Abstract
In prion diseases, the misfolded protein aggregates are derived from cellular prion protein (PrPC). Numerous ligands have been reported to bind to human PrPC (huPrP), but none to the structured region with the affinity required for a pharmacological chaperone. Using equilibrium dialysis, we screened molecules previously suggested to interact with PrP to discriminate between those which did not interact with PrP, behaved as nonspecific polyionic aggregates or formed a genuine interaction. Those that bind could potentially act as pharmacological chaperones. Here we report that a cationic tetrapyrrole [Fe(III)-TMPyP], which displays potent antiprion activity, binds to the structured region of huPrP. Using a battery of biophysical techniques, we demonstrate that Fe(III)-TMPyP forms a 1∶1 complex via the structured C terminus of huPrP with a Kd of 4.5 ± 2 μM, which is in the range of its IC50 for curing prion-infected cells of 1.6 ± 0.4 μM and the concentration required to inhibit protein-misfolding cyclic amplification. Therefore, this molecule tests the hypothesis that stabilization of huPrPC, as a principle, could be used in the treatment of human prion disease. The identification of a binding site with a defined 3D structure opens up the possibility of designing small molecules that stabilize huPrP and prevent its conversion into the disease-associated form.
Keywords: biophysics, drug discovery, protein folding, structural biology
Protein-folding disorders are a diverse set of diseases which share one common feature—the accumulation of misfolded proteins inside or outside the cell. These include many well-known neurodegenerative disorders such as Alzheimer’s disease, amyotrophic lateral sclerosis, Parkinson disease, and prion diseases. Although diverse, disease-specific therapeutic strategies are being investigated, some approaches could be generally applicable to all protein-misfolding disorders. For example, pharmacological chaperones have shown promise in a number of disease areas (1) including lysosomal storage diseases (2), cancer (3), and neurodegeneration (4). In the case of prion diseases, it has been proposed that stabilizing the native folded state of these proteins should reduce the rate of their misfolding and therefore slow or even stop disease (5, 6).
Prion diseases are fatal, untreatable, neurodegenerative protein-misfolding disorders, including Creutzfeldt–Jakob disease (CJD) in humans, and scrapie, bovine spongiform encephalopathy (BSE), and chronic wasting disease in animals, all of which are associated with the misfolding of a normal host-encoded protein, the cellular prion protein (PrPC) (7). Prions are naturally or experimentally transmissible agents and, according to the widely accepted protein-only hypothesis (8), consist solely or largely of the misfolded-protein isoform, designated PrPSc (9). The arrival of the epizootic bovine prion disease BSE to which there was widespread dietary exposure of the UK population, and the confirmation that it was transmissible to humans causing variant CJD (vCJD) (10–12), has led to major efforts to develop therapeutics for prion infection. Human prion diseases can also occur sporadically, as inherited conditions associated with PrP mutations, and be transmitted iatrogenically by medical and surgical procedures including, in the case of vCJD, by blood transfusion (7, 13).
In spite of the fact that prion diseases are so strongly linked to aberration of a single protein, much remains unknown, including the precise mechanism of autocatalytic protein misfolding and the nature of the toxic agent that actually leads to neuronal death (14). Although multiple roles for PrPC have been proposed, no clear consensus for its normal cellular function has emerged. Despite these uncertainties, PrPC has been firmly validated as a target for antiprion therapeutics. Mice lacking PrP expression are healthy (15), resistant to prion infection, and do not undergo neurodegeneration (16). Furthermore, targeting neuronal PrP expression during established neuroinvasive prion infection prevents development of clinical disease and results in reversal of early pathological changes and behavioral deficits (17, 18). Passive immunization of mice with an antibody selective for PrPC reduces PrPSc to undetectable levels in the spleen and allows the mice to remain healthy for over 300 d after infection (19). Preventing misfolding of the structured region of PrP therefore offers a potential means of stopping, or even reversing, prion disease even in the absence of a detailed understanding of the pathobiology (6).
The structured region of PrP, encompassing approximately residues 124–231, contains three α-helices and a small two-strand antiparallel β-sheet (20, 21). There is a large unstructured region at the N terminus and three unstructured residues at the C terminus. H/D exchange experiments revealed that the structured domain of PrP unfolds in a single transition without populating any significant kinetic intermediate and as such PrP is required to unfold in order to misfold (22). Fundamental principles of free energy dictate that any small molecule which selectively binds to, and stabilizes the structured region of PrPC, will be able to inhibit misfolding and thus retard prion propagation. However, designing molecules that accomplish this is not trivial. The fact that the biological role of PrP remains unclear (23) precludes the use of screens for compounds that alter its activity. The recently reported crystal structure of human PrPC (24) and also the NMR structures of human PrPC show no obvious clefts for ligand binding (21). In contrast, a number of molecules have been demonstrated to bind to the unstructured region of PrP including pentosan polysulfate (25), Congo red (26), and a number of anionic tetrapyrroles (27, 28). Congo red and anionic tetrapyrroles can stack in solution, and are believed to behave like polyanions and bind with low specificity to the N-terminal domain (29–31). Indeed, the ability of anionic tetrapyrroles to stack in solution correlated well with their antiprion activity (31). In contrast, the cationic porphyrin Fe(III)-TMPyP [Fe(III) meso-tetra (N-methyl-4-pyridyl) porphine] has good antiprion activity in cell culture (32) and animal models (33) in spite of its preference for the monomeric state (34) and its positive charge, suggesting this porphyrin may have a different mode of action.
We wished to use equilibrium dialysis to screen ligands previously suggested to bind to PrP to differentiate between those which did not bind to human PrP, those that bound as colloidal polyanions to the unstructured region, and those that bound to the structured domain and could potentially act as pharmacological chaperones. We found that Fe(III)-TMPyP bound to the structured domain of human PrPC (huPrP) and herein describe the biophysical and structural analysis of the PrP∶Fe(III)-TMPyP interaction. Given that this molecule binds to the structured region of the cellular conformation of huPrP and is able to act as a pharmacological chaperone for its folding, it can be used to test the hypothesis that stabilization of huPrPC, as a principle, could be used in the treatment of human prion disease or other protein misfolding disorders.
Results
Energetics and Binding Site of PrP∶Fe(III)-TMPyP Interaction.
To identify compounds that interact with PrP without behaving as colloidal polyions, we utilized equilibrium dialysis. We placed compound in one chamber and huPrP in another separated by a 5,000 molecular weight cutoff semipermeable membrane and allowed the sample to reach equilibrium, allowing us to differentiate between three classes of compounds. First, compounds that do not interact with PrP would equilibrate in both chambers. Second, by setting protein and ligand concentrations at 100 μM, we would be able to detect any therapeutically relevant interaction with compound concentrating in the protein-containing chamber. Third, compounds which form large, colloidal aggregates with molecular weight greater than 5,000 would fail to traverse the membrane. Compounds in the second class could be used as a basis to design drug-like small molecules capable of binding to PrP, whereas those in the other classes can be discarded as leads.
We used this method to probe the interaction of ligands previously suggested to bind to PrP. After 2 d equilibration time, we found compounds that fell into each of the three classes mentioned above (Table 1). Quinacrine, tetracycline, and GN8 all failed to bind to huPrP91–231. Congo red and all anionic tetrapyrroles behaved as aggregated polyanions, and so were considered unsuitable to function as pharmacological chaperones, demonstrating how this equilibrium dialysis-based screen is a unique way to detect aggregated ligands even in the absence of biological activity (35). The two cationic porphyrins tested, Fe(III)-TMPyP and Fe(III)-TAP [Fe(III) meso-tetra (4-N,N,N-trimethylanilinium) porphine], localized mainly in the PrP-containing chamber. In the case of Fe(III)TAP, aggregated material was visible in the PrP-containing chamber, so Fe(III)-TMPyP was chosen for more rigorous characterization.
Table 1.
Compounds previously suggested to interact with PrP tested by equilibrium dialysis
| Compound active | (Y/N) |
| Fe(III) meso-tetra (N-methyl-4-pyridyl) porphine | Yes |
| Fe(III) meso-tetra(4-N,N,N-trimethylanilinium) porphine | Yes |
| Fe(III) meso-tetra(4-sulfonatophenyl) porphine | Colloid |
| Fe(III) deuteroporphyrin IX 2,4 bis ethylene glycol | Colloid |
| Fe(III) phthalocyanine tetrasulfonic acid | Colloid |
| Hemin | Colloid |
| Congo red | Colloid |
| Quinacrine | No |
| Tetracycline | No |
| GN8 | No |
Yes signifies the majority of compound passed to the protein-containing side. Colloid signifies the compound failed to cross the membrane due to an apparent molecular weight greater than 5,000. No signifies compounds that distributed evenly between the two chambers.
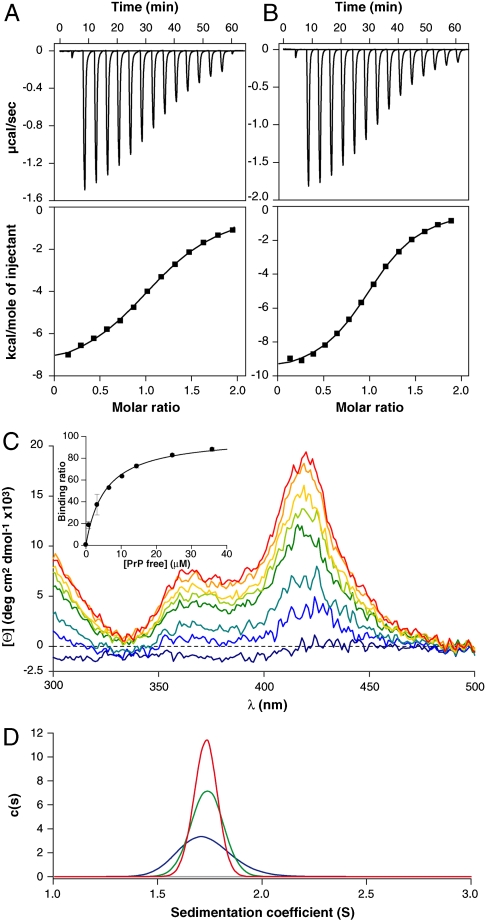
We used isothermal titration calorimetry (ITC) to test the binding of Fe(III)-TMPyP to the structured region of PrP (huPrP119–231) and to a longer construct (huPrP91–231), which also contains the part of the unstructured region found in the proteinase-resistant core of PrPSc (36). Fig. 1 A and B show example titrations of Fe(III)-TMPyP into solutions containing huPrP91–231 and huPrP119–231. Fe(III)-TMPyP binds to both proteins at a 1∶1 ratio with a dissociation constant of 3.5 ± 1.0 μM for huPrP119–231 (n = 1.04 ± 0.07, ΔH = -8.9 ± 1.4 kcal mol-1, TΔS = -1.5 ± 1.5 kcal mol-1) and 8.1 ± 2.0 μM for huPrP91–231 (n = 1.0 ± 0.1, ΔH = -7.4 ± 0.5 kcal mol-1, TΔS = -0.5 ± 0.5 kcal mol-1) in a process that is almost entirely enthalpically driven. Thus, we showed that the unstructured N terminus is not required for Fe(III)-TMPyP binding and that this molecule binds to the structured C terminus of PrP. Reversing the ITC experiment by titrating concentrated huPrP91–231 into Fe(III)-TMPyP gave physical parameters in good agreement with standard conditions (n = 0.98 ± 0.09, ΔH = -8.5 ± 0.7 kcal mol-1, and Kd = 7.4 ± 2.4 μM), confirming the 1∶1 stoichiometry and validating the binding affinity. Further experiments in the presence of 150 mM NaCl showed a slightly reduced affinity (n = 0.92 ± 0.01, ΔH = -5.4 ± 0.1 kcal mol-1, and Kd = 11.2 ± 1.0 μM), but suggest that weak electrostatic interactions were not the main source of the interaction at 25 mM NaCl (Fig. S1). Fe(III)-TAP also showed an interaction dominated by the structured region, but appeared to cause self-association of PrP preventing derivation of meaningful thermodynamic parameters.
Fig. 1.
Biophysical characterization of the huPrP∶Fe(III)-TMPyP interaction. Isotherms for the titration of Fe(III)-TMPyP (429 μM) into huPrP: (A) huPrP91–231, (B) huPrP119–231. (C) CD spectra of Fe(III)-TMPyP (blue) with increasing additions of huPrP119–231, in blue through to red, as indicated. (Inset) The binding isotherm as derived from the average CD signal between 418–423 nm. (D) Sedimentation coefficient distributions derived from sedimentation velocity experiments of huPrP91–231 in the presence of Fe(III)-TMPyP. Distributions of c(s) for the huPrP91–231∶Fe(III)-TMPyP complex with both 278 nm (green) and 420 nm (red) detection, as well as for huPrP91–231 alone at 278 nm (blue) and 420 nm (gray).
We used induced CD and analytical ultracentrifugation (AUC) to quantify the Fe(III)-TMPyP∶huPrP interaction. The binding of Fe(III)-TMPyP to PrP caused an induced-CD signal around the Soret band of the porphyrin (Fig. 1C). Titration of huPrP119–231 induced a saturable CD signal around 420 nm (Fig. 1C, Inset) with a Kd of 5.9 ± 1.0 μM—in good agreement with that obtained by ITC of 3.5 ± 1.0 μM. The induced-CD signal indicates that the Fe(III)-TMPyP∶PrP complex restrains the porphyrin in a chiral environment. The huPrP91–231 construct gave a Kd of approximately 3 μM and a matching induced CD was observed (Fig. S2), confirming that the additional N-terminal residues are not required for the interaction with Fe(III)-TMPyP.
We used sedimentation velocity (SV) AUC to confirm the stoichiometry. HuPrP91–231 sedimented as a single species at 1.7 Svedberg on detection at 278 nm, as expected for a monomeric globular protein the size of PrP (Fig. 1D). Detection at a porphyrin-specific wavelength of 420 nm showed no observable species. Addition of Fe(III)-TMPyP did not alter the sedimentation coefficient of the species observed at 278 nm, which was now observed at 420 nm, confirming that the molecular size and shape of the PrP species was not significantly altered by the porphyrin association, and that the predominant protein species in the porphyrin complex is monomeric. A dose-response SV experiment using detection at 420 nm showed that this interaction was saturable with a calculated Kd of 3.6 ± 0.3 μM and a 1∶1 stoichiometry. A reverse titration gave similar results with a Kd of 3.2 ± 1.1 μM and a 1∶1.3 stoichiometry (Fig. S3).
Mapping the Binding Site of a Cationic Tetrapyrrole.
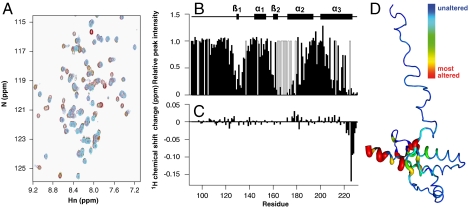
We used NMR to identify the location of the Fe(III)-TMPyP binding site on huPrP91–231 (Fig. 2A). Several PrP amide backbone resonances were severely attenuated on adding Fe(III)-TMPyP due to paramagnetic relaxation by the Fe(III). These residues are clustered at the C terminus of helix-3 in the region of the first β-sheet and in the few observable resonances in the loop between residues 160 and 180 (Fig. 2B). The binding site for the porphyrin must comprise residues in these regions because the effect of the paramagnetic relaxation is proportional to 1/r6, and many regions of PrP are unaffected. Titration of the iron-free porphyrin (apo-TMPyP) allowed us to monitor the porphyrin-induced chemical shift changes in the absence of the paramagnetism. We observe considerably up-field shifted amide resonances (Fig. 2 C and D) at the C terminus of helix-3 on addition of apo-TMPyP to huPrP91–231, indicating a face-on interaction of the end of helix-3 with the porphyrin plane. Thus we suggest that the apo- and Fe(III)-TMPyPs are binding to the same region of PrP.
Fig. 2.
NMR titration of huPrP91–231 with Fe(III)-TMPyP. (A) Overlay of heteronuclear single quantum coherence (HSQC) spectra of huPrP91–231 alone (black) and huPrP91–231 with increasing amounts of Fe(III)-TMPyP (red/green/blue/magenta/cyan), showing attenuation of some resonances. (B) Relative intensities of huPrP91–231 resonances between 0 and 0.6 molar equivalents of Fe(III)-TMPyP [gray bars indicate residues for which the effect cannot be mapped because of resonance overlap or due to the absence of an HSQC peak (for example due to conformational exchange in residues 167–171)]. (C) 1H amide chemical shift changes caused by the addition of 0.6 molar equivalents of apo-TMPyP to huPrP91–231. (D) Heat map showing the residues of huPrP91–231 for which backbone amide resonances are attenuated by the addition of Fe(III)-TMPyP, with red broad ribbon indicating strongest effect.
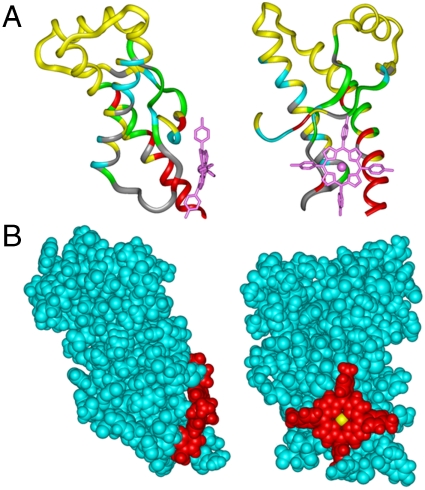
Using constraints derived from NMR, we modeled the structure of the complex between PrP and Fe(III)-TMPyP. The initial model, produced using the NMR intensity data displayed on the PrP backbone, was refined using in-house docking software (BUDE) (37–39).
The modeled structure suggests Fe(III)-TMPyP does indeed form a face-on interaction and fills a relatively shallow cleft of the surface of PrP, incorporating the C terminus of helix-3 and in close proximity to the β-strand that includes residue 129 (Fig. 3). Binding of ligands to this region of PrP suggests that this cleft may be a suitable target for the rational design of small molecules capable of stabilizing the ground state of PrPC.
Fig. 3.
Model of the structure of huPrP91–231 with bound Fe(III)-TMPyP. The location of the binding site for the ligand was mapped by the loss of N-H peak intensity caused by the proximity of the paramagnetic Fe(III) ion as described in Results and Materials and Methods. The initial binding position was refined by application of an automated docking routine.
Fe(III)-TMPyP as a Pharmacological Chaperone.
The ability of Fe(III)-TMPyP to stabilize the PrPC conformation was tested by monitoring its effect on thermal denaturation and aggregation. An apparent Tm of 66.1 ± 0.5 °C in the absence of Fe(III)-TMPyP, and 68.4 ± 0.2 °C in its presence, suggests the ligand is indeed capable of impeding the unfolding process even at highly elevated temperatures (Fig. S4). Although this is lower than some published pharmacological chaperones (40), the majority of these bind to biologically optimized clefts, whereas a pharmacological chaperone designed to fit into a surface cavity in p53 induced a Tm stabilization of approximately 2 °C (3).
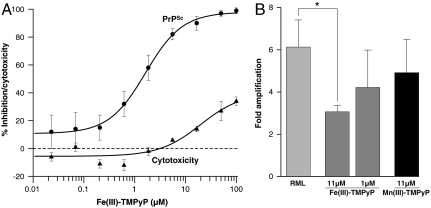
Compounds that bind to the ground state of PrPC should cure prion-infected cells by retarding the rate of PrPSc formation to below the rate of natural clearance. A dose response of Fe(III)-TMPyP in prion-infected PK1 cells (41) cured the cells with an IC50 = 1.6 ± 0.4 μM (Fig. 4A), well below the level of cytotoxicity. This ability of Fe(III)-TMPyP to retard prion formation was tested in a cell-free system using protein-misfolding cyclic amplification (PMCA) (42) (Fig. 4B). In the absence of porphyrin, substrate PrPC was efficiently converted to the proteinase-resistant form. In the presence of 11 μM Fe(III)-TMPyP, the conversion was inhibited by half (p = 0.0176), whereas Mn(III)-TMPyP, known not to bind to PrP or cure prion-infected cells (32), did not significantly inhibit the PMCA reaction. At a concentration of 1 μM, Fe(III)-TMPyP reduced the level of PMCA, but not significantly.
Fig. 4.
Effect of Fe(III)-TMPyP on PrP folding in a complex cellular environment. (A) Dose-response curve for the curing of prion-infected PK1 cells (●) by Fe(III)-TMPyP and the levels of cytotoxicity (▴). (B) Quantification of Western blot analyses of Rocky Mountain Laboratory seeded PMCA reactions in the absence of porphyrin, or in the presence of the stated concentrations of Fe(III)-TMPyP or Mn(III)-TMPyP.
Discussion
Locating amenable binding sites on PrP that can be targeted by pharmacological chaperones to stabilize PrPC is an important goal in the search for therapies for prion diseases. The compound GN8 was reported to bind to murine PrP with a dissociation constant of 4 μM (4) via the turn connecting helices 2 and 3. We have demonstrated that this molecule does not bind to human PrPC using equilibrium dialysis, a technique which should identify even nonspecific interactions. We also did not detect an interaction between GN8 and huPrP using NMR or ITC. This discrepancy may be because GN8 was shown to bind to murine PrP and our study used human PrP, although their sequences are identical around the GN8 binding site. Quinacrine has also been suggested to bind to huPrP with a dissociation constant of 4.6 mM (43), five orders of magnitude higher than is required to clear PrPSc from prion-infected cells in vitro (44). It is not effective therapeutically in human studies, suggesting its cellular effect may be off target (45).
The Fe(III)-TMPyP molecule is confirmed to bind to the structured region of huPrPC in the micromolar concentration range and, as such, is able to directly test if ground-state stabilization of huPrP is a valid strategy for producing therapeutic compounds for human prion disease. We have measured and characterized this interaction by several independent biophysical techniques. The binding site, monomeric structure, and positive charge of Fe(III)-TMPyP make it entirely different from polyanions and colloidal inhibitors, such as anionic tetrapyrroles, which appear to be nonspecific inhibitors of amyloid formation (29). As such this cationic tetrapyrrole represents a distinct class of human PrP-binding molecule with proven activity in prion-infected cells (32), animal models (33), and now in PMCA. Our cell-based data confirm that Fe(III)-TMPyP can effectively cure prion-infected cells without any cytotoxic response. The IC50 value obtained of 1.6 ± 0.4 μM is in good agreement with the Kd values obtained by ITC, induced CD, and AUC, suggesting the most likely mode of action of Fe(III)-TMPyP in cells is inhibiting prion propagation by binding to the ground state of PrPC and preventing its conversion to PrPSc. This contrasts with quinacrine, which is believed to concentrate in specific organelles (44) and where the cell curing is around 100,000-fold higher than the Kd calculated by NMR (43). The increased cell potency of Fe(III)-TMPyP may be related to its tendency to locate near anionic lipids (46) or may suggest that not all PrP needs to be stabilized to cure prion-infected cells, allowing curing below the Kd for the compound. PMCA confirmed that Fe(III)-TMPyP prevented misfolding of PrPC in a cell-free environment. These data suggest that the Fe(III)-TMPyP binding site is not obstructed in the crowded cellular environment and validate this site on PrP as a therapeutic target for human prion disease. Although Fe(III)-TMPyP is efficacious in animals when administered intraperitoneally (33), it would be unlikely to readily cross the blood–brain barrier of healthy individuals, although infected individuals may have compromised blood–brain barriers. This does not exclude its use as an antiprion therapeutic via head pump, given the invariably fatal nature of these diseases, but a more drug-like molecule would be highly desirable. As such, Fe(III)-TMPyP represents a starting point for the design of small molecules that bind to the structured region of huPrP. Its optical properties make it suitable as a binding probe or a control for high throughput screens and its micromolar affinity is ideal for displacement assays, for example by using equilibrium dialysis. Fe(III)-TMPyP binds to a region of PrP known to be prone to helix fraying (47) so might be particularly effective at stabilizing PrP. Furthermore, TMPyP compounds have been shown to be efficient photoinduced cross-linking reagents to study both protein–protein (48) and protein–DNA interactions (49). Given the association between PrP and Fe(III)-TMPyP, this technology may be particularly effective in identifying binding partners for PrP.
It is not yet known if PrP is a natural porphyrin binding protein, although hemin has been shown to bind and alter PrP subcellular localization (28). Porphyrins are known to interact with other natural receptors, such as benzodiazepine receptors, and it might be that Fe(III)-TMPyP is acting as an analogue for a natural ligand (50). Also, given the recent discovery that PrP binds to toxic amyloid beta oligomers (51), it is possible that compounds such as Fe(III)-TMPyP may be able to block the implied toxic signaling mediated by this interaction. If this binding event involves a structural reorganization of PrP, molecules such as Fe(III)-TMPyP, which hold PrP in its cellular conformation, could be of particular interest.
In summary, we have employed a distinct equilibrium dialysis screen to identify the cationic tetrapyrrole, Fe(III)-TMPyP, as a class of potential therapeutic in human prion disease and not a nonspecific colloidal aggregate, as previously assumed. Its monomeric structure and ability to stabilize the folded domain of human PrP mean it can act as a pharmacological chaperone for this protein. Furthermore, its spectroscopic and reactive properties make this class of molecule ideal as molecular probes in assays to discover protein and small molecule binding partners for PrP. The discovery of a defined binding site on huPrP is important in highlighting a potentially targetable region of PrP, which can be exploited in the design of drug-like pharmacological chaperones that can stabilize huPrPC.
Materials and Methods
Materials.
Tetrapyrroles were purchased from Frontier Scientific and other reagents from Sigma-Aldrich or Melford. All porphyrins were dissolved from solids in DMSO to 10 mM and then to the required concentration in buffer. The monomeric state of Fe(III)-TMPyP was confirmed spectroscopically by checking for the characteristic absorption maxima at 423 and 595 nm (52).
Methods.
Detailed methods can be found in SI Materials and Methods.
Protein Expression and Purification.
HuPrP91–231 and huPrP119–231 were expressed (53) and purified (54) as described previously.
Equilibrium Dialysis.
Seventy-five microliters of 100 μM compound and huPrP91–231 were placed in either chamber of a DispoEquilibrium DIALYZER (Harvard Appuratus) with a 5,000 molecular weight cutoff. Sample was left to equilibrate at room temperature for 2 d with gentle rocking. After this time, the compound concentrations were quantified using UV-visible spectroscopy with the appropriate buffer or protein backgrounds subtracted.
Isothermal Titration Calorimetry.
Titrations were performed on a VP-ITC (MicroCal) (55). HuPrP and Fe(III)-TMPyP were diluted to the required concentrations. Data were automatically collected and analyzed using a one-site model in the Origin software (version 7.0) provided by MicroCal with end point ligand dilution effects subtracted.
Circular Dichroism Spectroscopy.
Induced circular dichroism studies were performed on a Jasco J715 spectropolarimeter. Dissociation constants were calculated using the induced CD at 420 nm in Grafit 5 (Erithacus Software) by fitting a one-site Langmuir isotherm. CD denaturation experiments were performed with the aid of a Jasco peltier temperature control system with the sample heated at 1 °C/ min between 20–80 °C and signal measured at 235 nm. The Tm was calculated by fitting the curve to the Van’t Hoff equation.
Sedimentation Velocity Analytical Ultracentrifugation.
Experiments were performed on a Beckman XLI analytical ultracentrifuge. Samples were centrifuged at an average 193,000 × g at 20 °C with absorbance data collected at both 278 and 420 nm. Sedimentation velocity data were analyzed as described (56, 57). Dissociation constants were calculated for individual experiments in Grafit 5 (Erithacus Software) using a one-site Langmuir isotherm.
Nuclear Magnetic Resonance Spectroscopy.
Heteronuclear single quantum coherence spectra were acquired at 298 K on a Bruker DRX-600 spectrometer and data processed and analyzed on Linux Workstations using Felix 2004 (Accelrys) software.
Computer Modeling.
The NMR structure (1QLX) of residues 23–230 of the human prion protein was used for modeling the interaction between PrPc and Fe(III)-TMPyP. The initial model was refined using in-house docking software (37–39).
Cell-Based Prion Curing Assay.
Chronically prion-infected (58) N2a cells (subclone ‘‘PK1’’) (41) were cultured in OPTIMEM (Invitrogen) supplemented with 10% FCS (Perbio) and penicillin/streptomycin (Invitrogen) with Fe(III)-TMPyP at various concentrations for 3 d before assaying for levels of proteinase K-resistant. Half-maximal inhibition constants (IC50) were derived by fitting of the averaged and normalized data by using Grafit5 (Erithacus Software, Ltd.). Cytotoxicity was detected using the sensitive ATP-based cell titre-glo luminescence assay (Cell titre-glo, Promega) in duplicate on cells grown at the same time and under identical conditions.
Protein-Misfolding Cyclic Amplification.
PMCA was performed as described previously (42, 59). Briefly, 10% (wt/vol) PMCA substrate homogenates were prepared from Tg20 mice brains perfused with PBS containing 5 mM EDTA at the time of death. Brains were homogenized, clarified by centrifugation at 1000 × g for 45 s, and then stored at -70 °C without freeze-thawing until required. Rocky Mountain Laboratory (58) prion-infected homogenate was added to pooled PMCA substrate homogenate at a dilution of 500-fold. Samples were subjected to 85 cycles of PMCA consisting of a 20 s pulse of sonication at 70% power output using a Misonix S4000 sonicator with a microplate horn (Misonix) followed by incubation for 30 min at 35 °C. All samples were analyzed by PK digestion and Western blotting. Signal intensity of bands was quantified using ImageMaster 1D Elite software (GE Healthcare Life Sciences).
Supplementary Material
Acknowledgments.
We are grateful to our collaborators at GlaxoSmithKline for help and discussion including Mike Snowden, Jon Hutchinson, and Tony Dean. Thanks to Ray Young for the preparation of figures. This is an independent report commissioned and funded by the Policy Research Programme in the Department of Health, United Kingdom.
Footnotes
Conflict of interest statement: J.C. is a director and J.C., G.J., and A.R.C. are consultants to and shareholders of D-Gen Limited, an academic spin-out company in the field of prion diagnosis, decontamination, and therapeutics.
This article is a PNAS Direct Submission. B.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009062107/-/DCSupplemental.
References
- 1.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 2.Brooks DA. Getting into the fold. Nat Chem Biol. 2007;3:84–85. doi: 10.1038/nchembio0207-84. [DOI] [PubMed] [Google Scholar]
- 3.Boeckler FM, et al. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci USA. 2008;105:10360–10365. doi: 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuwata K, et al. Hot spots in prion protein for pathogenic conversion. Proc Natl Acad Sci USA. 2007;104:11921–11926. doi: 10.1073/pnas.0702671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallucci G, Collinge J. Rational targeting for prion therapeutics. Nat Rev Neurosci. 2005;6:23–34. doi: 10.1038/nrn1584. [DOI] [PubMed] [Google Scholar]
- 6.Nicoll AJ, Collinge J. Preventing prion pathogenicity by targeting the cellular prion protein. Infect Disord Drug Targets. 2009;9:48–57. doi: 10.2174/1871526510909010048. [DOI] [PubMed] [Google Scholar]
- 7.Collinge J. Molecular neurology of prion disease. J Neurol Neurosurg Psychiatry. 2005;76:906–919. doi: 10.1136/jnnp.2004.048660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffith JS. Self replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 9.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 10.Collinge J, et al. Molecular analysis of prion strain variation and the aetiology of “new variant” CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 11.Hill AF, et al. The same prion strain causes vCJD and BSE. Nature. 1997;389:448–50. doi: 10.1038/38925. [DOI] [PubMed] [Google Scholar]
- 12.Bruce ME, et al. Transmissions to mice indicate that “new variant” CJD is caused by the BSE agent. Nature. 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- 13.Wroe SJ, et al. Clinical presentation and pre-mortem diagnosis of variant Creutzfeldt-Jakob disease associated with blood transfusion: A case report. Lancet. 2006;368:2061–2067. doi: 10.1016/S0140-6736(06)69835-8. [DOI] [PubMed] [Google Scholar]
- 14.Collinge J, Clarke A. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 15.Bueler H, et al. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 16.Bueler H, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 17.Mallucci G, et al. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 18.Mallucci G, et al. Targeting cellular prion protein reverses early cognitive deficits and neurophysiological dysfunction in prion-infected mice. Neuron. 2007;53:325–335. doi: 10.1016/j.neuron.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 19.White AR, et al. Monoclonal antibodies inhibit prion replication and delay the development of prion disease. Nature. 2003;422:80–83. doi: 10.1038/nature01457. [DOI] [PubMed] [Google Scholar]
- 20.Riek R, et al. NMR structure of the mouse prion protein domain PrP (121–231) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 21.Zahn R, et al. NMR solution structure of the human prion protein. Proc Natl Acad Sci USA. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosszu LLP, et al. Structural mobility of the human prion protein probed by backbone hydrogen exchange. Nat Struct Biol. 1999;6:740–743. doi: 10.1038/11507. [DOI] [PubMed] [Google Scholar]
- 23.Westergard L, Christensen HM, Harris DA. The cellular prion protein (PrP(C)): Its physiological function and role in disease. Biochim Biophys Acta. 2007;1772:629–644. doi: 10.1016/j.bbadis.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antonyuk SV, et al. Crystal structure of human prion protein bound to a therapeutic antibody. Proc Natl Acad Sci USA. 2009;106:2554–2558. doi: 10.1073/pnas.0809170106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kocisko DA, et al. Potent antiscrapie activities of degenerate phosphorothioate oligonucleotides. Antimicrob Agents Chemother. 2006;50:1034–1044. doi: 10.1128/AAC.50.3.1034-1044.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caughey B, et al. Binding of the protease-sensitive form of prion protein PrP to sulfated glycosaminoglycan and Congo red. J Virol. 1994;68:2135–2141. doi: 10.1128/jvi.68.4.2135-2141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kocisko DA, et al. Identification of prion inhibitors by a fluorescence-polarization-based competitive binding assay. Anal Biochem. 2007;363:154–156. doi: 10.1016/j.ab.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Lee KS, et al. Hemin interactions and alterations of the subcellular localization of prion protein. J Biol Chem. 2007;282:36525–36533. doi: 10.1074/jbc.M705620200. [DOI] [PubMed] [Google Scholar]
- 29.Feng BY, et al. Small-molecule aggregates inhibit amyloid polymerization. Nat Chem Biol. 2008;4:197–199. doi: 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caughey B, et al. Prions and transmissible spongiform encephalopathy (TSE) chemotherapeutics: A common mechanism for anti-TSE compounds? Acc Chem Res. 2006;39:646–653. doi: 10.1021/ar050068p. [DOI] [PubMed] [Google Scholar]
- 31.Caughey WS, et al. Cyclic tetrapyrrole sulfonation, metals, and oligomerization in anti-prion activity. Antimicrob Agents Chemother. 2007;51:3887–3894. doi: 10.1128/AAC.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caughey WS, et al. Inhibition of protease-resistant prion protein formation by porphyrins and phthalocyanines. Proc Natl Acad Sci USA. 1998;95:12117–12122. doi: 10.1073/pnas.95.21.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Priola SA, Raines A, Caughey WS. Porphyrin and phthalocyanine antiscrapie compounds. Science. 2000;287:1503–1506. doi: 10.1126/science.287.5457.1503. [DOI] [PubMed] [Google Scholar]
- 34.Kurihara H, et al. Spectrophotometric and electrochemical investigations of [tetrakis(1-methylpyridinium-4-yl)porphine] iron(III) ion in aqueous-solution. Bull Chem Soc Jpn. 1982;55:3515–3519. [Google Scholar]
- 35.Feng BY, Shoichet BK. A detergent-based assay for the detection of promiscuous inhibitors. Nat Protoc. 2006;1:550–553. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peretz D, et al. A conformational transition at the N terminus of the prion protein features in formation of the scrapie isoform. J Mol Biol. 1997;273:614–622. doi: 10.1006/jmbi.1997.1328. [DOI] [PubMed] [Google Scholar]
- 37.Gibbs N, Clarke AR, Sessions RB. Ab initio protein structure prediction using physicochemical potentials and a simplified off-lattice model. Proteins. 2001;43:186–202. doi: 10.1002/1097-0134(20010501)43:2<186::aid-prot1030>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 38.Sessions RB, Thomas GL, Parker MJ. Water as a conformational editor in protein folding. J Mol Biol. 2004;343:1125–1133. doi: 10.1016/j.jmb.2004.08.105. [DOI] [PubMed] [Google Scholar]
- 39.Thomas GL, Sessions RB, Parker MJ. Density guided importance sampling: Application to a reduced model of protein folding. Bioinformatics. 2005;21:2839–2843. doi: 10.1093/bioinformatics/bti421. [DOI] [PubMed] [Google Scholar]
- 40.Vedadi M, et al. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci USA. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klohn P, et al. A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. Proc Natl Acad Sci USA. 2003;100:11666–11671. doi: 10.1073/pnas.1834432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castilla J, et al. Protein misfolding cyclic amplification for diagnosis and prion propagation studies. Methods Enzymol. 2006;412:3–21. doi: 10.1016/S0076-6879(06)12001-7. [DOI] [PubMed] [Google Scholar]
- 43.Vogtherr M, et al. Antimalarial drug quinacrine binds to C-terminal helix of cellular prion protein. J Med Chem. 2003;46:3563–3564. doi: 10.1021/jm034093h. [DOI] [PubMed] [Google Scholar]
- 44.Doh-ura K, Iwaki T, Caughey B. Lysosomotropic agents and cysteine protease inhibitors inhibit scrapie-associated prion protein accumulation. J Virol. 2000;74:4894–4897. doi: 10.1128/jvi.74.10.4894-4897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collinge J, et al. Safety and efficacy of quinacrine in human prion disease (PRION-1 study): a patient-preference trial. Lancet Neurol. 2009;2009:334–344. doi: 10.1016/S1474-4422(09)70049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yaffe O, Korin E, Bettelheim A. Interaction of Fe(III) tetrakis(4-N-methylpyridinium)porphyrin with sodium dodecyl sulfate at submicellar concentrations. Langmuir. 2008;24:11514–11517. doi: 10.1021/la802122q. [DOI] [PubMed] [Google Scholar]
- 47.Hosszu LL, et al. The residue 129 polymorphism in human prion protein does not confer susceptibility to CJD by altering the structure or global stability of PrPC. J Biol Chem. 2004;279:28515–28521. doi: 10.1074/jbc.M313762200. [DOI] [PubMed] [Google Scholar]
- 48.Kim K, et al. Photoinduced protein cross-linking mediated by palladium porphyrins. J Am Chem Soc. 1999;121:11896–11897. [Google Scholar]
- 49.Villanueva A, et al. Photodynamic induction of DNA-protein cross-linking in solution by several sensitizers and visible-light. Biopolymers. 1993;33:239–244. doi: 10.1002/bip.360330206. [DOI] [PubMed] [Google Scholar]
- 50.Verma A, Snyder SH. Characterization of porphyrin interactions with peripheral type benzodiazepine receptors. Mol Pharmacol. 1988;34:800–805. [PubMed] [Google Scholar]
- 51.Lauren J, et al. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature. 2009;457:1128–1132. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erdem SS. Spectral evidence for heterodimerization of water-soluble metalloporphyrins. J Porphyr Phthalocya. 1998;2:61–68. [Google Scholar]
- 53.Jackson GS, et al. Multiple folding pathways for heterologously expressed human prion protein. Biochim Biophys Acta. 1999;1431:1–13. doi: 10.1016/s0167-4838(99)00038-2. [DOI] [PubMed] [Google Scholar]
- 54.Zahn R, vonSchroetter C, Wuthrich K. Human prion proteins expressed in Escherichia coli and purified by high-affinity column refolding. FEBS Lett. 1997;417:400–404. doi: 10.1016/s0014-5793(97)01330-6. [DOI] [PubMed] [Google Scholar]
- 55.Wiseman T, et al. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 56.Schuck P, et al. Size-distribution analysis of proteins by analytical ultracentrifugation: Strategies and application to model systems. Biophys J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Horton JC, Rowe AJ, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, UK: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 58.Chandler RL. Encephalopathy in mice produced by inoculation with scrapie brain material. Lancet. 1961;1:1378–1379. doi: 10.1016/s0140-6736(61)92008-6. [DOI] [PubMed] [Google Scholar]
- 59.Tattum MH, et al. Discrimination between prion-infected and normal blood samples by protein misfolding cyclic amplification. Transfusion. 2010 doi: 10.1111/j.1537-2995.2010.02595.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.