Abstract
Oculo-skeletal dysplasia segregates in two canine breeds, the Labrador retriever and samoyed, in which the causative loci have been termed drd1 and drd2, respectively. Affected dogs exhibit short-limbed dwarfism together with severe ocular defects, and this phenotype is inherited as an autosomal recessive trait in both breeds. The clinical and pathological appearance resembles human hereditary arthro-ophthalmopathies such as Stickler syndrome, or Marshall Syndrome, although these human disorders are usually dominant. Linkage studies in drd1-informative pedigrees mapped the locus to canine chromosome 24, and led to the identification of an insertional mutation in exon 1 of the gene COL9A3 that cosegregates with the disease. The drd2 locus was similarly mapped to canine chromosome 15 and shown to cosegregate with a 1,267 bp deletion mutation in the 5′ end of COL9A2. Both mutations affect the COL3 domain of the respective gene. Northern analysis showed reduced RNA expression in affected retina compared to normal. These models offer potential for studies such as protein-protein interactions between different members of the collagen gene family; regulation and expression of these genes in retina and cartilage, and even opportunities for gene therapy.
Introduction
Oculo-skeletal Dysplasia (OSD) segregates in two canine breeds, the Labrador retriever (Carrig et al., 1977) and the samoyed (Meyers et al., 1983). Affected dogs have skeletal abnormalities characterized by short-limbed dwarfism and ocular defects including vitreous dysplasia, retinal detachment and cataracts, and this phenotype segregates as an autosomal recessive trait in both breeds. Obligate heterozygotes may exhibit a mild ocular phenotype that ranges from vitreal strands and or localized retinal dysplasia characterized by focal or multifocal retinal folds, to large plaques of dysplastic retinal tissue, but have a normal appendicular skeleton. Because cross breeding of an OSD-affected Labrador retriever to an OSD-affected samoyed resulted in non-dwarf progeny and established that these two disorders are non allelic (Acland and Aguirre, 1995; Du, 2000), the 2 loci have been termed drd1 (dwarfism with retinal dysplasia type 1) and drd2 (dwarfism with retinal dysplasia type 2), respectively.
The clinical and pathological phenotypes of skeletal and ocular defect in these dogs are similar to human hereditary arthro-ophthalmopathies such as Stickler syndrome (Stickler et al., 1965), Kniest dysplasia (Maumenee and Traboulsi, 1985) and Marshall syndrome (Ayme and Preus, 1984). There is considerable overlap, phenotypic variability, and genetic heterogeneity among these human disorders. Most commonly, however, for such human patients whose phenotype includes vitreous lesions, the trait is dominantly inherited and associated with mutations in the gene for collagen type II (COL2A1) or, less frequently, in COL11A1 (Annunen et al., 1999; Ritvaniemi et al., 1993; Snead and Yates, 1999). An autosomal recessive form of Stickler syndrome has been associated with inheritance of an R295X mutation in the COL9A1 gene in one human family (Van Camp et al., 2006). More commonly, however, mutations in the collagen type IX genes (COL9A1; COL9A2; COL9A3) have been implicated in dominantly inherited human multiple epiphyseal dysplasias without ocular involvement.
By linkage analysis, type II collagen (COL2A1) was excluded as a candidate for OSD in both Labrador retrievers and samoyeds despite a severe decrease of this protein in the vitreous of OSD affected Labrador retrievers (Du et al., 2000). As well, Opticin, a protein identified from bovine vitreous and associated with collagen fibrils was excluded from OSD in both breeds (Pellegrini et al., 2002). Other genes coding for fibrillar structure proteins have also been excluded from causal association with the disease, include collagen type XI (COL11A1, COL11A2), type V (COL5A2) and Vitrin (Du, 2000).
Having excluded the main candidate genes for OSD in both breeds, we undertook a genome wide scan of a set of experimental 3-generation pedigrees informative for OSD derived from Labradors (drd1) and from samoyeds (drd2). Linkage was detected for drd1 and drd2 to chromosomes 24 and 15, respectively. Positional candidate gene analysis identified an insertional mutation in COL9A3 cosegregating with drd1, and a deletion mutation in COL9A2 cosegregating with drd2. Both mutations are predicted to truncate the respective protein product.
Methods
Study animals
Colony dogs
The drd1 and drd2 strains of dog, are maintained as part of an NIH-sponsored project (EY006855) at the Retinal Disease Studies Facility (RDSF) in Kennett Square, PA.
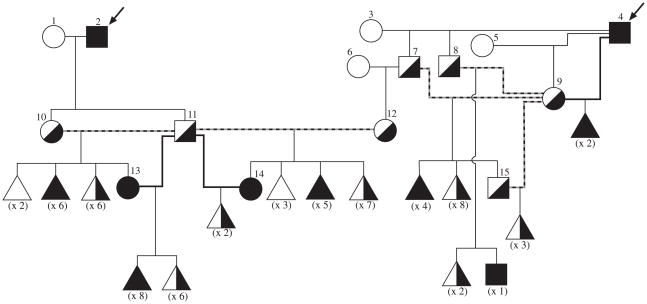
The drd1 strain was derived from two OSD-affected unrelated Labrador retriever dogs (Figure 1A, dogs 2 and 4, no recent ancestors in common). Both were bred to beagle-crossbred unrelated dogs and their heterozygous F1 progeny were then backcrossed to drd1-affected dogs or intercrossed to drd1-carriers to yield litters segregating the drd1 phenotype.
Figure 1.
drd1-(A) and drd2- (B) informative colony pedigrees used for genome wide scan and multimap analysis. Arrows point to probands. Dashed lines indicate intercross breedings, thick lines backcrosses. Black symbols represent OSD affected dogs, half-black symbols indicate carriers (heterozygotes), and white symbols identify homozygous normal dogs. Numbers in parentheses are the number of dogs in each category used in analysis.
The drd2 strain was derived from one OSD-affected samoyed (Figure 1B, dog 2) and an informative pedigree segregating the disease was established in the same manner as for the Labrador-derived drd1 strain.
Pedigreed dogs
Privately owned pedigreed dogs from breeds not known to segregate OSD, were screened for the mutations – these included 118 dogs from 22 breeds tested for the COL9A2 mutation, and 64 dogs from 17 breeds tested for the COL9A3 mutation (Supplement Table 1, A and B). Eight dogs from 4 breeds considered to be dwarf breeds (American bulldog, Pomeranian, Corgi and Dachshund) were also tested for both mutations (Supplement Table 1C). Non-dwarf dogs within each OSD-affected breed were tested as well: 59 unrelated Labrador retriever dogs (with no parents or grandparents in common), and 19 Labrador retrievers with clinically diagnosed retinal folds were tested for the COL9A3 mutation. Fifty-five unrelated and 6 related samoyeds were tested for the COL9A2 mutation.
Phenotypic evaluation of study dogs
Assignment of OSD phenotype was based on clinical and ophthalmoscopic examination, and morphologic studies. Animals from experimental pedigrees were examined ophthalmoscopically at approximately 8 and 12 weeks postnatal age. In selected cases, dog retinal tissues were collected postmortem and examined for routine histopathology or for higher resolution retinal morphologic examination by light microscopy. Evaluation of skeletal phenotype was based on physical examination at 8 and 12 weeks postnatal age and, in selected cases, by radiography. Dwarf dogs not retained for breeding purposes were euthanatized, and their skeletal phenotype confirmed at necropsy. All procedures involving animals were performed in accordance with the Association for Research in Vision & Ophthalmology (ARVO) Resolution for the use of Animal in Research.
DNA and RNA extraction
DNA was extracted from whole blood and spleen samples as previously described (Goldstein et al, 2006).
Retina samples from colony reference dogs were frozen in liquid nitrogen immediately after euthanasia in RNase-free conditions and stored at −70°C until used. Total RNA was extracted from retinas from a 7.6 weeks old drd1-affected dog and a 12 weeks old drd2-affected dog, as well as from two obligate heterozygotes (10.9 and 12.4 weeks old) and three homozygous normal dogs (4, 7, and 10 weeks old), using Trizol extraction, following the manufacturer’s protocol, and protected with RNase inhibitor (Roche Applied Science).
Whole genome scan and linkage analysis
A genome wide scan was undertaken by the Mammalian Genotyping Service (Marshfield, WI) as previously described (Kukekova et al., 2006). A total of 150 DNA samples were analyzed, comprising 80 Labrador retriever-derived (Figure 1A) and 70 samoyed-derived colony dogs (Figure 1B). The linkage analysis was done on the two breeds separately. All genotypes that passed Marshfield quality control were analyzed using MultiMap (Matise et al., 1994) as previously described (Mellersh et al., 1997; Acland et al., 1998, 1999., Kukekova et al, 2006). Briefly, genotypes were checked for Mendelian segregation using the prepare option of MultiMap. Linkage between each of the drd loci and each marker was determined using the MultiMap best-twopoints function. Phenotype was scored as follow: normal outbred dogs were scored homozygous 2,2; dwarf dogs were scored homozygous 1,1; obligate heterozygotes (non-dwarf dogs in back-cross litters, thus presumed carriers) were scored heterozygous 1,2, and non-dwarf dogs in intercross litters were either scored homozygous 0,0 if no ocular lesions were observed or heterozygous 1,2 if retinal folds and/or vitreal strands were detected (see supplement Table 3 for details). The regions of the dog genome identified by two-point linkage analysis with LOD >3 were further investigated. The order of markers, on chromosomes yielding positive linkage to either disease, was determined by multipoint analysis of data from both drd1- and drd2-informative pedigrees, assigning each breed as control for the other. Comprehensive maps of chromosomes 15 and 24 were built at LOD 2 support. Haplotypes for chromosome 24 and chromosome 15 were analyzed for drd1 and drd2 pedigrees, respectively, using Cyrillic (Exeter Software, Setauket, NY), given the multipoint marker order. Further linkage analyses were undertaken using LINKAGE (Lathrop et al., 1984) to confirm linkage of the OSD phenotypes, and to test for linkage using folds as the disease phenotype in drd1- derived pedigrees. For the latter analyses, dwarf dogs were scored as unknown phenotype (zero), skeletally normal dogs without observed folds were scored 1, and skeletally normal dogs with folds were scored 2.
Candidate gene screening
cDNA was synthesized from normal and drd1- and drd2-affected retinal RNA using Thermoscript RT-PCR kit (Invitrogen) with Oligo-dT.
Primers were designed from the CanFam1 canine genome database (July 2004 assembly) and CanFam2 canine genome database (May 2005 assembly, <http://genome.ucsc.edu/cgi-bin/hgGateway>) to produce overlapping fragments of the coding regions under standardized amplification conditions selected for a Tm between 56°C and 68°C, and minimal risk of primer-dimer formation (Table 2). The reactions were carried out using GoTaq green master mix (Promega, Madison, WI) and 5% DMSO. DNA sequencing was performed using the Applied Biosystems Automated 3730 DNA Analyzer (Applied Biosystems, Foster City, CA). Sequences were analyzed, assembled and compared using Sequencher® 4.2.2 Software (Gene Codes Corporation, Ann Arbor, MI). Genomic regions were screened by comparing PCR products from unaffected and affected dogs.
Table 2.
Canine primer sequences used to amplify the complete coding region of A. COL9A2 (primer pairs 1 to 10) and B. COL9A3 (primer pairs 11 to 21) from retina cDNA of normal and affected dogs. C. Primers used to establish the 3′ ends of the genes. D. Primers used to validate the 3′ RACE products. E. Primers used to retrieve the correct 5′ end of the COL9A2 gene F. Primers used to generate probes for COL9A1 gene.
| Pair | Forward primer name | Forward primer sequence | Reverse primer name | Reverse primer sequence |
|---|---|---|---|---|
| A. COL9A2 primers used to amplify the coding sequence. | ||||
| 1 | COL9A2_5UTR_F4 | ccgccccgtccgagagcagc | COL9A2exon4R | gcttcccatcaggcccatctgg |
| 2 | COL9A2exon1F | gagcctccgccgcccgcatg | COL9A2exon4_5R | gctccagttagaccatcaatcc |
| 3 | COL9A2rg1F(exon1_2) | ctggcgcagatcagaggt | COL9A2rg1R(exon10_11) | cagttggtcggacacaagaa |
| 4 | COL9A2exon2F | cctggatccgacggcatcgac | COL9A2exon7_8R | gtggtccaggaggtccagcaaag |
| 5 | COL9A2exon4_5F | tgggattgatggtctaactgg | COL9A2exon22R | gggtccgatttctccttgag |
| 6 | COL9A2exon21F | ctccctggattctctggtcc | COL9A2exon26R | gcccttctcgcctttctctc |
| 7 | COL9A2exon24F | cttgccaggcatcaagggagac | COL9A2exon31R | ggtcacccttctctccacgtttt |
| 8 | COL9A2exon24_25F | atcaagggagacaagggcttcc | COL9A2exon30R2 | ctcacagcgacctctgccagt |
| 9 | COL9A2exon29F | cgaccagcacatcgtgaccgt | COL9A2exon32R | tccttgccgttgattgcctg |
| 10 | COL9A2exon30_31F2 | ccaagggaaaacgtggagagaag | ColA2_3UTR_R2 | gaaagctggcttcctggtctgag |
| B. COL9A3 primers used to amplify the coding sequence. | ||||
| 11 | COL9A3_5UTR_F | gcgcgcagagccgctgagag | COL9A3exon8R | attccaccggggacaccact |
| 12 | COL9A3rg1R(exon10) | ggaggacccctgggtgac | ||
| 13 | COL9A3exon12R2 | cgacctctccttgatctcctt | ||
| 14 | COL9A3rg1F(exon1_2) | gcagaaagtgggacctcaag | COL9A3exon8R | attccaccggggacaccact |
| 15 | COL9A3rg1R(exon10) | ggaggacccctgggtgac | ||
| 16 | COL9A3exon4F | aagccagggaagccaggaga | COL9A3exon8R | attccaccggggacaccact |
| 17 | COL9A3rg1R(exon10) | ggaggacccctgggtgac | ||
| 18 | COL9A3exon8F | agaggcaggagagagtggtgt | COL9A3exon12R | cttgccgacctctccttgat |
| 19 | COL9A3exon25_26R | aacgccctggtctcccttag | ||
| 20 | COL9A3exon20_21F | gaagggagaactgctggagag | COL9A3exon29_30R | gatttgttcgctgagcatcc |
| 21 | COL9A3exon26F | ctggggacaaaggagagctg | COL9A3_3UTR_R2 | cccgaggtacgatgttagagc |
| C. RACE-PCR primers | ||||
| 22 | A3_3′RACE_F1 | cggggaccgaggagacaaaggctccgc | ||
| 23 | A2_3′RACE_F9 | ccgggcaggcaatcaacggcaaggacg | ||
| D. Validation of the RACE products | ||||
| 24 | A2_exon27_28F | ccaagggacagcaaggagtc | A2_Cds_1R | accagcagtcacccagcaagt |
| 25 | A2_Cds_2R | accagcagtcacccagcaagtc | ||
| 26 | A2_Cds_4R | tacagaaaggctggggtggatctc | ||
| 27 | A3_exon26F | ctggggacaaaggagagctg | A3_3UTR_AR1 | tgttagattggggtcgctcgggtt |
| 28 | A3_3UTR_AR2 | tatcacaacaggtaatagcagtagactttcc | ||
| 29 | A2_3endgapF1 | ccccacccaccctgagat | A2_3endgapR1 | cagaacagaacaaaaggcagagg |
| 30 | A2_3endgapF2 | ggacttgctgggtgactgct | A2_3endgapR2 | cttcctgtggggagcctgat |
| 31 | A2_3endgapF3 | Caccttcctcccctggacttgct | A2_3endgapR3 | aaatgcttgggtctcacactgga |
| E. Primers used to retrieve the correct 5′ end of the COL9A2 gene | ||||
| 32 | Col9A2_5UTR_F2 | tgtggtaggctttcctgaaga | A2i2R2 | cccgacccaggttagagact |
| 33 | Col9A2_5UTR_F3 | aaaagcctgacctcctagctg | ||
| 34 | Col9A2_5UTR_F7 | ccggttctgtgctgctaagccag | ||
| 35 | A2_exon1_F | gagcctccgccgccccgcatg | A2int1R11 | cgcctaacccgaaaggagcac |
| 36 | A2_exon1_F2 | ctgctgctgctccaggggctc | ||
| 37 | A2_F11 | gcccggttctgtgctgctaag | ||
| 38 | A2_exon1_F2 | ctgctgctgctccaggggctc | A2int1R | gtgaatgggcaccattgtct |
| 39 | A2_F10 | ccggttctgtgctgctaag | A2int1R10 | cctaacccgaaaggagcac |
| F. Primers used to amplify COL9A1 long and short transcript probes. | ||||
| 40 | A1F1 | cacaaaagaaaaacgaatgggaaa | A1R1 | gggaatcaaacaaagaaggcaaat |
| 41 | A1F3 | aaagggtgttgctggtgaaaag | A1R3 | atctgttggtgctctacctgga |
RNA expression analysis
To determine the 5′ and 3′ ends of COL9A2 and COL9A3, 5′ and 3′ RACE were undertaken using BD SMART RACE cDNA amplification Kit (Clontech) on a 10.4 weeks old normal retina RNA. RACE-PCR was done with gene specific primers for the 3′ and the 5′ ends (Table 2C). Products were cloned and sequenced. The 3′ RACE products were verified by RT-PCR (Table 2D).
RNA membranes containing total-RNA from normal, carrier and affected retinas for drd1 and drd2 were generated as previously described (Goldstein et al, 2006). COL9A1-long transcript, COL9A1-short transcript, COL9A2 and COL9A3 probes were produced by amplification of cDNA using primer pairs 40, 41, 5, and 11 in Table 2, respectively, cloned (TOPO TA cloning kit, Invitrogen, CA), and used for blot hybridization. Hybridization was carried out using Ultrahyb solution (Ambion, TX) following the manufacturer’s protocol. Blots were exposed to X-ray film at −70°C for 1–4 days with two intensifying screens. Loading control was achieved by hybridizing a canine-specific beta-actin probe to the membranes under the same conditions and exposed to X-ray film for ___ hours. (still need to run the beta actin hyb).
COL9A2 and COL9A3 mutation screening in the population
COL9A2 mutation screening
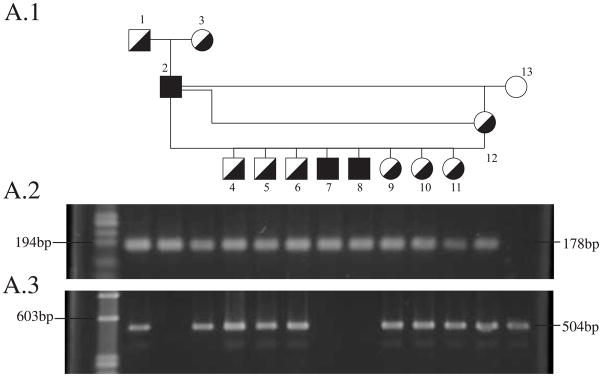
To identify drd2-affected, -carrier and -normal dogs, two sets of primer pairs were designed and used in two separate PCR reactions (primer pairs 1 and 2 in Table 3 and Supplement Figure 2B). The annealing temperature was 58°C for primer pair number 1, and 60°C for primer pair number 2. Primer pair number 1 amplifies a 1,445 bp product from homozygous normal samples, and a 178 bp product from affected dogs. Although heterozygous (carrier) dogs have both alleles, PCR of their DNA samples using primer pair 1 yields only the smaller product under the conditions used. A second PCR reaction was therefore designed, using primer pair number 2 in Table 3, to yield a 504 bp product from the normal allele, but no product from the mutated allele because the reverse primer binds within the deletion. Joint analysis of results from the two amplification reactions identifies all three genotypes (Table 3). PCR products were visualized on a 1.8% agarose gel stained with Ethidium Bromide using standard protocols (Figure 3A).
Table 3.
OSD genomic primers and expected PCR product sizes, designed to identify affected, carrier and normal dogs for COL9A2 and COL9A3 mutations.
| # | Forward primer name | Forward primer sequence | Reverse primer name | Reverse primer sequence | Expected size in wt | Expected size in carrier | Expected size in affected |
|---|---|---|---|---|---|---|---|
| 1 | COL9A2_Part11F | gctgaccttgtggattttctcc | COL9A2intron1R | gtgaatgggcaccattgtct | 1,445 bp | 178 bp | 178 bp |
| 2 | COL9A2_Part10F | catctctccctcactccctcct | COL9A2_Part10R | tcacccctctcccagtctcttag | 504 bp | 504 bp | No band |
| 3 | COL9A3test1F | gctgccactgggctcctttcttcg | COL9A3test1R | agcaggagcagggccagcgtg | 248 bp | 248 bp and 249 bp | 249 bp |
Figure 3. OSD genetic test results for drd2 (A) and drd1 (B).
A1- A subset of the colony pedigree used for the linkage analysis. A2- PCR product from the drd2 COL9A2 mutant allele on a 1.8% agarose gel. A3- PCR product from the COL9A2 normal allele on a 1.8% agarose gel. A normal dog (dog number 13) yields only the normal allele product, an affected dog (dogs number 2, 7, 8) only the mutant product and carrier dogs (remainder) yield both.
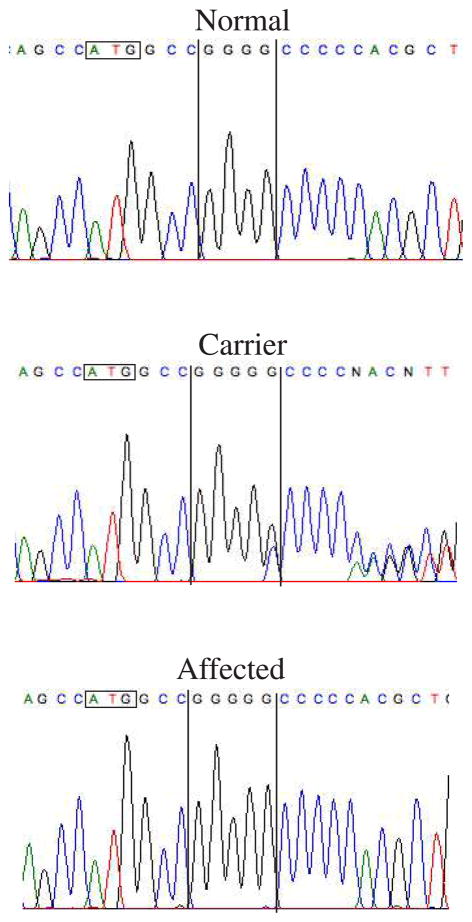
B- Chromatograms of normal, drd1-carrier and drd1-affected dogs. Four Guanines are observed in the normal sequence, 5 in affected, and overlapping peaks are observed in sequence from a carrier dog representing the two alleles.
COL9A3 mutation screening
To identify drd1-affected, -carrier and -normal dogs, genomic DNA was amplified using a primer pair flanking the mutation (Table 3, primer pair 3, and Supplement Figure 2A). PCR products were treated with ExoSAP-IT (USB) for 15 minutes at 37°C followed by inactivation at 80°C for 15 minutes, or purified with PCR purification kit (Qiagen). Direct sequencing of the PCR products with the forward primer identified the genotypes of the dogs (Figure 3B).
Results
A. Phenotypic Evaluation
In litters informative for either drd1 or drd2 (i.e. either backcross or intercross), affected (dwarf) pups were distinguishable from their non-dwarf littermates from approximately 4 to 6 weeks postnatal. The earliest and most easily recognized sign in such pups was an excessive doming of the cranium, together with a more pronounced facial stop and moderate exotropic strabismus. Foreshortening, curvature and varus/valgus deformity of the forelimbs became evident shortly thereafter. Ophthalmoscopic examination revealed a range of defects, the most consistent of which were radial cortical equatorial cataracts, and vitreal liquefaction. Complete retinal detachments were frequently present at first ophthalmoscopic examination, at approximately 6 weeks, and if not present at that age, usually developed subsequently.
Retinal detachments and cataracts were not observed in non-dwarf dogs in informative litters examined ophthalmoscopically. Retinal folds were observed in a minority of these dogs, as were vitreal abnormalities including partial liquefaction, and condensed vitreal strands. Such lesions were seen in some but not all dogs that were obligate heterozygotes for either drd1 or drd2. In Labrador retriever-derived OSD-informative litters, 14 of 34 obligate heterozygotes had retinal folds observed when examined between 6 weeks and 3 months of age and 20 were ophthalmoscopically normal. In samoyed-derived OSD-informative litters examined at the same age a higher proportion of obligate heterozygotes had retinal folds observed compared to drd1-heterozygotes, but a large minority of these drd2-heterozygotes were ophthalmoscopically normal at the same age.
B. Whole genome wide scan and linkage analysis
To map drd1 and drd2, experimental three-generation pedigrees were developed. Eight related colony-dog families segregating drd1 included 5 intercross breedings with 50 progeny and three backcross breedings with 18 progeny, for a total of 68 individuals in informative generations. Five drd2-informative pedigrees included 2 intercross breedings with 42 progeny and three backcrosses with 21 progeny, for a total of 63 individuals in informative generations (Supplement Table 2).
The drd1 and drd2 experimental pedigrees (150 individuals) were genotyped with the canine 249 microsatellite marker set at Marshfield Laboratories. Nine markers, 4 Labrador-derived and 3 samoyed-derived dogs were excluded from the study after quality control analysis. Initial linkage mapping and fine mapping of both diseases followed similar procedures: first, two-point linkage analysis between each disease and 240 markers was performed to map disease loci, then disease intervals were identified using multipoint analysis, and, finally, disease loci were refined using haplotype analysis.
From drd1–informative (Labrador-derived) pedigrees, a maximum lod score of 3.589 was obtained on chromosome 24 (CFA24) for marker FH2079 at theta = 0.142, when all unaffected (i.e. non dwarf) dogs from intercrosses were designated as drd1–carriers (heterozygotes) (Table 1A, linkage analysis II); a lower lod score of 2.225 at theta = 0.19 was achieved when these nonaffected dogs were designated as of unknown status (Table 1A, linkage analysis I). The order of the CFA24 markers and the relative position of the drd1 locus were established by multipoint linkage analysis at LOD 2 support. Five markers mapped to unique positions and their order was in agreement with CanFam2. The drd1 locus was mapped to the telomeric end of CFA24, 12 cM distal to FH2079. The order of CFA24 markers established by haplotype analysis of drd1 informative pedigrees was in agreement with the marker order identified by multipoint analysis. Haplotype analysis identified 13 affected dogs, and a few heterozygotes, as recombinants between FH2079 and the disease locus (data not shown) and confirmed the mapping of the drd1 locus achieved by multipoint analysis, suggesting that drd1 was most likely distal to FH2079 in the interval telomeric to CFA24: 39,338,870.
Table 1.
Linkage analysis results for the drd1 and drd2 diseases in the colony models.
| A. drd1 linkage analysis results | ||||
|---|---|---|---|---|
| Marker | Location on chromosome 24 | Linkage analysis I a (Lod score, theta) | Linkage analysis II b (Lod score, theta) | Linkage analysis III c (Lod score, theta) |
| FH3750 | 3,633,845-3,634,019 | 0.409, 0.36 | 0.369, 0.366 | 0.29, 0.414 |
| AHTH138 | 9,244,217-9,244,422 | 0.119, 0.413 | 0.119, 0.411 | 0.069, 0.451 |
| FH2168 | 11,080,590-11,080,803 | 0.135, 0.416 | 0.148, 0.404 | 0.218, 0.418 |
| FH2261 | 21,330,554-21,330,729 | 0.709, 0.25 | 0.756, 0.24 | 0.725, 0.267 |
| AHT125 | 29,857,622-29,857,723 | 1.463, 0.188 | 1.463, 0.193 | 1.472, 0.216 |
| FH2079 | 39,338,601-39,338,870 | 2.225, 0.19 | 3.589, 0.142 | 3.565, 0.197 |
| COL9A3 mutation | 49,699,847-49,699,850 | 25.889, 0.0 | ||
| B. drd2 linkage analysis results | ||||
| Marker | Location on chromosome 15 | Linkage analysis IVd (Lod score, theta) | Linkage analysis Ve (Lod score, theta) | |
| FH3802 | 4,372,086-4,372,350 | 2.382, 0.077 | 2.382, 0.077 | |
| COL9A2-mutation | 5,641,441-5,642,420 | 23.179, 0.0 | ||
| REN06C11 | 21,168,041-21,168,127 | 16.461, 0.031 | 17.915, 0.0 | |
| FH2535 | 31,159,141-31,159,256 | 11.448, 0.076 | 10.136, 0.104 | |
| RVC1 | 40,767,046-40,767,209 | 8.395, 0.12 | 7.734, 0.162 | |
| FH2088 | 53,905,651-53,905,779 | 2.053, 0.274 | 2.722, 0.373 | |
| FH2360 | 56,517,563-56,517,873 | 0.719, 0.357 | 1.485, 0.42 | |
Initial analysis was done when all carriers from intercrosses scored 0,0 (n=30)
Initial analysis was done when all carriers from intercrosses scored 1,2 (n=30)
Final analysis was done when all carriers from intercross scored as genotyped (25=1,2; 5=2,2)
Initial analysis was done when 26 unaffected animals from intercrosses were used and scored (23=1,2; 3=2,2).
Final analysis was done when all 29 unaffected from intercrosses were used and scored according to genotype (25=1,2; 4=2,2).
For drd2–informative (samoyed-derived) pedigrees, a maximum lod score of 16.461 was obtained on chromosome 15 (CFA15) with marker REN06C11 at theta= 0.031 (Table 1B, linkage analysis IV). Multipoint analysis assigned drd2 to the centromeric end of CFA15, proximal to REN06C11 with theta 0.0. The relative order of five markers mapped on CFA15 and the drd2 locus was established by multipoint analysis. Four CFA15 markers and drd2 mapped to unique positions at LOD 2 support and the order of mapped markers was in agreement with CanFam2. Because FH3802 was of low informativeness in the samoyed-derived pedigrees, it could not be assigned to a unique map position. According to CanFam2, however, FH3802 is located proximal to REN06C11. The drd2 locus was thus assigned to the centromeric end of CFA15, approximately 3.1 cM proximal to REN06C11. Haplotype analysis identified two affected dogs as recombinant between marker REN06C11 and drd2. Because FH3802 was so poorly informative, no recombination between the disease locus and this marker was recognizable. All 26 unaffected dogs from intercross breedings had haplotypes consistent with their phenotypic scores, thus supporting the mapping of the drd2 disease locus to the interval centromeric to CFA15:21,168,041.
These mapping results strongly suggested COL9A2 on CFA15 (5,651,564–5,665,086) and COL9A3 on CFA24 (49,700,412–49,713,993), as positional candidates for drd2 and drd1, respectively, because they code for collagen proteins, as do other genes previously implicated in human oculoskeletal dysplasias.
C. Candidate gene evaluation
C.1 COL9A2 evaluation as a candidate gene for OSD in the samoyed
C.1.1. Canine COL9A2 mRNA
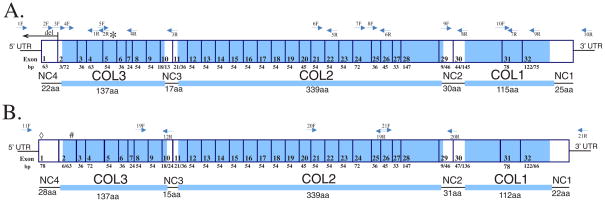
The human COL9A2 (NM_001852.3) has 32 exons and is 2,831bp long. Blasting this sequence against CanFam2 identified 30 predicted exons but fail to identify exons 1 and 26. Alignment of NM_001852.3 against the CanFam1 assembly identified a predicted exon 1 and a canine EST (DN364641) to support this prediction, but not exons 26 or 32. Ten primer pairs were designed based on the CanFam1 and CanFam2 assemblies to amplify overlapping cDNA fragments covering the complete coding sequence of canine COL9A2 (Table 2A). All ten successfully amplified the expected target fragments from retinal cDNA of normal dogs aged 4 weeks old to 12 weeks old. PCR products were sequenced and the canine COL9A2 mRNA was assembled using Sequencher software (Accession #___, Supplement Figure 1B). The structure of this canine mRNA was highly similar to the human mRNA sequence NM_001852.3 (Figure 2A). The length of each exon was identical to that of the corresponding human one, including the junction exons (i.e. exons that are junctional between the NC and COL domains), with one exception -- canine exon 1 is 12 bp shorter than the human, and encodes only 21 amino acids compared to 25 in human. The canine protein is predicted to have a very similar structure overall to the human with four non-collagenous domains (NC1 to NC4) and three collagenous domains (LOC1 to LOC3) (Figure 2A). The canine NC4 domain has 22 amino acids compared to 26 in human (NP_001843.1).
Figure 2.
Schematic illustrations of COL9A2 (A) and COL9A3 (B) mRNAs and their corresponding protein domains. Shaded areas in the mRNA correspond to the COL domains; unshaded to NC domains. Gray arrows indicate binding sites of primers used to amplify the cDNA.
A. The long black arrow and the asterisk (*) indicate the location of the drd2 COL9A2 mutation (deletion of exon 1) and its putative result (a first methionine in exon 5), respectively.
B. The diamond symbol (◇) and pound symbol (#) indicate the location of the drd1 COL9A3 mutation (insertion in exon 1) and its result (premature stop codon in exon 2) respectively.
The 5′ and 3′ ends of the normal transcript were determined by RACE. 3′ RACE-PCR used primer A2_3′RACE_F9 located on the last exon (Table 2C, primer 23). One transcript was identified with a poly A tail. This product extended 587 bp after the stop codon, and included a polyadenylation signal preceding the start of the poly A tail by 30 bp. This sequence is missing from both CanFam1 and CanFam2. To validate the authenticity of the poly A tail, genomic DNA from several normal dogs was amplified using primers located in sequence flanking both sides of the gap (primer pairs 29–31, Table 2) to retrieve the missing sequence (Accession #_____). The absence of a polyadenosine stretch from the sequenced products was taken as a validation of the 3′ RACE product, which was further validated by RT-PCR amplification of normal retinal cDNA using a forward primer located on exon 27–28 and reverse primers located just upstream to the poly A tail (primer pairs 24–26, Table 2). 5′ RACE experiments failed, presumably because of the GC-richness of the 5′ end of this transcript.
C.1.2. Establishing the canine 5′ end genomic sequence of COL9A2
Comparison of CanFam1 and CanFam2 revealed differences in the 5′ end, exon 1 and intron 1 of COL9A2 between the two assemblies. Forward primers were designed based on 5′ end sequence shared between the two assemblies, reverse primers were designed from intron 2 of the gene, and further primers were subsequently designed based on newly derived sequences to cover the entire region (Primer pairs 32–39, Table 2E). Genomic DNA from a nonaffected dog was amplified and the correct sequence determined (Accession #____, Supplement Figure 1A) as similar but not identical to that in CanFam1.
C.1.3. Evaluation of COL9A2 in drd2 affected samoyed
The COL9A2 gene was amplified from retinal cDNA from a 12 week old drd2-affected dog. Except for exon 1, the gene amplified successfully and showed only one difference from the normal mRNA sequence: a polymorphism (C/T transition) in exon 3 (chr15: 5,652,893, in CanFam2), with the C allele homozygous in all drd2-affected individuals. This SNP changes an amino acid (Proline to Serine) but is not causally associated with the disease because the C allele was also observed in dogs homozygous normal for the disease. Because cDNA amplification failed for primer pairs 1, 2 and 3 (Table 2, Figure 2), a sequence difference was suspected in the 5′ end of the gene in affected dogs. Once the correct sequence for the 5′ end of the region was established from an unaffected animal, primers were designed to amplify a) the genes SMAP1L and ZNF643 that flank COL9A2, from both retinal cDNA and genomic DNA (supplement Table 4, primer pairs 1–5); b) the approximately 31 Kb region between SMAP1L and COL9A2- exon2 in 11 distinct amplicons evenly distributed (Supplement Table 4, primer pairs 6 to 21); and c) exon 2 of COL9A2 and its flanking donor-acceptor sites (Supplement Table 4, primer pair 22).
Genomic sequence of COL9A2 exon 2 revealed no differences between drd2-affected and normal DNA, including the splice acceptor site of intron 1 and splice donor site of intron 2. Two genes (SMAP1L, ZNF643) located 5′ to COL9A2, amplified successfully from both normal and affected retinal cDNA, suggesting that the mutation does not involve these regions. Six of the 11 primer pairs designed to amplify fragments located between SMAP1L and intron 2 of COL9A2 yielded identically sized products from normal and affected substrates. Five pairs, however, yielded different results for affected compared to normal samples: primer pairs 15 and 17 failed to amplify an affected product, and amplicons from pairs 16, 18 and 19 were smaller for affected compared to normal. Sequence analysis of these products revealed a 1,267bp deletion in affected dogs. This mutation deletes all of exon 1 and parts of both the 5′ UTR and intron 1 (Supplement Figure 1A and 2B).
C.2 COL9A3 evaluation as a candidate gene for OSD in the Labrador retrievers
C.2.1. Canine COL9A3 mRNA
The human COL9A3 gene (NM_001853.2) has 32 exons and is 2,511 bp long. Blasting this sequence against CanFam2 identified 29 of the predicted canine exons but failed to identify exons 16, 17 and 32. Thirteen primers were designed from CanFam2 sequence to amplify 11 different overlapping cDNA fragments covering the complete canine COL9A3 coding sequence (Table 2B, primer pairs 11 to 21). A 3′ UTR primer was designed from CanFam2 sequence downstream to the gap where exon 32 is presumably located.
Canine COL9A3 fragments were amplified successfully from normal retinal cDNAs and the products sequenced. A consensus sequence for canine COL9A3 was assembled using Sequencher (Accession #___, Supplement Figure 1C). The structure of the canine mRNA is identical to that of human mRNA NM_001853.3 (Figure 2). Comparison of the predicted protein to human COL9A3 (NP_001844.3) also suggests a completely identical structure.
The 5′ and 3′ ends of the transcript were determined by RACE. 3′ RACE-PCR used primer COL9A3_RACE_F1 located on exon 31 (Table 2C, primer 22). Products were cloned and sequenced. Two polyadenylated-transcripts were identified: one extended 458 bp after the stop codon and included a polyadenylation signal preceding the start of the poly A tail by 11 bases; the second extended 591 bp after the stop and included a polyadenylation signal preceding the poly A tail by 16 bases (Supplement Figure 1C). The corresponding genomic 3′ end of the gene had no such stretches of adenosine. The 3′ RACE products were further validated by RT-PCR of cDNA from unaffected retina using a forward primer located on exon 26, and 2 reverse primers each located just upstream to the polyA tail (Primer pairs 27 and 28 in Table 2 for the short and long transcripts, respectively). 5′ RACE experiments failed presumably because of GC-richness in the 5′ end of this transcript.
Four single nucleotide polymorphisms were found in the normal transcript within the coding sequence, none of which cosegregate with the disease: G/A in exon 4 (silent), G/A in exon 5 (arginine/glutamine), C/T in exon 28 (silent) and A/G in exon 31 (silent), (nucleotides 49,702,320; 49,702,704; 49,712,359; 49,713,957 respectively in CanFam2). Seven more SNPs were identified in the 3′ UTR (Supplement Figure 1C).
C.2.2. Evaluation of COL9A3 in drd1 affected Labradors
To evaluate the role of COL9A3 in the Labrador drd1 disease, retinal cDNA from a 7.6 weeks old drd1-affected dog was amplified using gene-specific primers. Sequence of the PCR products revealed a one-base insertion (guanine) in exon 1 that changes a string of 4 guanines (chr24:49,699,847–49,699,850, CanFam2;) to a string of 5 guanines. This shifts the reading frame and introduces a premature stop codon after 48 codons. This insertion is linked to a G-G-T-A haplotype corresponding to the 4 SNPs found in the coding region (Supplement Figure 1D).
D. Segregation evaluation and population study
D.1. OSD in the samoyed (drd2)
To identify drd2-affected, carrier and normal dogs in the samoyed-derived pedigrees and confirm co-segregation of the COL9A2 mutation with the disease, genomic DNA of the 70 samoyed-derived dogs used in the linkage analysis was amplified using two primer pairs designed for screening (Table 3 and Figure 3A, see methods). Analysis of the PCR products showed that all outbred normal dogs were homozygous wild type; all affected dogs were homozygous for the deletion; and all drd2-obligate heterozygotes were heterozygous for the deletion. The 29 unaffected dogs from intercross litters genotyped as follows: 25 as carriers (23 of which had been scored 1,2 in the multimap analysis, 1 scored 2,2 and 1 not used), and 4 as normal (2 not used and 2 scored 2,2), (supplement Table 3). Two-point analysis to measure linkage between the mutation and the disease phenotype gave a lod score of 23.179 at theta 0.0 (Table 1B, linkage analysis V).
55 unrelated pedigreed samoyed dogs were screened for the deletion using the above screening test. One was identified as a carrier of the deletion. Six relatives of this dog were then tested and one of these was also identified as a carrier. The affected allele frequency is thus very roughly estimated as about 0.01 in the samoyed population. Another 126 dogs from 26 additional breeds considered to be normal for OSD were screened for the deletion, including 8 dogs from 4 breeds considered to be dwarf breeds (American bulldog, Pomeranian, Corgi and Duchshund); all were homozygous wildtype (supplement Table 1, A and C).
D.2 OSD in Labradors (drd1)
To identify drd1-affected, carrier and normal dogs in the Labrador-derived pedigrees and confirm cosegregation of the COL9A3 mutation with the disease, genomic DNA of the 80 Labrador-derived dogs used in the linkage analysis were amplified using primer pairs COL9A3test1F and COL9A3test1R (Table 3, Figure 3B, see methods). Sequencing of the resulting amplicon with the forward primer identified normal animals (4 Gs) or affected animals (5 Gs) while an overlapping sequence was observed at the insertion point in chromatograms from carrier dogs. All outbred normal dogs were homozygous for the 4 Gs haplotype, all affected dogs were homozygous for the insertion (5 Gs), and all obligate drd1-heterozygotes were heterozygous for the insertion (producing overlapping chromatograms). Of the 30 nonaffected dogs in intercross litters, 5 genotyped homozygous normal (4 Gs) and 25 were heterozygous for the insertion (Supplement Table 3). Because all 30 dogs had been scored as carriers in the previous multimap analysis, two-points analysis was rerun to measure linkage between the mutation and the disease phenotype. That gave a lod score of 25.889 at theta 0.0 (Table 1A, linkage analysis III). This was confirmed by analysis using LINKAGE, which yielded a lod score of 18.72 at theta 0.0.
Fifty-nine unrelated Labrador dogs (i.e. with no parents or grandparents in common) considered to be phenotypically normal for OSD, and 19 Labrador retrievers with ophthalmoscopically diagnosed retinal folds were tested for the COL9A3 mutation. One dog with retinal folds was found to be homozygous for the mutation. Subsequent enquiry determined that this dog also had orthopedic abnormalities consistent with short-limbed dwarfism. From these results we can very roughly estimate the frequency of the affected allele in the Labrador population as about 1% (0.012). Seventy- eight dogs from 21 additional breeds, all considered to be normal for OSD were also screened; among them were 8 dogs from 4 breeds considered to be dwarf breeds (American bulldog, Pomeranian, Corgi and Dachshund). All 78 dogs proved homozygous wildtype at the COL9A3 locus (Supplement Table 1, B and C).
E. RNA expression analysis
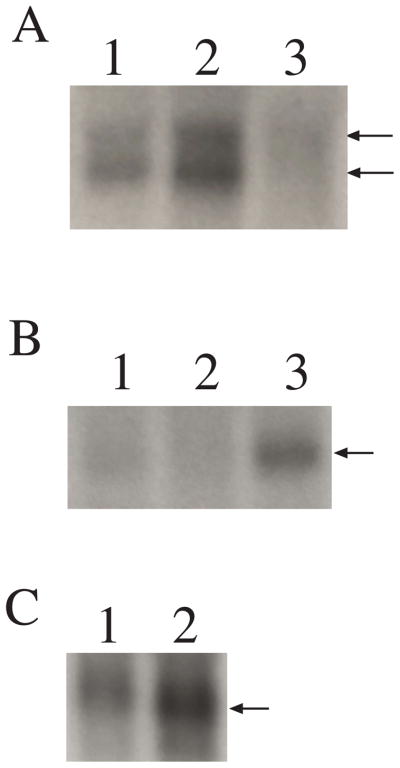
Northern blot analysis shows two transcripts of COL9A2, approximately 3.0 Kb and 3.8 Kb long, in drd2-nonaffected retinas (Figure 4A, lanes 1 and 2). Both bands were absent from a 12 weeks old drd2-affected retina (Figure 4A, lane 3). The same affected retina shows expression of COL9A3 (Figure 4B, lane 3), suggesting that the absence of the COL9A2 mRNA is due not to cell death but to other regulatory mechanisms. The low mRNA expression levels may result from loss of the promoter region which is normally present in the 5′ region of the gene that is deleted in the mutant allele. A similar expression pattern is observed in drd1: only one COL9A3 transcript (approximately 3.0 Kb) is observed in drd1-nonaffected retina (Figure 4B, lanes 1 and 3), and this is absent in a 7.6 weeks old drd1-affected retina (Figure 4B, lane 2). The same retina does express COL9A2 (Figure 4A, lane 2) which suggests that the absence of the COL9A3 mRNA is due not to cell death but rather to other regulatory mechanisms. In this case, since the promoter region is not mutated, it is likely that nonsense-mediated-decay is the mechanism responsible.
Figure 4. RNA expression of COL9A2 (A), COL9A3 (B), and COL9A1(C) in the dog retina.
A. COL9A2 is expressed in the retina of a dog not affected with OSD (6.9 weeks old, lane 1) and in drd1-affected retina (7.6 weeks old, lane 2). Two bands are observed of approximately 3.0 Kb and 3.8 Kb long. In drd2-affected retina, COL9A2 is at undetectable levels (12 weeks old, lane 3).
B. COL9A3 is expressed in the retina of a dog not affected with OSD (6.9 weeks old, lane 1) and in drd2-affected retina (12 weeks old, lane 3). One band is observed of approximately 3.0 Kb long. In drd1-affected retina, COL9A3 is at undetectable levels (lane 2).
C. COL9A1 (short transcript) is expressed in normal retina (10.4 weeks- lane C1 and 4.3 weeks old-lane 2). One band of approximately 3.0 Kb is observed. The long transcript was not detectable in the normal retina (data not shown). Beta actin was used as a loading control.
COL9A1 expression in normal retina was evaluated using two probes, one specific for the long variant of this collagen IX subunit (5′ UTR to exon 5) and another that is common to the long and short forms (exon 26 to exon 34/35), (Table 2F). Results indicate that COL9A1 is expressed in the normal retina in its short form (approximately 3.0 kb, Figure 4C) while its long form is not expressed (data not shown).
E. Folds evaluation
Since folds were observed in some but not all drd1-carriers, we re-evaluated the genome wide scan data for the drd1-OSD pedigrees to determine whether a locus linked to the folds phenotype might be identifiable. A simulation using LINKAGE to estimate the power of the pedigree yielded a lod score of 6.7 with theta 0.0 assuming a recessive mode of inheritance. Analysis of all 240 markers revealed no significant linkage to the folds phenotype in these pedigrees (all lod scores less than 3.0, data not shown). Similarly, LINKAGE analysis to evaluate cosegregation of the folds phenotype with the drd1 COL9A3 locus (i.e. the mutation) revealed no significant evidence of linkage (Lod score −3.882e-6 at theta=0.5).
Discussion
In humans, mutations in COL9A2 and COL9A3 have been associated with multiple epiphyseal dysplasia (MED, EDM2 and EDM3 for COL9A2 and COL9A3 mutations, respectively), (Bonnemann et al, 1999; Paasilta et al, 1999; Holden et al, 1999; Lohiniva et al, 2000; Fiedler et al, 2002; Nakashima et al, 2005a; Nakashima et al, 2005b; Takahashi et al, 2006). MED is one of the common forms of skeletal dysplasia, characterized by a delay in the appearance of the epiphyses, irregular epiphyseal formation, mild short stature, and early-onset osteoarthiritis (Spranger et al, 2002). All known mutations in humans cause exon 3 skipping, resulting in the predicted consequence of a 12 amino acid deletion within the COL3 domain of the α2(IX) and α3(IX) chains. One MED-affected human was found to be heterozygous for a COL9A1 gene mutation (Czarny-Ratajczak et al, 2001). This was a splice-site mutation that resulted in an in-frame deletion of exon 8 and/or exon 10 and lead to an in-frame deletion of 25, 21 or 46 amino acids in the COL3 domain of alpha1(IX) chain. Since COL9A1 contains six additional exons compared to COL9A2 and COL9A3, exon 9 of COL9A1 corresponds to exon 3 in the other two COL9 genes. This remarkable consistency of type IX collagen mutations, clearly suggests that the COL3 domain is of critical functional importance in type IX collagen. It has been suggested that deletions in the COL3 domain disrupt interaction with other components or that the orientation of the NC4 domain relative to type II/XI collagen fibril is changed by the COL3 mutations (Van Camp et al, 2006).
Here we report mutations in the COL3 domain of COL9A2 and COL9A3 associated with OSD in samoyed and Labrador retriever dogs, respectively. In drd2-affected samoyeds, the mutation is a 1,267 bp deletion in the 5′ end of the gene. This deletion includes the entire exon 1, and the predicted protein would be missing the complete NC4 domain and 69 amino acids from the COL3 domain. In drd1-affected Labradors, the mutation is an insertion of a guanine in exon 1, causing a +1 frame-shift and resulting in an introduction of a premature stop codon and predicted truncated protein with only 48 amino acids, of which only the first three are identical to the functional peptide. In both diseases, the complete phenotype, including skeletal and ocular defects, is transmitted as an autosomal recessive trait, but ophthalmoscopic findings have suggested a possibly semi-dominant mechanism in which heterozygous dogs, although skeletally normal, can either show no ophthalmoscopic abnormalities or have multiple retinal folds, vitreal membranes or vitreous degeneration. Since in both disorders it is predicted that the alpha peptide is truncated and completely nonfunctional, it is possible that the effects of haploinsufficiency are not significant in cartilage, but can be apparent in vitreous. Further analysis of the mRNA expression levels in the carriers, and protein expression studies is needed to evaluate these issues.
Although attempts to amplify the 5′ UTR of COL9A2 cDNA failed, sequence analysis of the corresponding genomic region reveals several promoter elements in the normal chromosome. Sp1 elements (GGGCGG) are located at −153 bp, −326 bp and −607 bp upstream to the ATG. A TATA box is located at −975 bp upstream from the ATG start codon. These elements are missing from the affected chromosome and yet the mutated COL9A2 mRNA is still expressed at least sufficiently enough to allow amplification by RT-PCR. This suggests that a different upstream promoter region is active in affected chromosomes. A TATA box at −1,287 bp and −1409 bp upstream to the ATG, as well as CCAAT-enhancer biding site at −1,402 bp, are recognizable and are not deleted from the affected chromosome. RT-PCR with forward primers located immediately 5′ to the deletion and reverse primers located on several of the exons, show positive transcripts (data not shown). Thus these upstream TATA boxes may be alternatively activated in affected chromosomes. Since very weak expression is observed at the RNA level, it might be that the alternative promoter is not regulated or that those mRNA molecules are degraded very quickly by nonsense-mediated decay.
In humans, a recessive form of Stickler Syndrome has been reported in a family with a nonsense mutation (R295X) in exon 9 of COL9A1 (Van Camp et al, 2006). It is postulated that, in this family, Collagen IX is absent in affected members. These patients had ophthalmologic findings that differed from those affected with the more usual autosomal dominant form of Stickler Syndrome. Affected patients had degenerative vitreous changes, including progressive gel liquefaction, that resembled those in aged eyes. This is similar to the ophthalmologic findings in the affected Labrador retrievers and samoyeds, although the abnormalities are much more severe in homozygous dogs. The R295X-affected members also showed a moderate-to-severe hearing impairment, with a mildly down-sloping audiogram. In OSD-affected Labrador and samoyed dogs, no obvious signs of hearing impairment has been recognized, though specific quantitative hearing tests have not yet been undertaken. Nevertheless, these canine diseases provide valuable models for Stickler Syndrome in its recessive mode.
Dogs homozygous for either drd1 or drd2 can usually be recognized upon physical examination by an experienced veterinarian, particularly an experienced veterinary ophthalmologist, or an experienced veterinary orthopedist, and sometimes by experienced dog breeders. However, recognition of affected dogs is not sufficient to allow adequate selection pressure to be applied to significantly reduce the gene frequency of the mutation in the population. It has been widely held that dogs heterozygous for either drd1 or drd2, can usually be recognized upon physical examination by the presence of retinal folds or retinal dysplasia. However, we find no evidence to support this hypothesis in the current study. Although one Labrador retriever dog with an ophthalmoscopically diagnosed retinal fold genotyped homozygous for the drd1 mutation, 18 with retinal folds were wild-type normal at the locus (see results). In a purebred Labrador retriever pedigree segregating the drd1-OSD disease, several dogs had retinal folds but were homozygous for the wild-type allele, whereas others had no ophthalmoscopically recognized abnormalities but were heterozygous for the drd1-mutation (data not shown). In the colony-derived Labrador retrievers, 41% of the drd1-carrier dogs had folds and 51% did not show the folds phenotype. One samoyed dog was found to be a carrier for the COL9A2 mutation and had no reported ophthalmologic abnormalities.
It should be noted that several purebred Labrador retrievers, diagnosed clinically as affected with shortlimbed dwarfism were found to be homozygous normal when tested for the drd1 mutation (data not shown). n of these Labrador retrievers have also been genotyped for the recently identified mutation causing chondrodysplasia in multiple shortlegged breeds of dog (Parker et al., 2009) with the following results: [ORLY to add data asap]. These findings suggest that: (i) At least in Labrador retrievers there are likely to be further mutations yet to be found that account for phenotypes including retinal folds and or dwarfisms; (ii) retinal folds, if caused by OSD mutations, might be also influenced by other modifier loci or by environmental effects These findings emphasize the need for molecular genetic testing to obtain accurate genotypes of these dogs.
These OSD canine models can be used to further evaluate the relationships between the COL9 alpha chains A1, A2 and A3, their regulatory mechanism and their effect on folds and vitreal structure in the eye, as well as future treatment possibilities including, potentially, gene therapy.
Supplementary Material
References
- Acland GM, Aguirre GD. Oculoskeletal dysplasia in samoyed and Labrador retriever dogs: 2 nonallelic disorders akin to Stickler-like syndromes affecting humans. Presented at the 2nd International DOGMAP meeting; Cambridge, Great Britain. 1995. [Google Scholar]
- Acland G, Ray K, Mellersh C, Langston A, Rine J, Ostrander E, Aguirre G. A Novel Retinal Degeneration Locus Identified by Linkage and Comparative Mapping of Canine Early Retinal Degeneration. Genomics. 1999;59(2):134–142. doi: 10.1006/geno.1999.5842. [DOI] [PubMed] [Google Scholar]
- Acland GM, Ray K, Mellersh CS, Gu W, Langston AA, Rine J, Ostrander EA, Aguirre GD. Linkage analysis and comparative mapping of canine progressive rod-cone degeneration (prcd) establishes potential locus homology with retinitis pigmentosa (RP17) in humans. Proc Nat Acad Sc USA. 1998;95(6):3048–53. doi: 10.1073/pnas.95.6.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunen S, Korkko J, Czarny M, Warman ML, Brunner HG, Kaariainen H, Mulliken JB, Tranebjaerg L, Brooks DG, Cox GF, Cruysberg JR, Curtis MA, Davenport SL, Friedrich CA, Kaitila I, Krawczynski MR, Latos-Bielenska A, Mukai S, Olsen BR, Shinno N, Somer M, Vikkula M, Zlotogora J, Prockop DJ, Ala-Kokko L. Splicing mutations of 54-bp exons in the COL11A1 gene cause Marshall syndrome but other mutations cause overlapping Marshall/Stickler phenotypes. Am J Hum Genet. 1999 Oct;65(4):974–83. doi: 10.1086/302585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayme S, Preus M. The Marshall and Stickler syndromes: objective rejection of lumping. J Med Genet. 1984 Feb;21(1):34–8. doi: 10.1136/jmg.21.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnemann CG, Cox GF, Shapiro F, Wu JJ, Feener CA, Thompson TG, Anthony DC, Eyre DR, Darra BT, Kunkel LM. A mutation in the alpha 3 chain of type IX Collagen causes autosomal dominant multiple epiphyseal dyplasia with mild myopathy. PNAS. 1999;97:1212–1217. doi: 10.1073/pnas.97.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrig CB, MacMillan A, Brundage S, Pool RR, Morgan JP. Retinal dysplasia associated with skeletal abnormalities in Labrador retrievers. JAVMA. 1977;170:49–57. [PubMed] [Google Scholar]
- Czarny-Ratajczak M, Lohiniva J, Rogala P, Kozlowski K, Perälä M, Carter L, Spector TD, Kolodziej L, Seppänen U, Glazar R, Królewski J, Latos-Bielenska A, Ala-Kokko L. A mutation in COL9A1 causes multiple epiphyseal dysplasia: further evidence for locus heterogeneity. Am J Hum Genet. 2001;69:969–980. doi: 10.1086/324023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F, Acland GM, Ray J. Cloning and expression of type II collagen mRNA: evaluation as a candidate for canine oculo-skeletal dysplasia. Gene. 2000 Sep 19;255(2):307–16. doi: 10.1016/s0378-1119(00)00324-3. [DOI] [PubMed] [Google Scholar]
- Du F. PhD Thesis. Cornell University; 2000. Molecular and genetic studies of Oculo-Skeletal Dysplasia (OSD) in Dogs. [Google Scholar]
- Fiedler J, Stove J, Heber F, Brenner RE. Clinical Phenotype and Molecular Diagnosis of Multiple Epiphyseal Dysplasia with Relative Hip Sparing During Childhood (EDM2) Am J Med Genet. 2002;112:144–153. doi: 10.1002/ajmg.10554. [DOI] [PubMed] [Google Scholar]
- Goldstein O, Zangerl B, Pearce-Kelling S, Sidjanin DJ, Kijas JW, Felix J, Acland GM, Aguirre GD. Linkage disequilibrium mapping in the domestic dog breeds narrows the progressive rod-cone degeneration interval and identifies ancestral disease-transmitting chromosome. Genomics. 2006;88:541–550. doi: 10.1016/j.ygeno.2006.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden P, Canty EG, Mortier GR, Zabel B, Spranger J, Carr A, Grant ME, Loughlin JA, Briggs MD. Identification of Novel pro-alpha2(IX) Collagen Gene Mutations in Two Families with Distinctive Oligo-epiphyseal Form of Multiple Epiphyseal Dyplasia. Am J Hum Genet. 1999;65:31–38. doi: 10.1086/302440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukekova AV, Nelson J, Kuchtey RW, Lowe JK, Johnson JL, Ostrander EA, Aguirre GD, Acland GM. Linkage Mapping of Canine Rod Cone Dysplasia Type 2 (rcd2) to CFA7, the Canine Orthologue of Human 1q32. IOVS. 2006;47:1210–1215. doi: 10.1167/iovs.05-0861. [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J. Strategies for multilocus linkage analysis in humans. Proc Nat Acad Sci USA. 1984;81:3443–3446. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohiniva J, Paassilta P, Seppanen U, Vierimaa O, Kivirikko S, Ala-Kokko L. Splicing Mutations in the COL3 Domain of Collagen IX Causes Multiple Epiphyseal Dyplasia. Am J Med Genet. 2000;90:216–222. [PubMed] [Google Scholar]
- Matise TC, Perlin M, Chakravarti A. Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat Genet. 1994 Apr;6(4):384–90. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]
- Maumenee IH, Traboulsi EI. The ocular findings in Kniest dysplasia. Am J Ophthalmol. 1985 Jul 15;100(1):155–60. doi: 10.1016/s0002-9394(14)74998-0. [DOI] [PubMed] [Google Scholar]
- Mellersh CS, Langston AA, Acland GM, Fleming MA, Ray K, Wiegand NA, Francisco LV, Gibbs M, Aguirre GD, Ostrander EA. A linkage map of the canine genome. Genomics. 1997 Dec 15;46(3):326–36. doi: 10.1006/geno.1997.5098. [DOI] [PubMed] [Google Scholar]
- Meyers VN, Jezyk PF, Aguirre GD, Patterson DF. Short-limbed dwarfism and ocular defects in the samoyed dog. JAVMA. 1983;183:975–979. [PubMed] [Google Scholar]
- Nakashima M, Ikegawa S, Ohashi H, Kimizuka M, Nishimura G. Double- Layered Patella in Multiple Epiphyseal Dysplasia Is Not Exclusive to DTDST Mutation. Am J Med Genet. 2005a;133A:106–107. doi: 10.1002/ajmg.a.30481. [DOI] [PubMed] [Google Scholar]
- Nakashima M, Kitoh H, Maeda K, Haga N, Kosaki R, Mabuchi A, Nishimura G, Ohashi H, Novel Ikegawa S. COL9A3 Mutation in a Family with Multiple Epiphyseal Dysplasia. Am J Med Genet. 2005b;132A:181–184. doi: 10.1002/ajmg.a.30411. [DOI] [PubMed] [Google Scholar]
- Paasilta P, Lohiniva J, Annunem S, Bonaventure J, Le-Merrer M, Pai L, Ala-Kokko L. COL9A3: A Third Locus for Multiple Epiphyseal Dysplasia. Am J Hum Genet. 1999;64:1036–1044. doi: 10.1086/302328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, Vonholdt BM, Quignon P, Margulies EH, Shao S, Mosher DS, Spady TC, Elkahloun A, Cargill M, Jones PG, Maslen CL, Acland GM, Sutter NB, Kuroki K, Bustamante CD, Wayne RK, Ostrander EA. An Expressed Fgf4 Retrogene Is Associated with Breed-Defining Chondrodysplasia in Domestic Dogs. Science. 2009 Jul 16; doi: 10.1126/science.1173275. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini B, Acland GM, Ray J. Cloning and characterization of opticin cDNA: evaluation as a candidate for canine oculo-skeletal dysplasia. Gene. 2002 Jan 9;282(1–2):121–31. doi: 10.1016/s0378-1119(01)00842-3. [DOI] [PubMed] [Google Scholar]
- Ritvaniemi P, Hyland J, Ignatius J, Kivirikko KI, Prockop DJ, Ala-Kokko L. A fourth example suggests that premature termination codons in the COL2A1 gene are a common cause of the Stickler Syndrome: Analysis of the COL2A1 Gene by Denaturing Gradient Gel Electrophoresis. Genomics. 1993;17:218–221. doi: 10.1006/geno.1993.1306. [DOI] [PubMed] [Google Scholar]
- Snead MP, Yates JR. Clinical and Molecular genetics of Stickler syndrome. J Med Genet. 1999 May;36(5):353–9. [PMC free article] [PubMed] [Google Scholar]
- Spranger JW, Brill PW, Poznanski AK. Multiple epiphyseal dysplasia. In: Spranger JW, Brill PW, Poznanski AK, editors. Bone dysplasia: an atlas of genetic disorders of akeletal development. Oxford Univesrity Press; New York: 2002. pp. 141–146. [Google Scholar]
- Stickler GB, Belau PG, Farrell FJ, Jones JD, Pugh DG, Steinberg AG, Ward LE. Hereditary progressive arthro-ophthalmopathy. Mayo Clin Proc. 1965;40:433–455. [PubMed] [Google Scholar]
- Takahashi M, Matsui Y, Goto T, Nishimura G, Ikegawa S, Ohashi H, Yasui N. Intrafamilial phenotypic diversity in multiple epiphyseal dysplasia associated with the COL9A2 mutation (EDM2) Clin Rheumatol. 2006;25:591–595. doi: 10.1007/s10067-005-0034-z. [DOI] [PubMed] [Google Scholar]
- Van Camp G, Snoeckx RL, Hilgert N, Ende J, Fukuoka H, Wagetsuma M, Suzuki H, Smets RME, Vanhoenacker F, Declau F, Van De Heyning P, Shin-ichi Usami. A New Autosomal Recessive Form of Stickler Syndrome Is Caused by a Mutation in the COL9A1 Gene. Am J Hum Genet. 2006;79:449–457. doi: 10.1086/506478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.