Self) (lane 2), Oct-1 consensus (Oct-1) (lane 3), and −1642/−1623 of GnRH-E1 (−1642) (lane 4). Supershift assays were performed with mouse (Ms) and rabbit (Rb) IgG controls (lanes 5 and 7, respectively) and antibodies (Ab) recognizing Msx1 (lane 6) and Dlx2 (lane 8). Numbered arrows denote complexes of interest. Arrowheads denote supershifted complexes. C, EMSA with GT1-7 nuclear extracts using radiolabeled −2913/−2889 probe. Competition assays were performed with unlabeled oligonucleotides containing mutations in the Oct-1 binding site: −2913/−2889 (Self) (lane 2), mOct (lane 3), m4a (lane 4), and m4b (lane 5). D, Luciferase reporters with RSVp alone, or GnRH-E3, the GnRH-E3 critical region (−3952/−3895), or GnRH-E2, upstream of RSVp, were cotransfected into GT1-7 cells with either empty or Dlx2 expression plasmid. Data are presented as percent luciferase/β-galactosidase activity, relative to empty expression vector control, mean ± sd. *, Significantly different from empty vector by Student’s t test and significantly different from RSVp by one-way ANOVA and post hoc Tukey-Kramer HSD (P < 0.05).
EMSA was then performed with radiolabeled −2913/−2889 probe. Seven specific complexes formed on this probe (Fig. 7B, lanes 1 and 2). These complexes likely contained Q50 homeodomain proteins in addition to Oct-1, because almost all of these complexes were competed by both the Oct-1 consensus and −1642/−1623 probes (Fig. 7B, lanes 3 and 4). To determine whether Msx1 and Dlx2 bound these regions, supershift assays were performed using the Msx1 and Dlx2 antibodies. Addition of the Msx1 antibody resulted in a supershifted band (upper arrowhead) and inhibition of the formation of complexes 1, 3, and 4 (Fig. 7B, lanes 5 and 6). Addition of the Dlx2 antibody also resulted in a supershifted band (lower arrowhead) and prevented the formation of complexes 6 and 7 (Fig. 7B, lane 8), indicating the presence of Dlx2 in these complexes. Complex 7 was also competed by the −1642/−1623 probe (Fig. 7B, lane 4), but complex 6 was instead competed only by the Oct-1 consensus probe (Fig. 7B, lanes 3 and 4), suggesting that the Dlx2 in this complex could bind similar AT-rich sites in the Oct-1 consensus probe. These results indicate that Msx1 and Dlx2 bind portions of GnRH-E2. The mOct and m4b mutant probes either weakly competed or did not compete the binding of Msx1 or Dlx2-containing complexes 1, 3, 4, 6, and 7 (Fig. 7C), demonstrating the importance of the ATTA site for Msx1 and Dlx2 binding and that, similar to GnRH-E3, Msx1 and Dlx2 bind the same sites as Oct-1.
Dlx2 increases expression through GnRH-E2 and GnRH-E3
Msx1 represses, and Dlx2 enhances, cell-specific expression of the GnRH gene as these factors are expressed in GT1-7 cells but not immature GnRH neuronal cells or NIH3T3 cells (16). To determine whether Msx1 and Dlx2 can regulate expression through GnRH-E2 and GnRH-E3, transcriptional activity of luciferase reporter plasmids containing these regions was measured in the presence and absence of overexpressed Msx1 or Dlx2 in GT1-7 cells. Although overexpression of Msx1 strongly repressed expression through GnRH-E1, Msx1 did not have a significant effect on transcriptional activity through GnRH-E2 or GnRH-E3 (Supplemental Fig. 2). Overexpression of Dlx2 caused a significant 25–30% increase in expression of the full-length GnRH-E3, its critical region −3952/−3895, and GnRH-E2, compared with empty expression vector (Fig. 7D), confirming a role for Dlx2 in GnRH-E2 and GnRH-E3 enhancer function.
GnRH regulatory elements display chromatin signatures of active promoters and enhancers in GT1-7 cells
Active enhancers and promoters in the genome have been shown to be marked by distinct and predictive chromatin signatures (36). Promoters display high levels of the tri-methylated form [histone H3 lysine 4 (H3K4)-Me3], and low levels of the mono-methylated form of H3K4 (H3K4-Me1), whereas enhancers display low levels of H3K4-Me3 and high levels of H3K4-Me1. Because GnRH is in a relatively gene-rich region of the genome, and GnRH-E2 and GnRH-E3 were contained with the 3′ UTR of the upstream gene Kctd9, chromatin immunoprecipitation (ChIP) in GT1-7 cells was used to determine whether GnRH regulatory elements displayed chromatin signatures of promoters and/or enhancers. Chromatin was sonicated to generate fragments between 300 and 500 bp to distinguish regulatory elements of the endogenous mouse GnRH gene that are roughly 1000 bp or more apart, using PCR primers corresponding to GnRH-P and the three enhancer regions (Supplemental Table 1). In addition, GnRH intron 3, approximately 3.5 kb downstream from the TSS, was used as a control for a region of the GnRH gene that was neither a promoter nor an enhancer.
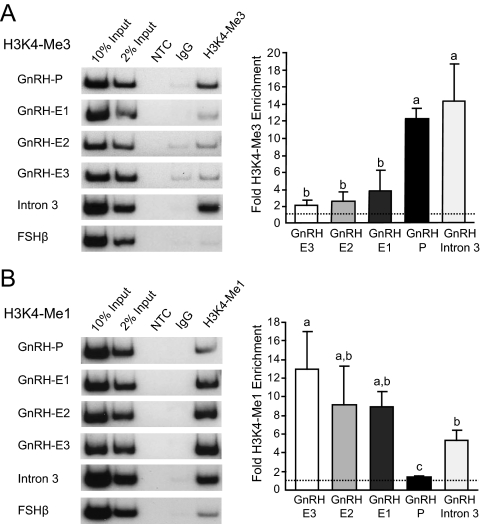
ChIP using an antibody to H3K4-Me3 followed by both conventional and qPCR analysis (Fig. 8A) showed that GnRH-P displayed significantly higher enrichment of H3K4-Me3 compared with GnRH-E regions. None of the three GnRH-E elements showed significant enrichment of this modification over the FSHβ negative control. GnRH intron 3 showed significant enrichment of this modification at levels similar to GnRH-P. ChIP using an antibody to H3K4-Me1 showed that GnRH-E regions were enriched for H3K4-Me1 at similar levels that were all significantly higher than GnRH-P, by both conventional and qPCR (Fig. 8B). GnRH intron 3 showed significant enrichment of H3K4-Me1, higher than those at GnRH-P, and at levels similar to GnRH-E regions.
Figure 8.
GnRH regulatory elements display chromatin signatures of active promoters and enhancers in GT1-7 cells. ChIP in GT1-7 cells was performed using antibodies specific to H3K4-Me3 (A) and H3K4-Me1 (B). Immunoprecipitated chromatin was subjected to conventional PCR (left) or qPCR (right) using primers specific to GnRH regulatory elements, intron 3, and the FSHβ negative control gene. qPCR data are presented as fold enrichment of the indicated histone modification relative to IgG, with values normalized to the FSHβ negative control gene (dotted line). NTC, No template control. Levels not connected by the same letter are significantly different by one-way ANOVA and post hoc Tukey-Kramer HSD with Box Cox Y Transformation (P < 0.05). P, Promoter; E, enhancer.
In summary, GnRH-P displayed high levels of H3K4-Me3 and low levels of H3K4-Me1. GnRH-E regions contained low levels of H3K4-Me3 but high levels of H3K4-Me1. These patterns were specific to the promoter and enhancers, because GnRH intron 3 showed enrichment of both H3K4-Me3 and H3K4-Me1. These results indicate that GnRH regulatory elements are endogenously marked in the genome as promoters and enhancers through specific histone modification signatures.
Discussion
The targeting of GnRH gene expression to approximately 800 highly specialized hypothalamic neurons requires a developmentally orchestrated program of gene regulation involving cis-regulatory elements grouped into complex enhancers and interactions with DNA-binding transcription factors and coregulatory proteins. This study identifies and characterizes novel, evolutionarily conserved enhancers of GnRH expression in the 3′ UTR of the neighboring upstream gene that use both common and tissue-specific regulators to mediate specification of GnRH transcription to differentiated GnRH neuronal cells.
Evolutionarily conserved regions within a neighboring gene function as enhancers of neuron-specific GnRH expression
The complete sequencing of many mammalian genomes has facilitated comparative genomic studies that have identified highly and ultraconserved elements in noncoding regions of the genome. Such elements have been shown to be functional cis-regulatory modules with critical roles in gene regulation during development (27,37,38). The completion of the rat genome allowed us to identify four highly conserved regions in previously unavailable sequence upstream of the GnRH gene. We show that two of these regions, defined here as GnRH-E2 and GnRH-E3, are involved in GnRH gene regulation as enhancers of neuron-specific GnRH expression but, remarkably, are also contained within the 3′ UTR of the neighboring upstream gene Kctd9. It is rare to find this high degree of conservation maintained in the 3′ noncoding sequences of genes, but recent comparative genomic studies have revealed that roughly 3–6% of conserved elements reside specifically in 3′ UTRs and suggest that these regions could have functional overlap with both a cis-regulatory as well as posttranscriptional function (38,39). The cell-specific expression of GnRH mRNA and transfected −5-kb GnRH reporter, in addition to the very low levels of Kctd9 mRNA in the three cell lines studied, indicates that these elements function mainly as enhancers of GnRH gene expression. There are precedents for the presence of cis-regulatory regions within neighboring genes, especially in gene clusters or gene-rich regions of the genome. For example, a long-range enhancer of sonic hedgehog expression is found in an intron of the upstream gene Lmbr1 (40), and an enhancer for the nicotinic acetylcholine receptor (nAchR)-α3 gene is present in the 3′ UTR of the upstream gene, nicotinic acetylcholine receptor-β4 (41). Moreover, it has been shown that regulatory regions located between two genes in close proximity are capable of enhancing both the upstream and downstream genes depending on the cellular context (41,42). Thus, although these elements could have a posttranscriptional function or regulate other genes in this gene-rich region in other cellular contexts, our data show that within the GnRH neuron, they function to enhance neuron-specific GnRH gene expression.
Histone modifications, in addition to evolutionary conservation, can identify and define functional regions of the genome, such as promoters, enhancers, and transcribed regions (36,43,44,45). The GnRH regulatory elements are not only highly conserved but also display chromatin signatures predictive of promoters and enhancers. The three GnRH-E regions displayed a high enrichment of the H3K4-Me1, but not H3K4-Me3, a pattern predictive of enhancers (36), whereas the GnRH promoter displayed the reciprocal pattern of high H3K4-Me3 and low H3K4-Me1, predictive of a promoter. Intron 3 displayed significant levels of both modifications, consistent with reports of histone modifications of transcribed genes (36,46,47,48). Our results not only demonstrate the evolutionarily conserved nature of these chromatin signatures first found to distinguish enhancers from promoters in human cells (36) but also provide further in vivo evidence that these regions of the genome are enhancers of GnRH gene regulation. The Kctd9 3′ UTR, where GnRH-E2 and GnRH-E3 are located, is only 3–4 kb upstream of the GnRH TSS, but nearly 26 kb downstream of the TSS of the Kctd9 mRNA. Regardless of the activity of the Kctd9 gene, these histone modifications are typically not enriched at that distance downstream from the TSS, with the majority of such modifications being nearly absent 10 kb downstream from the TSS especially at the 3′ end of the mRNA (44,47,48). Therefore, the chromatin signatures, in combination with the observation that these upstream sequences recapitulate the cell-specific expression pattern of endogenous GnRH mRNA, indicate that these regions are functional and endogenously marked as enhancers and promoters in the mouse genome and regulate neuron-specific GnRH expression.
GnRH-E2 and GnRH-E3 enhance neuron-specificity of GnRH expression via related mechanisms
Specification of GnRH expression to differentiated hypothalamic neurons is thought to involve coordinate regulation among GnRH regulatory elements. Although GnRH-E1 is a bona fide neuron-specific enhancer (4,9), GnRH-E2 and GnRH-E3 alone are more general enhancers that are regulated by transcription factors, at least some of which are expressed in all three cell lines studied, i.e. Oct-1. However, in the presence of GnRH-E1 or GnRH-P, GnRH-E2 or GnRH-E3 enhance GnRH expression only in mature GnRH neuronal cells, indicating that they increase cell specificity of GnRH expression through interactions with GnRH-E1 and GnRH-P. In GT1-7 cells, these elements cooperate to increase expression, but in GN11 and NIH3T3 cells, they cooperate to decrease or blunt expression. This may be mediated by a repressor protein expressed in GN11 or NIH3T3 cells but not in GT1-7 cells or, conversely, an activator protein, such as Dlx2, expressed in GT1-7 cells but not in GN11 or NIH3T3 cells. Therefore, this interacting complex of four regulatory regions act together to focus the narrow specificity of GnRH gene expression to the approximately 800 GnRH mature neurons in the adult hypothalamus.
GnRH-E2 and GnRH-E3 share related mechanisms in regulating neuron-specificity likely mediated by similar transcription factor complexes binding to both. This coordinate regulation is partially driven by the combination of a ubiquitously expressed transcription factor (Oct-1) and factors expressed more strongly in GT1-7 cells than in immature GnRH neuronal or NIH3T3 cells, such as Dlx2 (16). We demonstrate that Msx1, Dlx2, and a complex likely containing Oct-1 bind similar, highly conserved, AT-rich sites in GnRH-E2 and GnRH-E3 that are important for enhancer activity. Dlx2 is one of the activators involved in specifying expression to mature GnRH neurons through the three GnRH-E regions as cotransfection increases expression of both GnRH-E2 and GnRH-E3. Unlike GnRH-E1 (16), overexpression of Msx1 did not repress expression through GnRH-E2 or GnRH-E3. This could be due to high levels of endogenous Msx1 or the activity of these enhancers being significantly lower than GnRH-E1, making an observation of repression difficult. It is also possible that Msx1-mediated repression through GnRH-E2 and GnRH-E3 could occur in a different physiological situation, i.e. in development or in the presence of hormones and neurotransmitters, and/or may require additional cofactors. Previous studies demonstrate that neuron-specific expression through GnRH-E1 and GnRH-P is mediated by interactions between Oct-1, Msx1, and Dlx2 (16,22). Similar interactions are likely involved in regulation through GnRH-E2 and GnRH-E3, because the Oct-1 consensus EMSA probe competed almost all of the protein complexes shown to contain Msx1 and Dlx2.
GnRH-P and GnRH-E1 are sufficient to specify GnRH expression to differentiated GnRH neurons in model cell lines (4,9) and target expression exclusively to GnRH neurons in transgenic mice (9), though they do not target the entire population of GnRH neurons. Because GnRH-E2 and GnRH-E3 regulate expression through similar transcription factor complexes as both GnRH-E1 and GnRH-P, it is very likely that these regions enhance specificity driven by GnRH-E1 and GnRH-P by increasing overall expression to maximal levels to target a greater population of GnRH neurons, which is necessary for full reproductive axis development and function. This is supported by the observation that transgenic mice containing longer GnRH 5′ gene fragments that include GnRH-E2 and GnRH-E3 target expression to 85–97% of GnRH neurons, more than in mice using shorter transgenes lacking these regions (6,8,23,24,49), suggesting that the discrete enhancer regions that we have characterized are also critical for GnRH gene regulation in vivo. These secondary enhancers could also contribute to phenotypic robustness in the face of environmental and genetic variability, as has been recently reported in Drosophila development (50). Although they exhibit functional similarity in specifying expression to mature GnRH neuronal cells, GnRH-E2 and GnRH-E3 may also have distinct and unique functions in various other physiological contexts, perhaps during different stages of development, in hormonal and neurotransmitter regulation of GnRH expression, or in subpopulations of GnRH neurons.
The specialized phenotype of the GnRH neuron is defined by the cell-specific expression of the GnRH gene. We identify and characterize two novel enhancer regions contained within the 3′ UTR of the adjacent gene as important for the narrowly targeted specification of GnRH gene expression to the mature GnRH neuron. This work provides new insight into the mechanisms of neuron-specific gene expression critical for specification of the GnRH neuron during development and for attaining levels of GnRH transcription vital for adult reproductive function.
Materials and Methods
Comparative genomic analysis
Rat chromosome 15 sequence approximately 5 kb upstream of the GnRH TSS, and corresponding sequences for mouse chromosome 14, and human chromosome 8, were obtained through the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov). Corresponding sequences for dog and cow were obtained through the Ensembl Genome browser (http://www.ensembl.org) (51). Sequences were aligned by pairwise BLAST using the default parameters (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The low-resolution image was reproduced in Microsoft Office PowerPoint 2003. Further pairwise alignments were performed using Ensembl. Rat chromosome 15 (v. November 2004: 47,298,607–47,303,197) was used as base genome for VISTA Browser alignments (29) (http://genome.lbl.gov/vista/index.shtml) with corresponding mouse, human, dog, and chicken sequences, and also for UCSC Genome Browser (http://genome.ucsc.edu) (52) comparisons with available sequences from mammalian and nonmammalian species.
Plasmids
The −5-kb GnRH sequence (−4984 to +22 relative to the TSS) was isolated by PCR from rat genomic DNA. The fragment was cloned into the KpnI and XhoI sites of the pGL3-basic luciferase expression vector (Promega, Madison, WI) using T4 DNA Ligase (New England Biolabs, Ipswich, MA) to generate −5-kb-GnRH-luc. The 5′ truncations were made by PCR cloning into KpnI and XhoI restriction sites in pGL3 or by generating targeted deletions using the QuikChange Site Directed Mutagenesis system (Stratagene, La Jolla, CA). GnRH-P, GnRH-E1/GnRH-P, −2980/−2631-GnRH-P, −2980/−2631-GnRH-E1/GnRH-P, RSVp, GnRH-E1/RSVp, and −2980/−2631-GnRH-E1/RSVp in the pGL3 vector have been described (26,35). The −4199/−3895 and −3135/−2631 regions were subcloned upstream of known GnRH regulatory elements by PCR with Platinum Pfx DNA Polymerase (Invitrogen, San Diego, CA) and ligation with the FastLink DNA Ligation System (Epicentre Technologies, Madison, WI) into either the KpnI site or the KpnI and MluI sites in the previously described plasmids. GnRH-E2 and GnRH-E3 mutations and deletions were made using the QuikChange Site Directed Mutagenesis system (Stratagene). pCB6+, pCB6+-Dlx2, and pCB6+-Msx1 have been described (16).
Cell culture, transfections, and reporter assays
GT1-7, GN11 (kindly provided by Sally Radovick), and NIH3T3 (American Type Culture Collection, Manassas, VA) cell lines were cultured in DMEM with 4.5% glucose, 10% fetal bovine serum, and 1× penicillin-streptomycin in 5% CO2 at 37 C. GT1-7, GN11, and NIH3T3 cells were seeded into 24-well plates at 90,000, 50,000, and 30,000 cells per well, respectively, approximately 24 h before transfection. Transient transfections were performed using FuGENE 6 Transfection Reagent (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s recommendations. Cells were cotransfected, as indicated in figure legends, with 400 ng/well luciferase reporter plasmid and 200 ng/well expression plasmid (where indicated), as well as 100 ng/well thymidine kinase-β-galactosidase reporter plasmid as an internal control for transfection efficiency.
Cells were harvested 48 h after transfection in lysis buffer [100 mm potassium phosphate (pH 7.8) and 0.2% Triton X-100]. Luciferase assays were performed as previously described (16), and β-galactosidase assays were performed using the Galacto-Light Plus assay system as directed by the manufacturer (Applied Biosystems, Foster City, CA). Luciferase values were normalized to β-galactosidase values to control for transfection efficiency. Transient transfections performed on multiple cell lines were transfected in parallel with an RSV enhancer/RSV promoter-luciferase reporter plasmid (RSVe/RSVp-luc) (26). Values were normalized to pGL3, RSVe/RSVp-luc, RSVp, or empty expression plasmid as indicated in figure legends. Experiments comparing multiple cell lines were normalized to either RSVe/RSVp-luc to evaluate promoter activity and to control for overall differences in transfection efficiency among the cell lines or to RSVp to evaluate activity of the enhancers. Experiments performed only in GT1-7 cells were normalized to pGL3. Data represent the mean ± sd of at least three independent experiments. Statistical analyses were performed using either one-way ANOVA or two-way ANOVA as indicated in figure legends, followed by post hoc analysis by Tukey-Kramer honestly significant difference (HSD), with P < 0.05 to indicate significance.
Nuclear extracts and electrophoretic mobility shift assays
GT1-7 cells were scraped in hypotonic buffer [20 mm Tris-HCl (pH 7.4), 10 mm NaCl, 1 mm MgCl2, 10 mm NaF, 0.5 mm EDTA, and 0.1 mm EGTA] with protease inhibitor cocktail (Sigma, St. Louis, MO) and 1 mm phenylmethylsulfonyl fluoride and allowed to swell on ice. Cells were lysed by passing through a 25-gauge needle, and the nuclei were collected by centrifugation. Nuclear proteins were extracted on ice for 30 min in hypertonic buffer [20 mm HEPES (pH 7.9), 20% glycerol, 420 mm KCl, 2 mm MgCl2, 10 mm NaF, 0.5 mm EDTA, and 0.1 mm EGTA] with protease inhibitor cocktail (Sigma) and 1 mm phenylmethylsulfonylfluoride. Protein concentration was determined using the Bio-Rad Protein Assay (Bio-Rad, Hercules, CA). Oligonucleotide probes are listed in Supplemental Table 1. Oct-1 consensus (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) (14) and −1642/−1623 GnRH-E1 probes (16) have been described. All synthetic oligonucleotides were made by IDT (San Diego, CA). Annealed double-stranded oligonucleotides (1 pmol/μl) were end-labeled with T4 Polynucleotide Kinase (New England Biolabs) and [γ32P]ATP (7000 Ci/mmol; MP Biomedicals, Solon, OH). Probes were purified using Micro Bio-Spin 6 Chromatography Columns (Bio-Rad). Binding reactions contained 3 μg nuclear protein and 4 fmol of labeled probe in 10 mm HEPES (pH 7.9), 25 mm KCl, 2.5 mm MgCl2, 1% glycerol, 0.1% Nonidet P-40, 0.25 mm EDTA, 0.25% BSA, 1 mm dithiothreitol, and 0.0125 μg/μl poly dIdC. For competition experiments, 1000-fold molar excess competitor probes were added to the reaction and incubated for 10 min on ice, and for supershift experiments, 2.5 μl Msx1 Mouse Monoclonal Antibody (Covance MMS-261R; Covance, Emeryville, CA), 2.5 μg Dlx2 rabbit polyclonal antibody (AB5726; Millipore, Temecula, CA), or corresponding amount of Normal Mouse or Normal Rabbit IgG (Santa Cruz Biotechnology, Inc.) were added to the reaction and incubated for 25 min on ice, before the addition of labeled probe. Samples were then incubated for 10 min at room temperature before loading on a 5% nondenaturing polyacrylamide gel in 0.25× Tris-borate EDTA buffer. Gels were run for 2.5 h at 200–250 V, then dried under vacuum and exposed to film for 4–5 d.
Chromatin immunoprecipitation
ChIP assays were performed as previously described (53). Chromatin was sonicated to an average length of 300–500 bp using a Branson Sonifier 250 (Branson Ultrasonics Corp., Danbury, CT). Antibodies recognizing specific histone modifications were anti-H3K4-Me3 (07-473; Millipore) and anti-H3K4-Me1 (ab8895; Abcam, Cambridge, MA). Immunoprecipitated DNA and DNA from input chromatin were analyzed for sequences of interest by conventional PCR and qPCR using primers specific to GnRH regulatory elements (Supplemental Table 1). The mouse FSHβ promoter (53) was used as a negative control. DNA from immunoprecipitated samples was quantified relative to a standard curve representing percent of input chromatin. The fold enrichment of antibody signal over IgG was calculated for each primer set, and data from each independent experiment were normalized to the negative control gene. Data are presented as mean ± sd of at least three independent experiments. Statistical analyses were performed using one-way ANOVA with Box Cox Y Transformation as indicated in figure legends, followed by post hoc analysis by Tukey-Kramer HSD, with P < 0.05 to indicate significance.
PCR and qPCR
PCR of ChIP samples was performed using Illustra Taq polymerase (GE Healthcare, Piscataway, NJ). PCR products were labeled by the addition of [α-32P]dATP (MP Biomedicals) in the nucleotide mix and were resolved on a 5% acrylamide gel in 0.5× Tris-borate EDTA buffer. The gels were dried and subjected to autoradiography. qPCR was performed using the iQ5 Real-Time PCR Detection System and Software and iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer’s recommendations.
Reverse transcription-qPCR
Total RNA from GT1-7, GN11, and NIH3T3 cells was extracted using Trizol Reagent (Invitrogen) according to manufacturer’s recommendations. cDNA was obtained by reverse transcription of 2 μg RNA from each cell line using Oligo(dT)20 primer and Superscript III First Strand Synthesis System (Invitrogen). cDNA was subject to qPCR using primers specific to the coding sequences for mouse GnRH (54), and 3′ UTR of Kctd9 (GnRH-E3) (Supplemental Table 1), in addition to the coding sequence of cyclophilin B as a positive control (55). Standard curves were generated using serial dilutions of plasmid DNA containing the qPCR amplicons cloned into pCR2.1 (Invitrogen). Quantities of GnRH and Kctd9 were normalized to cyclophilin B. Data are represented as mean ± sd from three independent RNA samples from each cell line. Statistical analyses were performed using two-way ANOVA with post hoc Tukey-Kramer HSD with P < 0.05 to indicate significance.
Statistical analyses
JMP 8 (SAS Institute, Inc., Cary, NC) software was used for statistical analyses.
Supplementary Material
Acknowledgments
We thank James W. Posakony for assistance with bioinformatics analyses, Mark A. Lawson for assistance with statistical analyses, Bing Ren for valuable discussions, Sally Radovick for kindly providing GN11 cells, C. Abate Shen for Msx1 and Dlx2 expression plasmids, Anna Grove and Marjory Givens for additional assistance, Dan Clark for review of figures, Melissa Brayman and Christine Glidewell-Kenney for critical review of the manuscript, members of the Mellon laboratory for insightful discussions, and Susan L. Mayo and Jason D. Meadows for expert technical assistance.
Footnotes
Present address for N.L.G.M.: Moores Cancer Center, University of California, San Diego, La Jolla, California 92093-0803.
This work was supported by National Institutes of Health (NIH) R01 Grants DK044838 and HD020377 (to P.L.M.) and by the National Institute of Child Health and Human Development/NIH through a cooperative agreement (U54 HD012303) as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research (to P.L.M.). A.K.I. was partially supported by the National Institute of Child Health and Human Development (NICHD) Grant F32 HD058427 and the NIH Grant T32 DK007494. N.L.G.M. was partially supported by the NIH Grant T32 GM08666. K.Y. was partially supported by the Amgen Scholars Program, funded by the Amgen Foundation.
Disclosure Summary: The authors have nothing to disclose.
First Published Online July 28, 2010
Abbreviations: BLAST, Basic Local Alignment Search Tool; ChIP, chromatin immunoprecipitation; GnRH-E; GnRH-enhancer; GnRH-P, GnRH minimal promoter; H3K4, histone H3 lysine 4; H3K4-Me1, mono-methylated form of H3K4; H3K4-Me3, tri-methylated form of H3K4; HSD, honestly significant difference; Kctd9, potassium channel tetramerization domain containing 9; m, mutation; mOct, mutation of the Oct-1 binding site; mutAT, mutation AT; qPCR, quantitative real-time PCR; RSVe/RSVp-luc, RSV enhancer/RSV promoter-luciferase reporter plasmid; RSVp, Rous sarcoma virus promoter; TSS, transcription start site; 3′ UTR, 3′ untranslated region.
References
- Burbach JP 2002 Regulation of gene promoters of hypothalamic peptides. Front Neuroendocrinol 23:342–369 [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Pfaff DW 1989 Origin of luteinizing hormone-releasing hormone neurons. Nature 338:161–164 [DOI] [PubMed] [Google Scholar]
- Wray S, Grant P, Gainer H 1989 Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci USA 86:8132–8136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte DB, Lawson MA, Belsham DD, Eraly SA, Bond CT, Adelman JP, Mellon PL 1995 A neuron-specific enhancer targets expression of the gonadotropin-releasing hormone gene to hypothalamic neurosecretory neurons. Mol Endocrinol 9:467–477 [DOI] [PubMed] [Google Scholar]
- Kepa KJ, Wang C, Neeley CI, Raynolds MV, Gordon DF, Wood WM, Wierman ME 1992 Structure of the rat gonadotropin releasing hormone (rGnGH) gene promoter and functional analysis in hypothalamic cells. Nuc Acids Res 20:1393–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape JR, Skynner MJ, Allen ND, Herbison AE 1999 Transgenics identify distal 5′- and 3′-sequences specifying gonadotropin-releasing hormone expression in adult mice. Mol Endocrinol 13:2203–2211 [DOI] [PubMed] [Google Scholar]
- Skynner MJ, Slater R, Sim JA, Allen ND, Herbison AE 1999 Promoter transgenics reveal multiple gonadotropin-releasing hormone-I-expressing cell populations of different embryological origin in mouse brain. J Neurosci 19:5955–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter KJ, Song WJ, Sampson TL, Wuarin JP, Saunders JT, Dudek FE, Moenter SM 2000 Genetic targeting of green fluorescent protein to gonadotropin- releasing hormone neurons: characterization of whole-cell electrophysiological properties and morphology. Endocrinology 141:412–419 [DOI] [PubMed] [Google Scholar]
- Lawson MA, MacConell LA, Kim J, Powl BT, Nelson SB, Mellon PL 2002 Neuron-specific expression in vivo by defined transcription regulatory elements of the gonadotropin-releasing hormone gene. Endocrinology 143:1404–1412 [DOI] [PubMed] [Google Scholar]
- Simonian SX, Herbison AE 2001 Regulation of gonadotropin-releasing hormone (GnRH) gene expression during GnRH neuron migration in the mouse. Neuroendocrinology 73:149–156 [DOI] [PubMed] [Google Scholar]
- Seminara SB, Hayes FJ, Crowley Jr WF 1998 Gonadotropin-releasing hormone deficiency in the human (idiopathic hypogonadotropic hypogonadism and Kallmann’s syndrome): pathophysiological and genetic considerations. Endocrine Rev 19:521–539 [DOI] [PubMed] [Google Scholar]
- Mellon PL, Windle JJ, Goldsmith P, Padula CA, Roberts JL, Weiner RI 1990 Immortalization of hypothalamic GnRH neurons by genetically targeted tumorigenesis. Neuron 5:1–10 [DOI] [PubMed] [Google Scholar]
- Eraly SA, Mellon PL 1995 Regulation of GnRH transcription by protein kinase C is mediated by evolutionarily conserved, promoter-proximal elements. Mol Endocrinol 9:848–859 [DOI] [PubMed] [Google Scholar]
- Eraly SA, Nelson SB, Huang KM, Mellon PL 1998 Oct-1 binds promoter elements required for transcription of the gonadotropin-releasing hormone gene. Mol Endocrinol 12:469–481 [DOI] [PubMed] [Google Scholar]
- Rave-Harel N, Givens ML, Nelson SB, Duong HA, Coss D, Clark ME, Hall SB, Kamps MP, Mellon PL 2004 TALE homeodomain proteins regulate gonadotropin-releasing hormone gene expression independently and via interactions with Oct-1. J Biol Chem 279:30287–30297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givens ML, Rave-Harel N, Goonewardena VD, Kurotani R, Berdy SE, Swan CH, Rubenstein JL, Robert B, Mellon PL 2005 Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J Biol Chem 280:19156–19165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BJ, Cho GJ, Norgren Jr RB, Junier MP, Hill DF, Tapia V, Costa ME, Ojeda SR 2001 TTF-1, a homeodomain gene required for diencephalic morphogenesis, is postnatally expressed in the neuroendocrine brain in a developmentally regulated and cell-specific fashion. Mol Cell Neurosci 17:107–126 [DOI] [PubMed] [Google Scholar]
- Kelley CG, Lavorgna G, Clark ME, Boncinelli E, Mellon PL 2000 The Otx2 homeoprotein regulates expression from the gonadotropin-releasing hormone proximal promoter. Mol Endocrinol 14:1246–1256 [DOI] [PubMed] [Google Scholar]
- Nelson SB, Lawson MA, Kelley CG, Mellon PL 2000 Neuron-specific expression of the rat gonadotropin-releasing hormone gene is conferred by interactions of a defined promoter element with the enhancer in GT1-7 cells. Mol Endocrinol 14:1509–1522 [DOI] [PubMed] [Google Scholar]
- Clark ME, Mellon PL 1995 The POU homeodomain transcription factor Oct-1 is essential for activity of the gonadotropin-releasing hormone neuron-specific enhancer. Mol Cell Biol 15:6169–6177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson MA, Buhain AR, Jovenal JC, Mellon PL 1998 Multiple factors interacting at the GATA sites of the gonadotropin-releasing hormone neuron-specific enhancer regulate gene expression. Mol Endocrinol 12:364–377 [DOI] [PubMed] [Google Scholar]
- Rave-Harel N, Miller NLG, Givens ML, Mellon PL 2005 The Groucho-related gene family regulates the gonadotropin-releasing hormone gene through interaction with the homeodomain proteins MSX1 and OCT1. J Biol Chem 280:30975–30983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermantel TM, Campbell RE, Porteous R, Bock D, Gröne HJ, Todman MG, Korach KS, Greiner E, Pérez CA, Schütz G, Herbison AE 2006 Definition of estrogen receptor pathway critical for estrogen positive feedback to gonadotropin-releasing hormone neurons and fertility. Neuron 52:271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A, Divall S, Singh SP, Nikrodhanond AA, Baria AT, Le WW, Hoffman GE, Radovick S 2008 Temporal and spatial regulation of CRE recombinase expression in gonadotrophin-releasing hormone neurones in the mouse. J Neuroendocrinol 20:909–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H, Enquist LW, Dulac C 2005 Olfactory inputs to hypothalamic neurons controlling reproduction and fertility. Cell 123:669–682 [DOI] [PubMed] [Google Scholar]
- Givens ML, Kurotani R, Rave-Harel N, Miller NLG, Mellon PL 2004 Phylogenetic footprinting reveals functional upstream regions of the gonadotropin-releasing hormone gene that enhance cell-specific expression. Mol Endocrinol 18:2950–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Bristow J, Pennacchio LA 2007 Enhancer identification through comparative genomics. Semin Cell Dev Biol 18:140–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, DeRamo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M, Hawes A, Gill R, Celera, Holt RA, Adams MD, Amanatides PG, Baden-Tillson H, Barnstead M, Chin S, Evans CA, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee HM, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Li B, Liu Y, Qin X, Cawley S, Worley KC, Cooney AJ, D'Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler DA, Zhang Z, Bailey JA, Eichler EE, Tuzun E, Birney E, Mongin E, Ureta-Vidal A, Woodwark C, Zdobnov E, Bork P, Suyama M, Torrents D, Alexandersson M, Trask BJ, Young JM, Huang H, Wang H, Xing H, Daniels S, Gietzen D, Schmidt J, Stevens K, Vitt U, Wingrove J, Camara F, Mar Albà M, Abril JF, Guigo R, Smit A, Dubchak I, Rubin EM, Couronne O, Poliakov A, Hübner N, Ganten D, Goesele C, Hummel O, Kreitler T, Lee YA, Monti J, Schulz H, Zimdahl H, Himmelbauer H, Lehrach H, Jacob HJ, Bromberg S, Gullings-Handley J, Jensen-Seaman MI, Kwitek AE, Lazar J, Pasko D, Tonellato PJ, Twigger S, Ponting CP, Duarte JM, Rice S, Goodstadt L, Beatson SA, Emes RD, Winter EE, Webber C, Brandt P, Nyakatura G, Adetobi M, Chiaromonte F, Elnitski L, Eswara P, Hardison RC, Hou M, Kolbe D, Makova K, Miller W, Nekrutenko A, Riemer C, Schwartz S, Taylor J, Yang S, Zhang Y, Lindpaintner K, Andrews TD, Caccamo M, Clamp M, Clarke L, Curwen V, Durbin R, Eyras E, Searle SM, Cooper GM, Batzoglou S, Brudno M, Sidow A, Stone EA, Venter JC, Payseur BA, Bourque G, Lopez-Otin C, Puente XS, Chakrabarti K, Chatterji S, Dewey C, Pachter L, Bray N, Yap VB, Caspi A, Tesler G, Pevzner PA, Haussler D, Roskin KM, Baertsch R, Clawson H, Furey TS, Hinrichs AS, Karolchik D, Kent WJ, Rosenbloom KR, Trumbower H, Weirauch M, Cooper DN, Stenson PD, Ma B, Brent M, Arumugam M, Shteynberg D, Copley RR, Taylor MS, Riethman H, Mudunuri U, Peterson J, Guyer M, Felsenfeld A, Old S, Mockrin S, Collins F 2004 Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428:493–521 [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I 2004 VISTA: computational tools for comparative genomics. Nucleic Acids Res 32:W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radovick S, Wray S, Lee E, Nicols DK, Nakayama Y, Weintraub BD, Westphal H, Cutler GB Jr, Wondisford FE 1991 Migratory arrest of gonadotropin-releasing hormone neurons in transgenic mice. Proc Natl Acad Sci USA 88:3402–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR 2007 Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176 [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T 2005 MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21:2933–2942 [DOI] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA 1998 Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 26:362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplan C, Theis J, O'Farrell PH 1988 The sequence specificity of homeodomain-DNA interaction. Cell 54:1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley CG, Givens ML, Rave-Harel N, Nelson SB, Anderson S, Mellon PL 2002 Neuron-restricted expression of the rat gonadotropin-releasing hormone gene is conferred by a cell-specific protein complex that binds repeated CAATT elements. Mol Endocrinol 16:2413–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B 2007 Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39:311–318 [DOI] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA 2008 Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet 40:158–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, Walter K, Abnizova I, Gilks W, Edwards YJ, Cooke JE, Elgar G 2005 Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol 3:e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D 2005 Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15:1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Horikoshi T, Heaney SJ, van Baren MJ, van der Linde HC, Breedveld GJ, Joosse M, Akarsu N, Oostra BA, Endo N, Shibata M, Suzuki M, Takahashi E, Shinka T, Nakahori Y, Ayusawa D, Nakabayashi K, Scherer SW, Heutink P, Hill RE, Noji S 2002 Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci USA 99:7548–7553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough J, Deneris E 1997 β43′: an enhancer displaying neural-restricted activity is located in the 3′-untranslated exon of the rat nicotinic acetylcholine receptor β4 gene. J Neurosci 17:2273–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyama K, Irvine SQ, Stock DW, Weiss KM, Kawasaki K, Shimizu N, Shashikant CS, Miller W, Ruddle FH 2002 Genomic structure and functional control of the Dlx3–7 bigene cluster. Proc Natl Acad Sci USA 99:780–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Qu C, Van Calcar S, Trinklein ND, Cooper SJ, Luna RM, Glass CK, Rosenfeld MG, Myers RM, Ren B 2005 Direct isolation and identification of promoters in the human genome. Genome Res 15:830–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon G, Wang W, Ren B 2009 Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol 5:e1000566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Ren B 2009 Predictive chromatin signatures in the mammalian genome. Hum Mol Genet 18:R195–R201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas 3rd EJ, Gingeras TR, Schreiber SL, Lander ES 2005 Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169–181 [DOI] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaöz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I 2007 The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res 17:691–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA 2006 Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol 26:9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Wolfe A, Smith GR, Tobet SA, Radovick S 2002 Promoter sequences targeting tissue-specific gene expression of hypothalamic and ovarian gonadotropin-releasing hormone in vivo. J Biol Chem 277:5194–5202 [DOI] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL 30 May 2010 Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 10.1038/nature09158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, Coates G, Fairley S, Fitzgerald S, Fernandez-Banet J, Gordon L, Graf S, Haider S, Hammond M, Holland R, Howe K, Jenkinson A, Johnson N, Kahari A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Rios D, Schuster M, Slater G, Smedley D, Spooner W, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wilder S, Zadissa A, Birney E, Cunningham F, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Kasprzyk A, Proctor G, Smith J, Searle S, Flicek P 2009 Ensembl 2009. Nucleic Acids Res 37:D690–D697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D 2002 The human genome browser at UCSC. Genome Res 12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coss D, Jacobs SB, Bender CE, Mellon PL 2004 A novel AP-1 site is critical for maximal induction of the follicle-stimulating hormone β gene by gonadotropin-releasing hormone. J Biol Chem 279:152–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder R, Mellon PL 2009 Otx2 induction of the gonadotropin-releasing hormone promoter is modulated by direct interactions with Grg co-repressors. J Biol Chem 284:16966–16978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller NL, Wevrick R, Mellon PL 2009 Necdin, a Prader-Willi syndrome candidate gene, regulates gonadotropin-releasing hormone neurons during development. Hum Mol Genet 18:248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.