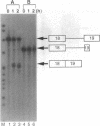
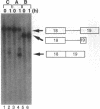
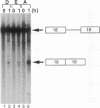
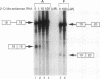
Abstract
Molecular analysis of dystrophin Kobe showed that exon 19 of the dystrophin gene bearing 52-bp deletion was skipped during splicing, although the known consensus sequences at the 5' and 3' splice sites of exon 19 were maintained (Matsuo, M., T. Masumura, H. Nishio, T. Nakajima, Y. Kitoh, T. Takumi, J. Koga, and H. Nakamura. 1991. J. Clin. Invest. 87:2127-2131). These data suggest that the deleted sequence of exon 19 may function as a cis-acting element for exact splicing for the upstream and downstream introns. To investigate this potential role of exon 19, an in vitro splicing system using artificial dystrophin mRNA precursors (pre-mRNAs) was established. Pre-mRNA containing exon 18, truncated intron 18, and exon 19 was spliced precisely in vitro, whereas splicing of intron 18 was almost completely abolished when the wild-type exon 19 was replaced by the dystrophin Kobe exon 19. Splicing of intron 18 was not fully reactivated when dystrophin Kobe exon 19 was restored to nearly normal length by inserting other sequences into the deleted site. These results suggest that the presence of the exon 19 sequence which is lost in dystrophin Kobe is more critical for splicing of intron 18 than the length of the exon 19 sequence. Characteristically, the efficiency of splicing of this intron seemed to correlate with the presence of polypurine tracks within the downstream exon 19. Moreover, an antisense 31-mer 2'-O-methyl ribonucleotide complementary to the 5' half of the deleted sequence in dystrophin Kobe exon 19 inhibited splicing of wild-type pre-mRNA in a dose- and time-dependent manner. This first in vitro evidence that dystrophin pre-mRNA splicing can be modulated by an antisense oligonucleotide raises the possibility of a new therapeutic approach for Duchenne muscular dystrophy.
Full text
PDF
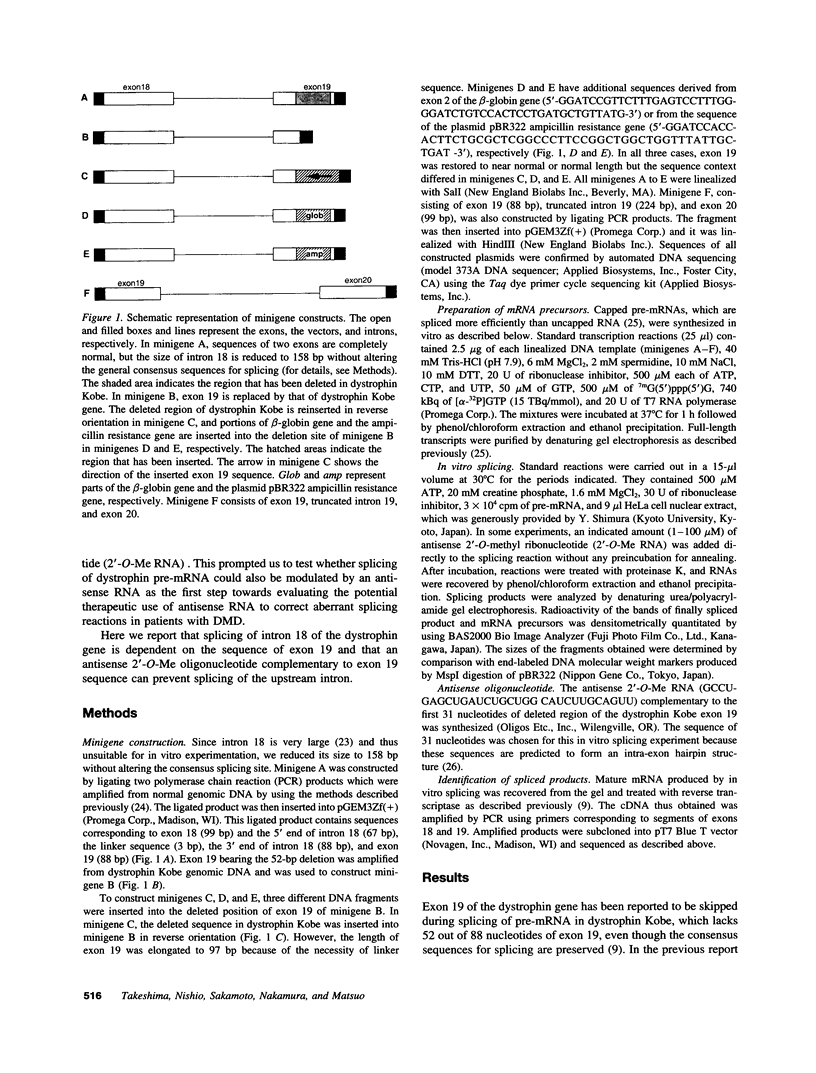
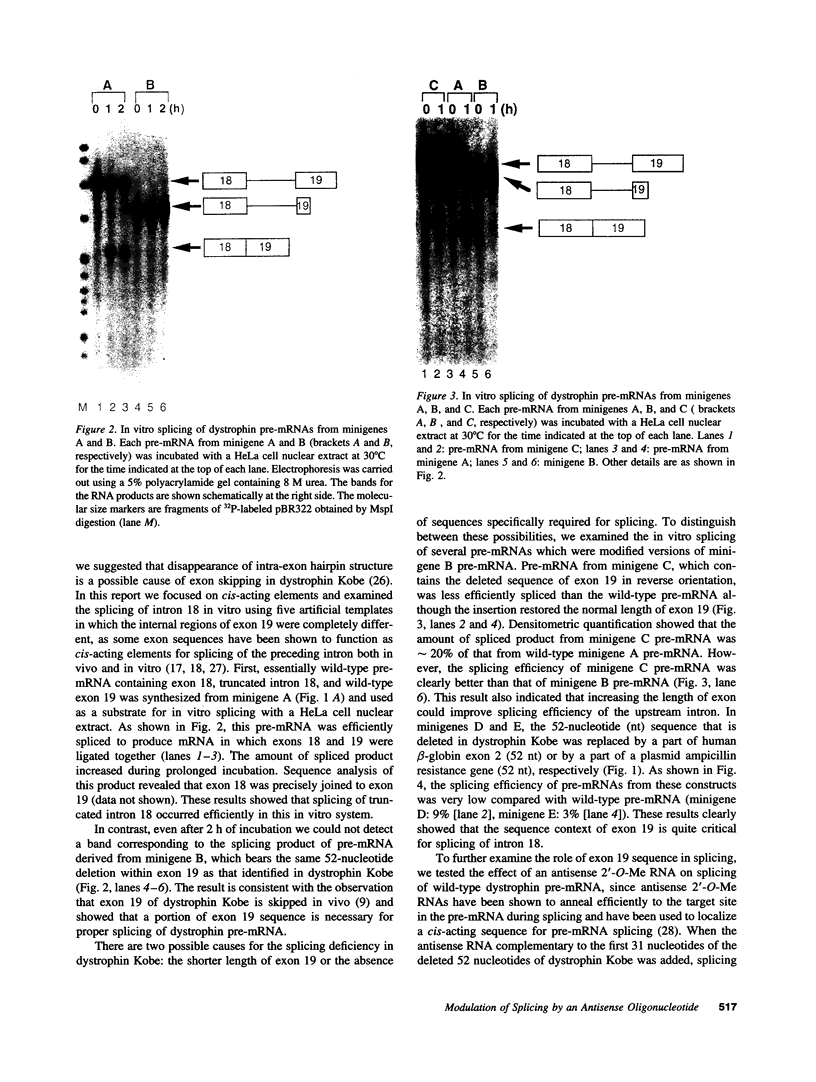



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahn A. H., Kunkel L. M. The structural and functional diversity of dystrophin. Nat Genet. 1993 Apr;3(4):283–291. doi: 10.1038/ng0493-283. [DOI] [PubMed] [Google Scholar]
- Barabino S. M., Sproat B. S., Lamond A. I. Antisense probes targeted to an internal domain in U2 snRNP specifically inhibit the second step of pre-mRNA splicing. Nucleic Acids Res. 1992 Sep 11;20(17):4457–4464. doi: 10.1093/nar/20.17.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M., Casari G., Guenzi S., Tagliabue R., Sidoli A., Melo C. A., Baralle F. E. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 1994 Mar 25;22(6):1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feener C. A., Koenig M., Kunkel L. M. Alternative splicing of human dystrophin mRNA generates isoforms at the carboxy terminus. Nature. 1989 Apr 6;338(6215):509–511. doi: 10.1038/338509a0. [DOI] [PubMed] [Google Scholar]
- Furdon P. J., Kole R. The length of the downstream exon and the substitution of specific sequences affect pre-mRNA splicing in vitro. Mol Cell Biol. 1988 Feb;8(2):860–866. doi: 10.1128/mcb.8.2.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillard E. F., Chamberlain J. S., Murphy E. G., Duff C. L., Smith B., Burghes A. H., Thompson M. W., Sutherland J., Oss I., Bodrug S. E. Molecular and phenotypic analysis of patients with deletions within the deletion-rich region of the Duchenne muscular dystrophy (DMD) gene. Am J Hum Genet. 1989 Oct;45(4):507–520. [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hagiwara Y., Nishio H., Kitoh Y., Takeshima Y., Narita N., Wada H., Yokoyama M., Nakamura H., Matsuo M. A novel point mutation (G-1 to T) in a 5' splice donor site of intron 13 of the dystrophin gene results in exon skipping and is responsible for Becker muscular dystrophy. Am J Hum Genet. 1994 Jan;54(1):53–61. [PMC free article] [PubMed] [Google Scholar]
- Hoffman E. P., Kunkel L. M. Dystrophin abnormalities in Duchenne/Becker muscular dystrophy. Neuron. 1989 Jan;2(1):1019–1029. doi: 10.1016/0896-6273(89)90226-2. [DOI] [PubMed] [Google Scholar]
- Hoshijima K., Inoue K., Higuchi I., Sakamoto H., Shimura Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science. 1991 May 10;252(5007):833–836. doi: 10.1126/science.1902987. [DOI] [PubMed] [Google Scholar]
- Koenig M., Beggs A. H., Moyer M., Scherpf S., Heindrich K., Bettecken T., Meng G., Müller C. R., Lindlöf M., Kaariainen H. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet. 1989 Oct;45(4):498–506. [PMC free article] [PubMed] [Google Scholar]
- Koenig M., Hoffman E. P., Bertelson C. J., Monaco A. P., Feener C., Kunkel L. M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987 Jul 31;50(3):509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Koenig M., Monaco A. P., Kunkel L. M. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988 Apr 22;53(2):219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- Krawczak M., Reiss J., Cooper D. N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992 Sep-Oct;90(1-2):41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- Matsuo M., Masumura T., Nakajima T., Kitoh Y., Takumi T., Nishio H., Koga J., Nakamura H. A very small frame-shifting deletion within exon 19 of the Duchenne muscular dystrophy gene. Biochem Biophys Res Commun. 1990 Jul 31;170(2):963–967. doi: 10.1016/0006-291x(90)92185-3. [DOI] [PubMed] [Google Scholar]
- Matsuo M., Masumura T., Nishio H., Nakajima T., Kitoh Y., Takumi T., Koga J., Nakamura H. Exon skipping during splicing of dystrophin mRNA precursor due to an intraexon deletion in the dystrophin gene of Duchenne muscular dystrophy kobe. J Clin Invest. 1991 Jun;87(6):2127–2131. doi: 10.1172/JCI115244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo M., Nishio H., Kitoh Y., Francke U., Nakamura H. Partial deletion of a dystrophin gene leads to exon skipping and to loss of an intra-exon hairpin structure from the predicted mRNA precursor. Biochem Biophys Res Commun. 1992 Jan 31;182(2):495–500. doi: 10.1016/0006-291x(92)91759-j. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Hayase Y., Inoue H., Ohtsuka E., Ohshima Y. Surveying cis-acting sequences of pre-mRNA by adding antisense 2'-O-methyl oligoribonucleotides to a splicing reaction. J Biochem. 1990 Sep;108(3):399–405. doi: 10.1093/oxfordjournals.jbchem.a123213. [DOI] [PubMed] [Google Scholar]
- Mayeda A., Helfman D. M., Krainer A. R. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol. 1993 May;13(5):2993–3001. doi: 10.1128/mcb.13.5.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco A. P., Bertelson C. J., Liechti-Gallati S., Moser H., Kunkel L. M. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988 Jan;2(1):90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- Narita N., Nishio H., Kitoh Y., Ishikawa Y., Ishikawa Y., Minami R., Nakamura H., Matsuo M. Insertion of a 5' truncated L1 element into the 3' end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J Clin Invest. 1993 May;91(5):1862–1867. doi: 10.1172/JCI116402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M., Sakamoto H., Shimura Y. Preferential excision of the 5' proximal intron from mRNA precursors with two introns as mediated by the cap structure. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5187–5191. doi: 10.1073/pnas.84.15.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prockop D. J., Colige A., Helminen H., Khillan J. S., Pereira R., Vandenberg P. Mutations in type 1 procollagen that cause osteogenesis imperfecta: effects of the mutations on the assembly of collagen into fibrils, the basis of phenotypic variations, and potential antisense therapies. J Bone Miner Res. 1993 Dec;8 (Suppl 2):S489–S492. doi: 10.1002/jbmr.5650081311. [DOI] [PubMed] [Google Scholar]
- Roberts R. G., Coffey A. J., Bobrow M., Bentley D. R. Determination of the exon structure of the distal portion of the dystrophin gene by vectorette PCR. Genomics. 1992 Aug;13(4):942–950. doi: 10.1016/0888-7543(92)90005-d. [DOI] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt T. G., Vulliamy T., Dubowitz V., Sewry C. A., Strong P. N. Exon skipping and translation in patients with frameshift deletions in the dystrophin gene. Am J Hum Genet. 1993 Nov;53(5):1007–1015. [PMC free article] [PubMed] [Google Scholar]
- Steingrimsdottir H., Rowley G., Dorado G., Cole J., Lehmann A. R. Mutations which alter splicing in the human hypoxanthine-guanine phosphoribosyltransferase gene. Nucleic Acids Res. 1992 Mar 25;20(6):1201–1208. doi: 10.1093/nar/20.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Mayeda A., Hampson R. K., Krainer A. R., Rottman F. M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993 Dec;7(12B):2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Watakabe A., Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol. 1994 Feb;14(2):1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volloch V., Schweitzer B., Rits S. Inhibition of pre-mRNA splicing by antisense RNA in vitro: effect of RNA containing sequences complementary to exons. Biochem Biophys Res Commun. 1991 Sep 30;179(3):1593–1599. doi: 10.1016/0006-291x(91)91756-3. [DOI] [PubMed] [Google Scholar]
- Watakabe A., Tanaka K., Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993 Mar;7(3):407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- Winnard A. V., Jia-Hsu Y., Gibbs R. A., Mendell J. R., Burghes A. H. Identification of a 2 base pair nonsense mutation causing a cryptic splice site in a DMD patient. Hum Mol Genet. 1992 Nov;1(8):645–646. doi: 10.1093/hmg/1.8.645. [DOI] [PubMed] [Google Scholar]