Abstract
Aneuploidy, or the abnormal number of chromosomes, adversely effects cell growth, but it is also linked with cancer and tumorigenesis. Now Torres et al. (2010) help resolve this paradox by demonstrating that aneuploid yeast cells can evolve mutations in the proteasome protein degradation pathway that alleviate imbalances in protein production and increase the cell’s proliferative capacities.
During mitosis, duplicated chromosomes are equally distributed to daughter cells so that the total number of chromosomes is preserved throughout many generations. Errors in chromosomal segregation can lead to the loss or gain of chromosomes in daughter cells, a condition known as aneuploidy. Aneuploidy is a hallmark of cancer cells (Albertson et al, 2003), but the causality of the relationship between aneuploidy and tumorigenesis remains highly complex and controversial (Schvartzman et al, 2010). Aneuploidy can either promote or suppress tumor formation, and the outcome depends on the genetic and cellular context, including the specific genes on the abnormal chromosome, the extent of the aneuploidy, the already accumulated genetic errors, and specific features unique to the cell type (Holland and Cleveland, 2009). Paradoxically, despite its association with uninhibited cell growth in cancer, aneuploidy itself has adverse effects on the growth of organisms and their individual cells. The most straight forward reconciliation of these contrasting properties is that aneuploidy initially inhibits growth, but then the acquisition of additional mutations or chromosomal shuffling increases the fitness of cells. In this issue of Cell, Torres et al. (2010) demonstrate that this is indeed true for aneuploid yeast cells. The authors find that a general feature of aneuploidy is proteomic stress caused by an imbalance in protein synthesis for the genes encoded on the extra chromosome; in several cases, mutations in a deubiquitination enzyme can alleviate this stress and enhance cellular growth and fitness.
Earlier work by Torres and colleagues (2007) described the physiological consequences of yeast cells having an extra copy of one or more chromosome. The authors generated these disomic strains by attempting to mate haploid yeast cells carrying a mutation that prevents fusion of the nuclei (i.e., karyogamy), leading to unsuccessful or abortive matings (Hugerat et al., 1994). In these experiments, one chromosome of the parental yeast strains also contained a selection marker, such as a gene that supports growth in the absence of an essential amino acid histidine (HIS) or one that confers resistance against G418 (also known as Geneticin), an aminoglycoside that interferes with protein synthesis elongation (Bar-Nun et al., 1983). During the abortive matings, the marked chromosomes were occasionally transferred between two nuclei, and the chromosomal markers allowed for the selection of disomic clones on G418- containing and histidine-deficient media. Most of the aneuploid strains isolated possessed a growth defect on a nonselective medium, and this deficiency was enhanced on the selective medium. Furthermore, the growth defects were due primarily to a delay in the G1 phase of the cell cycle.
As anticipated, analysis of the transcripts in these disomic yeast strains revealed that most genes on the extra chromosome are transcribed at twice the rate as the rest of the genome. On the other hand, expression levels of a small number of proteins, especially those that are subunits of multiprotein complexes, are not elevated. All the disomic strains also displayed increased energy requirements and enhanced sensitivity to conditions that interfere with protein synthesis, folding, and degradation. These findings led the authors to propose that proteotoxic stress due to imbalanced protein expression might be responsible for the reduced fitness of disomic yeast cells (Figure 1, top). Furthermore, the cells’ enhanced sensitivity to proteasome inhibitors may reflect an increased reliance on protein degradation to restore proteomic balance in the disomic yeast cells.
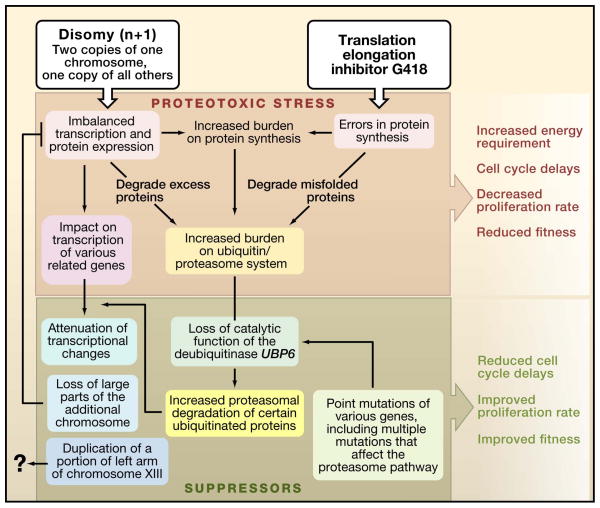
Figure 1. Aneuploidy induces proteotoxic stress.
(Top) An extra copy of an individual yeast chromosome, or disomy, causes imbalanced expression of the proteins encoded on that chromosome. Adding an inhibitor of protein synthesis, such as G418 (Geneticin), increases the errors in translation and enhances the proteotoxic stress. This stress reduces fitness and inhibits cell growth primarily during the G1 phase. (Bottom) Suppressers of proteotoxic stress, including mutations in components of ubiquitin/proteasome pathway, can ameliorate the proteomic imbalance and restore fitness (Torres et al. 2010). For example, disrupting the deubiquitinase UPB6 can increase the growth rate of aneuploid cells by triggering more rapid protein degradation by the proteasome.
Now in their new study, Torres and colleagues (2010) examined 13 different haploid yeast strains, each with an extra copy of one of the 16 yeast chromosomes. They grew the disomic strains over several generations in the selective medium. Initially the doubling times of these strains were significantly longer than the control cells. However, after a variable number of generations, 11 of the cultures sped up their doubling times. The authors isolated individual clones from these “evolved” cultures to identify the basis of their improved growth rates (Figure 1, bottom). Comparative genome hybridization analyses showed that descendants of 3 disomic strains had lost large parts of their additional chromosome. These deletions alone may have accounted for the improved proliferation of the descendants. Interestingly, however, 3 independent clones possessed the same duplication of a 183kb fragment from the short arm of chromosome XVIII, suggesting that genes located in this fragment may also play a role in increasing the proliferative ability of these aneuploid yeasts.
To identify point mutations that increased the fitness of the aneuploid yeast, Torres and colleagues then sequenced several of the evolved isolates from 6 of the disomic strains that retained the extra chromosome. Strikingly, they found that four genes of the ubiquitin/proteasome pathway, UBP6 (a deubiquitinase), RPT1 (an ATPase of the proteasome), RSP5 (E3 ubiquitin ligase), UBR1 (E3 ubiquitin ligase), were mutated in the descendents of 5 different disomic strains. Two independent strains (disome V and disome IX) contained distinct truncations in a gene encoding the deubiquitinating enzyme Ubp6 that interacts with the proteasome. Both truncations impair the deubiquitinase catalytic activity of Ubp6 but not its association with the proteasome (Leggett et al., 2002).
The authors next tested if mutations in the Ubp6 deubiquitinase alone could directly help aneuploid yeast recover more normal growth rates. Indeed, in some cases it did. Mutating UBP6 increased the fitness of two disomic strains in selective medium and two strains (disome V and disome XI) in both selective and non-selective media. This latter finding is especially important because a gain of fitness in only the selective medium could reflect a suppressive function of the UBP6 mutation against the action of the elongation inhibitor G418. A final cautionary note is that the effect of mutating UBP6 was not consistent across the different disomic strains; in fact, it decreased the fitness of two disomic strains.
How could losing the deubiquitinating activity of Ubp6 increase the growth rate of aneuploid yeast cells? Ubp6 has been shown to reduce the activity of the proteasome although this function of Ubp6 apparently does not require the deubiquitinase catalytic activity (Hanna et al., 2006). Nevertheless, mutating Ubp6 may restore proteomic balance in the cell by generally boosting protein degradation by the proteasome or by increasing the proteasome’s activity on selective substrates.
To distinguish between these two possibilities, Torres and colleagues deleted UBP6 in the disomic strains and then used a combination of mass spectrometry and SILAC (i.e., stable isotope labeling with amino acids in cell culture) to analyze the effects of the deletion on the yeast proteome. They chose two disomic strains for these experiments: disome V, in which deletion of UBP6 improves fitness; and disome XIII, in which the mutation has no effect. As expected, adding the extra chromosome increased the average abundance of proteins encoded on the chromosome by nearly 2 fold. This mutation also caused a significant change in the relative abundance of a number of proteins across the whole proteome; whereas some proteins increased in concentration by nearly 2 fold, others decreased to nearly half the levels of haploid cells. For disome V, deleting UBP6 substantially attenuated these changes in protein abundance, and protein levels approached those of haploid cells. In particular, loss of UBP6 in disome V downregulates proteins with relatively high expression levels without affecting their transcription but transcriptionally upregulates proteins with relatively low expression levels. Curiously, deleting UBP6 in disome XIII does not increase transcription of proteins with relatively low expression levels, and this difference may explain why mutating UBP6 does not enhance the fitness of disome XIII.
The implication from the new findings by Torres and colleagues is that extra chromosomes generally increase proteomic stress by elevating the cost of protein synthesis, folding, and degradation due to the imbalance of proteins produced (Figure 1). Thus, although each additional chromosome creates an altered abundance of a different set of encoded proteins, any extra chromosome leads to a growth disadvantage.
As the authors note, these new results raise the possibility that aneuploid cancer cells are under profound proteotoxic stress and thus must rely on the increased activity of the ubiquitin/proteasome pathway to maintain their proliferative state. This hypothesis provides an elegant rationale for extending the use of proteasome inhibitors (such as Velcade) to treating many types of cancers with aneuploid cells; currently these inhibitors are clinically approved for treating only the overproduction of immunoglobulin synthesis in multiple myeloma. In this regard, the next step is to determine the extent to which tumor cells with chromosomal instability experience proteotoxic stress and then to test if increasing this stress with proteasome inhibitors controls their growth.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Albertson DG, Collins C, McCormick F, Gray JW. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Bar-Nun S, Shneyour Y, Beckmann JS. Biochim Biophys Acta. 1983;741:123–127. doi: 10.1016/0167-4781(83)90018-0. [DOI] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugerat Y, Spencer F, Zenvirth D, Simchen G. Genomics. 1994;22:108–117. doi: 10.1006/geno.1994.1351. [DOI] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Schvartzman JM, Sotillo R, Benezra R. Nat Rev Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Cell. 2010 doi: 10.1016/j.cell.2010.08.038. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]