Abstract
Concentrated solutions of duplex-forming DNA oligomers organize into various mesophases among which is the nematic (N∗), which exhibits a macroscopic chiral helical precession of molecular orientation because of the chirality of the DNA molecule. Using a quantitative analysis of the transmission spectra in polarized optical microscopy, we have determined the handedness and pitch of this chiral nematic helix for a large number of sequences ranging from 8 to 20 bases. The B-DNA molecule exhibits a right-handed molecular double-helix structure that, for long molecules, always yields N∗ phases with left-handed pitch in the μm range. We report here that ultrashort oligomeric duplexes show an extremely diverse behavior, with both left- and right-handed N∗ helices and pitches ranging from macroscopic down to 0.3 μm. The behavior depends on the length and the sequence of the oligomers, and on the nature of the end-to-end interactions between helices. In particular, the N∗ handedness strongly correlates with the oligomer length and concentration. Right-handed phases are found only for oligomers shorter than 14 base pairs, and for the sequences having the transition to the N∗ phase at concentration larger than 620 mg/mL. Our findings indicate that in short DNA, the intermolecular double-helical interactions switch the preferred liquid crystal handedness when the columns of stacked duplexes are forced at high concentrations to separations comparable to the DNA double-helix pitch, a regime still to be theoretically described.
Keywords: cholesteric, RNA
Understanding how the physicochemical details of macromolecules affect their interactions and their self-ordering properties represents one of the major challenges in the physics of soft and biological matter. In particular, because most of the biologically relevant molecules are chiral—i.e., they lack mirror symmetry—the relevant biointeractions are typically highly sensitive to the molecular handedness. One important effect of chiral interactions is the propagation of chirality from the molecular structure to the supramolecular assemblies such as aggregates or ordered phases. Often, in this phenomenon, minor molecular modifications are amplified to produce remarkable changes in the macroscopic ordering, as for DNA supercoiling and its biological role (1). The propagation of chirality has been studied in various systems, including thermotropic liquid crystals (LCs) and lyotropic assemblies of viruses, DNA, polymers, and dye aggregates. Despite the generality of the phenomenon, its understanding is still poor. Predicting, only on the basis of the molecular structure, the most basic parity of chiral LC ordering, the right- vs. left-handedness, is still a challenge (2–4). One would expect the problem to become easier when restricted to molecular structures with a clean cylindrical shape decorated with regular helical structures and charges, such as DNA, G-quartets, and filamentous viruses. However, even in this limited frame, experimental and theoretical results appear difficult to fit coherently in a common scenario. Experiments indicate that there is no simple rule connecting the handedness of the individual helices to that of the phase. There are examples of right-handed (RH) helices yielding RH chiral nematic (N∗) phases, as in the case of the four stranded guanosine columnar aggregates (G-wires) formed by homooligomeric chains with 3 or more guanosines (5); left-handed (LH) helices yielding LH N∗ phases, as in the case of G-wires formed by the aggregation of the single nucleotide guanosine-monophosphate (5); RH helices yielding LH N∗ phases, such as long DNA double strands (6, 7), filamentous viruses (8,10), and G-wires made of diguanosines (5).
On the theoretical side, the situation is not much clearer and a unifying model is lacking. The classic Straley model (2, 11), based on steric interaction of helices, predicts that the RH DNA helices should yield RH N∗ phases, opposite to observations. More recent models incorporate the effect of electrostatic interactions, believed to bring a significant contribution. Typically, these approaches calculate the energy of two long helices as a function of their mutual twist angle, which is in general also dependent on the azimuthal orientation of the helices about their long axes. More specifically, these calculations have been pursued according to two basic different strategies: either describing the helices as clean cylinders with helical lines of charge accounting for phosphate groups and adsorbed counterions [Kornyshev–Leikin model (KL)] (12), or modeling the helices as cylinders decorated with protruding charged spheres having a steric volume [Tombolato–Ferrarini model (TF)] (13). The two yield opposite handedness for B DNA. The current implementation of the KL model to the N∗ phase of DNA predicts RH N∗, whereas a LH N∗ phase is correctly predicted by the TF model, where the interhelical interactions, and especially the coupling of azimuthal angles, include steric repulsion. A possible change of sign in the twist is not completely ruled out in the KL model (14), because its application to DNA requires simplifying assumptions possibly masking a richer behavior implied by the general formulation of the theory.
The dependence of the N∗ helical pitch (p, p > 0 is right-handed) on DNA concentration (c) is also complex, investigations revealing a nonmonotonic behavior of p(c) in DNA N∗ phases, where, as c is increased, p at first decreases—A behavior that can be interpreted as due to the increased strength of the chiral interactions between molecules closer together, whereas, at larger c, it increases again, indicating that as the columnar phase is approached, the parallel packing of hard rods begins to dominate (15). This nonmonotonic behavior of p is accounted for, although with a sign of p, opposite that predicted by KL model.
We have recently shown that concentrated solutions of ultrashort fragments of DNA double helices [“nano-DNA” (nDNA)], of length ranging from N = 6 to N = 20 base pairs (bp; N ≡ bp number) can develop macroscopic LC order, including a chiral nematic phase and, at higher concentration, a columnar (COL) phase. Based on the variety of sequences accounted for in the Nucleic Acid Database (16), we expect the nDNA duplexes studied previously and here to be the basic right-handed B-DNA structure. Also evident in the database are a variety of sequence dependent variations in the B-DNA structure, such as bend of the double-helix axis, which introduce variability in the LC ordering in the oligomers studied here.
LC ordering has been observed in two main categories of nanooligonucleotides: “blunt-ended” (BE) (17) and “sticky-dangling” end (SD) duplexes (18). BE are fully mutually or self-complementary sequences that upon hybridizing form duplexes in which all bases are paired, therefore terminating with a blunt end. These duplexes interact through the stacking interactions at their terminals, forming reversible linear aggregates (17). SD are instead partially self-complementary sequences, where the pairing takes place in a shift-register type scheme, leaving a few unpaired bases at each end of the duplex. If such unpaired bases are chosen so as to pair with one another, they become sticky and through their pairing the duplexes assemble end-to-end (18).
Both the stacking of BE and the pairing of SD duplexes promote living polymerization, hence giving rise to linear aggregates whose average length ℓ grows with the duplex concentration and decreases with temperature (T). The solutions develop nematic order when T is sufficiently low and c sufficiently large to make ℓ match the Onsager criterion ϕ ≈ 4D/ℓ, where D is the diameter of the double helix and ϕ is the nDNA duplex volume fraction. An analysis of the phase diagram enables estimation of the interduplex interaction energy, ∼3 kcal/mol (or ∼5kBT) for the BE stacking (17) and in the range 3–5 kcal/mol for the SD interactions with 2-base-long sticky ends (18). The broken mirror symmetry of the DNA helices gives rise, within the nematic phase, to a chiral torque Kt that requires an additional term in the standard Frank–Oseen free energy density for uniaxial nematics 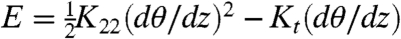 where dθ/dz is the twist reorientation per length of the N∗ director. The nematic phase hence becomes chiral, characterized by a helical progression of the nematic director. The pitch of the N∗ phase, p ≡ 2π/q where q is the associated helix lattice wavevector, is determined by the balance of the chiral torque with the Frank elastic constant for twist K22, q = Kt/K22 and p = 2πK22/Kt.
where dθ/dz is the twist reorientation per length of the N∗ director. The nematic phase hence becomes chiral, characterized by a helical progression of the nematic director. The pitch of the N∗ phase, p ≡ 2π/q where q is the associated helix lattice wavevector, is determined by the balance of the chiral torque with the Frank elastic constant for twist K22, q = Kt/K22 and p = 2πK22/Kt.
In our previous studies of nDNA solutions, we observed N∗ pitch in the range 0.3 μm < p < 5 μm, but we did not focus on the handedness of the phase, taking for granted that it matched the one of long DNA solutions. Quite surprisingly, this is not at all the case. We find that the N∗ phase of nDNA displays either RH or LH order, subtly depending on the oligomer sequences and on their mutual end-to-end interactions.
Results
Sequences Explored in this Work.
We report the characterization of the N∗ phase for the set of DNA sequences listed in Table 1 with their nicknames and commented in SI Text. These oligos enable study of the influence of the following sequence properties on N∗ structure: length; end-to-end coupling; self vs. mutual complementarity; terminal or center sequence composition; relative content of adenine (A)–thymine (T) and cytosine (C)–guanine (G), base pairs (AT and CG); and terminal phosphorylation.
Table 1.
Properties of the investigated DNA oligomers
| Sequence | Nickname | N (bp) | CIN (mg/mL) | |p| range (μm) | H | |p| VS T | |p| VS c | sets | ||
| 1 | CGAATTCG | 8sc1 | 8 | BE | 610 | 1–2 | L | INC | — | 1 |
| 2 | CGCATGCG | 8sc2 | 8 | BE | 720 | 0,45–0,8 | L | DEC | INC | — |
| 3 | CGCAATTGCG | 10sc | 10 | BE | 650 | 0,4–∞ | R | INC | INC | 1 |
| 4 | ATAAATTTAT | 10allAT | 10 | BE | — | 1–3 | R | — | — | — |
| 5 | CGCGAATTCGCG | DD | 12 | BE | 730 | 0,7–3 | R | INC | — | 1,2 |
| 6 | pCGCGAATTCGCG | pDD | 12 | BE | 670 | 0,35–1 | R | DEC | — | 2 |
| 7 | CGCGAATTCGCGp | DDp | 12 | BE | — | 0,3–1 | R | DEC | INC | 2 |
| 8 | GCGCTTAAGCGC | antiDD | 12 | BE | — | 1–2 | R | — | INC | — |
| 9 | GGAGTTTTGAGG + CCTCAAAACTCC | 12mc | 12 | BE | 770 (32) | 0,7–2 | R | — | — | — |
| 10 | ACCGAATTCGGT | ACC | 12 | BE | 850 | 1–3 | R | — | INC | 2,3 |
| 11 | AACGAATTCGTT | AAC | 12 | BE | 620 | ∞ | A | — | — | 3 |
| 12 | AATGAATTCATT | AAT | 12 | BE | 500 | 1–2 | L | — | — | 3 |
| 13 | AATAAATTTATT | allAT | 12 | BE | 600 | 0,5–1 | L | INC | — | 3 |
| 14 | CCGGCGCGCCGG | allCG1 | 12 | BE | 650 | 1–3 | L | — | — | 3 |
| 15 | CGCGCCGGCGCG | allCG2 | 12 | BE | 570 | 0,3–∞ | L,R | INC, DEC | HI | — |
| 16 | GCGCGAATTCGC | sDD | 12 | SD | 400 | 0,3–1 | L | — | INC | 2 |
| 17 | ACGCGAATTCGCGT | 14Aterm | 14 | BE | — | 1–3 | L | — | — | — |
| 18 | CGCGAAATTTCGCG | 14sc | 14 | BE | 440 (17) | 1–3 | L | — | INC | 1 |
| 19 | pCGCGAAATTTCGCG | p14sc | 14 | BE | — | 1–3 | L | — | — | — |
| 20 | GCCGCGAATTCGCG | GC-DD | 14 | SD | 400 | 0.35–1 | L | INC | — | — |
| 21 | CGCGAATTCGCGGC | DD-GC | 14 | SD | 350 | 1–3 | L | — | — | — |
| 22 | CGCGCGAATTCGCG | CG-DD | 14 | SD | 470 | 1–3 | L | INC | INC | — |
| 23 | ATCGCGAATTCGCG | AT-DD | 14 | SD | 430 | 1–3 | L | — | — | — |
| 24 | ACGCAGAATTCTGCGT | 16Aterm | 16 | BE | 440 (17) | 1–4 | L | INC | INC | — |
| 25 | CGCGAAAATTTTCGCG | 16sc | 16 | BE | — | 1–4 | L | — | — | 1 |
| 26 | AACGCAAAGATCTTTGCGTT | 20sc | 20 | BE | 220 (17) | 1–4 | L | — | INC | — |
| 27 | CGCGAAUUCGCG | DD-RNA | 12 | BE | 900 (31) | 0.3–1 | R | — | DEC | — |
| 28 | — | long DNA | 150 | — | 160 | 2–4 | L | INC | DEC-INC | — |
A, C, G, T refer to DNA bases, adenosine, cytosine, guanine, and thymine, respectively. For each sequence, we give the short name used in the text, the length N (base pair number), and the type of end-to-end aggregation (BE or SD). cIN is the critical concentration for the Isotropic-(Chiral) Nematic transition at room temperature and recalculated with an improved calibration of refractive index-concentration relationship). H is the observed handedness, LH, RH, or achiral nematic. The range of absolute values of the cholesteric pitch is determined within the concentration range of the N∗ phase at room temperature. The slope of the dependence of |p| on T (discussed in SI Text) and c is either positive (INC) or negative (DEC), and Handedness Inversion (HI) can occur. The last column highlights the sets of sequences described and discussed in the text. The missing quantities or dependencies have not been determined within this work.
N∗ Ordering of nDNA.
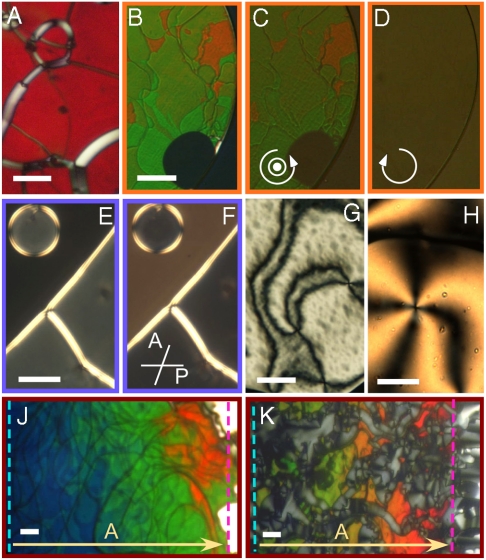
All the oligos in Table 1 order into the nematic phase. Fig. 1 shows some of the textures obtained with optical microscopy from nematic samples. The pitch varies significantly from sequence to sequence: some of them yield chiral nematic phases with pitch in the visible range. In this case, the N∗ is immediately recognizable in polarized transmission optical microscopy (PTOM), as shown in Fig. 1, for pDD and sDD, respectively, identified in Table 1. Both images show a typical Grandjean texture, where in the colored regions the axis of the chiral nematic helix is perpendicular to the cell surfaces (i.e., parallel to the direction of the propagating light), whereas the white line defects are the so called “oily streaks,” in which the helical axis lays in the plane of the cell, giving rise to an effective birefringence having the largest refractive index for polarization normal to the defect line. Colors arise because of the “selective reflection” effect, the strong back-reflection of the radiation having peak wavelength λ0 and circular polarization matching, respectively, pitch and handedness of the LC. In this condition, λ0/2n = p/2, and n is the average refractive index (a green texture, λ ∼ 500 nm, as in Fig. 1A, corresponds to p ∼ 330 nm) (19). Selective back-reflection is demonstrated in Fig 1 C and D, obtained by viewing the cell in reflected light (illumination through the objective). Illumination with, respectively, LH and RH circularly polarized light (for sign conventions, see SI Text) demonstrates that the N∗ phase of sDD is LH. Within the selective reflection band, transmitted light is predominantly circularly polarized with opposite handedness, and is thus visualized, with a much better contrast, in transmission through crossed linear polarizers, as in Fig. 1B. The increased contrast is due to the elimination of background, seen by comparing the bottom right corner of Fig 1 B–D where a region with the sealing oil (at the very corner of the picture) and a region of isotropic solution (having an approximately round shape) are visible.
Fig. 1.
Typical PTOM microscopic textures of the nematic phases of nDNA duplexes, between crossed polarizers, unless otherwise indicated. (A, B) Oily streaks texture of the N∗ phase of pDD (A) and of sDD (B). (C, D) The L handedness of the sDD N∗ phase is confirmed by observing the reflected intensity when illuminating the sample by LH (C) and RH (D) circular light. (E) N∗ phase of DD: The droplet and the region in the lower-right part can only be extinguished by decrossing the analyzer clockwise (F), revealing RH optical rotation. The black region surrounding the droplet is isotropic fluid. (G, H) N∗ “Schlieren” textures of AAC (G) and of a 1∶1 mixture of 10sc and 20sc (H). The difference in colors between the G and H is caused by the different cell thicknesses. (J, K) Dependence of the N∗ pitch on c in cells with c gradient for the LH sDD (J) and the RH 10sc (K). The dashed lines mark the isotropic-N∗ (blue) and N∗-COL boundaries (pink). Bars are 30 μm.
In some of the compounds, the N∗ phase has a pitch longer than the visible wavelengths, as for the case of DD reported in the PTOM Fig. 1E taken through crossed polarizers (uniform alignments are found in the grayish areas within the circular domain and in the bottom right part of the figure, the black area being instead isotropic fluid). The transmitted light can be extinguished by an appropriate decrossing of the analyzer from the polarizer, as in Fig. 1F, thus revealing optical rotation. In a few instances, we also observed solutions with a LC helix pitch that was so long that the nematic appeared to be nonchiral, as revealed by the typical nematic Schlieren textures. Examples are shown in Fig. 1G for the AAC sequence and in Fig. 1H for a 1∶1 mixture of 10sc and 20sc. Various cells were prepared for each sequence. Careful sealing enabled a good stabilization of the cell properties, allowing quantitative characterization through: (i) micro-interferometry to determine the nDNA concentration (17) (see SI Materials and Methods); and (ii) quantitative analysis of the polarization of the transmission spectra to measure pitch and handedness of the N∗ phase, as detailed next.
Characterization of N∗ Phases by Transmission Spectra.
The handedness of DNA N∗ phase has been usually assigned through the study of specific bands in circular dichroism spectra (6, 7), a method not free from ambiguity and artifacts, and that requires substantial amounts of sample. Pitch and handedness of thermotropic LCs are instead typically extracted via optical methods (19). These have, however, limited applicability for DNA lyotropic LCs because of the poor control of boundary conditions. Fig. 1 A and B are clear examples of this: domains where the N∗ helix is perpendicular to the cell plates and whose color indicate definite structure; i.e., which have clear optical signatures but are surrounded by the birefringent oily streaks. We show here that a suitable analysis of the spectra of the transmitted intensity IT(λ) vs. the orientation of the analyzer, enables a full characterization of the N∗ helical structures with pitch in the optical range, even in the presence of defects and irregular ordering.
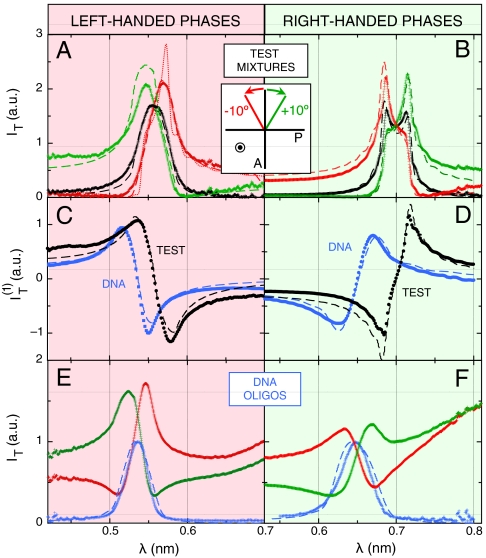
Fig. 2 A and B show IT(λ) for two test cells filled with thermotropic N∗ LC mixtures: the LH mixture M1 of cholesteryl nonanoate, cholesteryl chloride, and cholesteryl oleyl carbonate (∼1∶1∶2 weight ratio) (20), and the RH mixture M2 of CCN47 + CB15 (∼2∶1 weight ratio) (21). Mixtures with low-birefringence were chosen to better approximate that of the DNA samples. IT(λ) has been measured with crossed polarizer and analyzer (ϑ = 0°, black lines) and with the analyzer rotated to ϑ = +10° (green lines) and ϑ = -10° (red lines). The transmission spectra of N∗ phases of opposite handedness are modified in opposite ways upon equally rotating the analyzer.
Fig. 2.
Experimental transmission spectra through samples of test thermotropic cholesteric mixtures, LH M1 (A) and RH M2 (B), and through nDNA N∗ phases, LH sDD (E) and RH pDD (F). Black and blue curves give spectra of light passing through crossed polarizers, whereas green and red curves represent spectra obtained by decrossing the analyzer of 10° respectively clockwise and counterclockwise (light is propagating toward the observer). (C, D) Linear term of the expansion of the transmitted intensity (Eq. 1), giving the handedness of the N∗ phase for thermotropic (black) and nDNA (blue) samples. The dashed lines are calculated from the propagation model described in SI Text. The dotted line in (A) is obtained from the same model, assuming no deviation in the N∗ pitch—see SI Text.
Spectra calculated through the propagation model, IT(λ,ϑ|p,d,Δn), described in SI Text, were fit to measured spectra IT(λ,ϑ) by varying p, cell thickness d, and local nematic birefringence Δn, with the results shown in Fig. 2 A and B (dashed lines). The fitted values of Δn (0.05 for M1, 0.06 for M2) and d agree well with the birefringence of the mixtures and the actual cell thickness. Most importantly, the measured handedness matches the one expected from the literature (20, 21).
This analysis can be extended to cells with a defected structure, as those in Fig. 1 A and B, by including the dependence on ϑ in the approach. Both the experimental and the calculated transmission spectra can be expanded as a function of ϑ:
| [1] |
It is then easy to show that the contribution of nearly isotropically distributed birefringent regions, such as the oily streaks surrounding the uniform domains, affects only 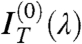 and
and 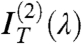 , the even terms of the expansion, whereas the coefficient of the linear term
, the even terms of the expansion, whereas the coefficient of the linear term 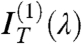 is basically unperturbed. As expected by symmetry,
is basically unperturbed. As expected by symmetry, 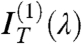 , quantifying the odd dependence on ϑ, carries the relevant information on the optical chirality and can be fit to effectively extract the parameters p, d, and Δn. Therefore, the N∗ structure in defected cells can be obtained by performing the analysis in Eq. 1, disregarding
, quantifying the odd dependence on ϑ, carries the relevant information on the optical chirality and can be fit to effectively extract the parameters p, d, and Δn. Therefore, the N∗ structure in defected cells can be obtained by performing the analysis in Eq. 1, disregarding 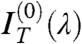 and
and 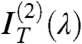 and focusing on
and focusing on 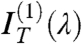 to extract, through a fitting to the model, pitch length, birefringence, and handedness. Fig. 2 C and D show the spectral dependence of
to extract, through a fitting to the model, pitch length, birefringence, and handedness. Fig. 2 C and D show the spectral dependence of 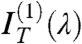 for M1 and M2, respectively (black dots). The opposite handedness of the N∗ phase of the LH and RH test mixtures is unmistakable. Dashed lines are the best fit to
for M1 and M2, respectively (black dots). The opposite handedness of the N∗ phase of the LH and RH test mixtures is unmistakable. Dashed lines are the best fit to 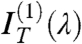 calculated from the model; i.e. obtained with the same values of p, d and Δn as in Fig. 2 A and B. The agreement validates our modeling.
calculated from the model; i.e. obtained with the same values of p, d and Δn as in Fig. 2 A and B. The agreement validates our modeling.
All sequences in Table 1 displaying optical pitch were analyzed with this procedure. Fig. 2 E and F show the transmittance spectra IT(λ) for sDD and pDD, respectively, whereas Fig. 2 C and D show the asymmetric component 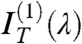 (blue dots). IT(λ,ϑ = 0) and
(blue dots). IT(λ,ϑ = 0) and 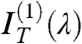 are satisfactorily fit by the model (dashed lines in the figure). The local Δn is found to be, in both cases, Δn = 0.038 ± 0.005, in agreement with the value for long B-DNA, once properly rescaled for c (22). Data in Fig. 2 demonstrate that nDNA LC may organize in either RH or LH phases. This is reported in Table 1, where we also indicate the range of p values at room T and the handedness H. When the pitch of the N∗ phase is in the μm range, the measurement of spectra in the visible range is not as informative as for shorter pitch. The spectra are in this case quite featureless, but measurements of the intensity vs. ϑ are useful because they enable determining the optical rotation, in turn related to handedness, Δn, d, and p. We have hence measured the optical rotation of DNA N∗ phase for all sequences that organize with a long pitch, as in the example given by Fig. 1 E and F, to extract the phase handedness, as better described in SI Text.
are satisfactorily fit by the model (dashed lines in the figure). The local Δn is found to be, in both cases, Δn = 0.038 ± 0.005, in agreement with the value for long B-DNA, once properly rescaled for c (22). Data in Fig. 2 demonstrate that nDNA LC may organize in either RH or LH phases. This is reported in Table 1, where we also indicate the range of p values at room T and the handedness H. When the pitch of the N∗ phase is in the μm range, the measurement of spectra in the visible range is not as informative as for shorter pitch. The spectra are in this case quite featureless, but measurements of the intensity vs. ϑ are useful because they enable determining the optical rotation, in turn related to handedness, Δn, d, and p. We have hence measured the optical rotation of DNA N∗ phase for all sequences that organize with a long pitch, as in the example given by Fig. 1 E and F, to extract the phase handedness, as better described in SI Text.
Effects of Concentration.
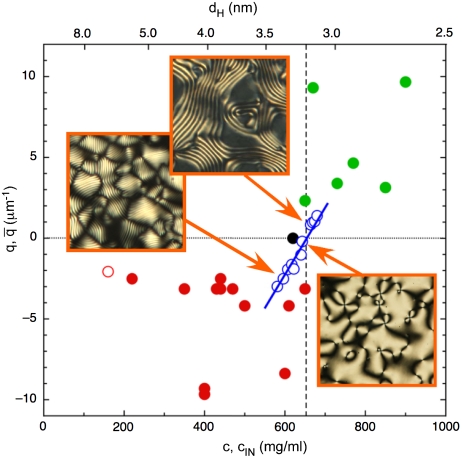
In the large majority of the sequences, the pitch monotonically grows with c regardless of the handedness, as reported in Table 1. Fig. 1 J and K visually shows such behavior for representative cases of short pitch N∗, both LH and RH (respectively sDD, Fig. 1J, and 10sc, Fig. 1K). The increase of |p| with c is evident from the change in the selectively reflected colors. The increase of |p| with c appears to be universal for all LH N∗ phases. In one of the sequences—the 12-mer allCG2—such increase produces a continuous handedness inversion. Specifically, we find a LH N∗ ordering at lower c and a progressive growth of |p| with c that leads to the unwinding of N∗ into a nonchiral nematic phase, followed, upon further increasing c, by the formation of a RH N∗ having p decreasing with c. This helical inversion (“HI” in Table 1) is quantitatively documented in Fig. 3 (open blue dots), where for convenience we plot q = 2π/p vs. c. The figure also reports, in the upper x-axis, dH, the mean center-to-center distance of neighboring duplexes, corresponding to each given concentration. Helical inversion leads to atypical p vs. c behavior, because the N∗ in the RH branches of these sequences winds-up as c increases, whereas non-HI sequences behave oppositely. The effect of other parameters, such as ionic strength and T, is described in SI Text.
Fig. 3.
Dependence of the N∗ helix wave vector q on nDNA concentration c and corresponding interaxial distance dH (upper x axis). Open blue dots: allCG2 behavior. Handedness inversion occurs at cINV ∼650 mg/mL (vertical dashed line), corresponding to dH ∼3.2 nm between neighboring helices. Pictures show typical textures of N∗ phase, LH at low c and RH at high c, and nematic phase at intermediate c. Pictures side ∼80 μm. Red and green dots: average helical period 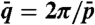 vs. transition concentration cIN for each of the sequences. Red dots: LH sequences. Green dots: RH sequences. Black dot: achiral AAC sequence. Open red dot: long DNA.
vs. transition concentration cIN for each of the sequences. Red dots: LH sequences. Green dots: RH sequences. Black dot: achiral AAC sequence. Open red dot: long DNA.
Discussion
Table 1 summarizes the observed behavior for all the sequences, described in the standard notation A, C, G, T for the DNA bases adenosine, cytosine, guanine, and thymine, respectively. Its inspection reveals that minimal changes in the sequences—even of a single base—can result in remarkably different collective behavior. The chiral macroscopic ordering of nDNA appears to be determined by a combination of various different elements, as it can be recognized by selecting, within the ensemble of explored oligomers, specific subsets. Accordingly, in the last column of Table 1 we label with the same number the sequences to be considered together in various parts of the discussion. The general aim of this discussion is to emphasize features unifying the large variety of observations. Clearly, as the properties of nDNA aggregates approximate those of long double helices, the nDNA solutions have to match the phase behavior and properties of the long DNA (23). This is what happens either changing length—making BE duplexes longer—or changing end-to-end interactions—making them stronger through SD coupling, such as when moving from DD to sDD. In both cases, the handedness of the long DNA and the basic dependence on T and c are retrieved, as expected.
Factors Affecting N∗ Behavior.
Oligomer length.
All the oligomers longer than 12 bp order in N∗ phases with LH symmetry, the same as long (N≥150) DNA. This is verified regardless of the sequence, the terminals or the aggregation mechanism. For shorter duplexes, instead, a more complex behavior sets in. Dodecamers with SD coupling are LH, whereas BE 12-mers show the greatest variability, with LH, RH, or nonchiral nematic phases. The BE decamers investigated here only show RH N∗ phase, whereas for BE 8-mers again we find LH cholesteric phase, indicating that length, a crucial parameter in determining the nDNA behavior, is not the only relevant quantity. As an example of the length effect, in Subset 1 we compare 8sc1, 10sc, DD, 14sc, and 16sc, which share the same core (AATT), the same terminals (BE with CG at both ends), the same basic pattern of CG/AT nucleotides and a similar CG fraction (varying between 0.5 and 0.67). The set displays handedness inversion: 10sc and DD are RH, whereas the shorter 8sc1 and the longer 14sc and 16sc are LH. It is intriguing that the oligomer length at which this variability of behavior shows up corresponds to the typical pitch of the double-helix structure (10 bp in B-form, 11 in A-form).
Dependence on oligomers length is also found when the pitch length is considered. By inspecting Table 1 it can be recognized that shorter oligomers generally have shorter N∗ pitch, well within the visible range, corresponding to twist angles between helices as high as 5°, whereas sequences longer than 12 bp only have pitch of 1 μm or above. Notable exceptions are the μm-scale pitch in 8sc and the sub-μm-scale colored textures in GC-DD.
Short pitches, with reflections in the in the visible range, appear to be a distinctive attribute of short nDNA (a relevant exception is reported in ref. 24 and discussed in SI Text). The basic expectation for the dependence of pitch on effective oligomer length L can be obtained from Straley’s model of interacting chiral hard rods (11) (see SI Text), that predicts p to be shorter in systems of shorter rods. This would explain the appearance of shorter pitches in shorter DNAs. However, Straley’s model offers a simple argument only. It cannot account properly for the interhelical chiral interactions and it cannot justify any change in the sign of p. Moreover, experiments present significant exceptions to this behavior, such as the longer pitch of the 8-mers. A behavior inverse with respect to Straley’s model is also found by studying rod-like viruses of various contour lengths, where the pitch shortens with the growing of virus length (25).
Oligomer sequence.
Oligo terminal structure strongly affects the N∗ helix. This is paradigmatically exemplified by Subset 2, which highlights the effects of terminal interactions in otherwise identical sequences. pDD and sDD, the former aggregating via BE stacking and the latter via SD strands, have the very same sequence pattern along the linear aggregates, as is shown in the SI. Both sequences show very short pitches, well within the visible range, but with opposite handedness. The chemical analogy of pDD and sDD makes it clear that the critical property determining such opposite behavior has not to be directly ascribed to compositional differences, but rather to the conformational properties of the aggregate. SD sequences aggregate not only with a stronger coupling energy, but they also arrange so to produce a continuous phosphate chains (26), a condition in which the persistence of the same N∗ helical sign as long DNA is not surprising. Aggregates of BE oligomers, on the other hand, are structurally different because the azimuthal coupling of end-to-end interacting duplexes is much weaker, being determined by base stacking interactions free of covalent links. In chemically continuous DNA, base stacking and covalent bonding produce an almost constant helical advance of the azimuthal orientation α per base along the chain: +30° < α < +40° (27, 28). At blunt end interfaces between aggregated oligomers, the duplex terminals can adjust α either to maximize stacking or to better accommodate interactions with helices in neighboring aggregates. This can yield quite different values of α—as for example α = -22° found in highly hydrated crystals of 12-mer duplexes terminating with AT (29)—and hence affect the periodicity of the helical pattern along the aggregate, as also found in G-quartets (5). Terminal structure also determines the differences between DD on the one side and pDD and DDp on the other (same handedness but different p). Furthermore, sequences with a phosphate at both ends (such as “pDDp”) do not form LCs (17), thus confirming the significance of phosphates in end-to-end interactions.
We also investigated the effect of gradual changes in the sequence—and thus slight changes in the structure—as among the oligomers in Subset 3. ACC, AAC, and AAT share the same core and the same terminal bases, and differ only in two or four nucleotides. However, their mesoscopic behavior is strikingly different: whereas AAT N∗ is LH, ACC N∗ is RH and AAC, intermediate between the two, does not show any chirality in its nematic phase in a wide range of temperatures (see Fig. 1G). The explanation for this behavior must be in the differences between their helical structures. Structural information is available for only DD and ACC, and X-ray diffraction crystal structures are not fully reliable for solution conformation. However, a few clues suggest that the structural variability among sequences can play a significant role. The reported structure of DD has an appreciable bend of the helical axis (19), which is even larger for the ACC structure (30), indicating that oligomeric duplexes are not necessarily straight. Both sequences are RH whereas their concentration at the isotropic–nematic transition (cIN) is rather high when compared to the whole set of data in Table 1. ACC, which is more curved, has a larger cIN than DD. Same handedness and an even larger cIN is found for the RNA analog of the DD (31). In the RNA case, given the A-type arrangement of the double-helix, there is a very significant inclination of the terminal bases. We speculate that sequences whose helical conformation has a sufficient degree of bending and/or tilt of the terminal bases have an RH N∗ phase. This effect may take place either directly—the helix geometry affecting the structure of the linear aggregate and hence the helical interactions—or indirectly—the geometry affecting the concentration at which the N∗ can be found and hence the symmetry of the interaction.
Oligomer concentration.
The helix inversion of allCG2 can be described with a concentration dependent Kt(c) of the form Kt(c) ∝ (c - 650 mg/mL); i.e., with a change of handedness at cINV ≈ 650 mg/mL. Inspection of Table 1 suggests that handedness strongly correlates with cIN. Sequences with large cIN, close to cINV or higher, organize into RH N∗ phases, whereas the opposite handedness is found for sequences with cIN < cINV. The AAC sequence, with cIN ≈ 620 mg/mL, features an achiral nematic. This behavior can be better appreciated in Fig. 3 (full dots), where we summarize the results in Table 1 by plotting the points 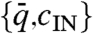 for each of the sequences. Here
for each of the sequences. Here 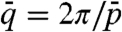 , where
, where 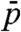 is the average value of the interval of pitches observed for each given sequence. Even though the experimental points are scattered, it is quite clear that the handedness depends on concentration, the RH ordering being preferred at large cIN. In Fig. 3 we also plot points referring to the achiral AAC (black dot), the inversion point for allCG2 (blue open dot), the long DNA (red open dot). It is thus tempting to speculate that the factors introduced and discussed so far—length, terminals, helical conformation—all combine to determine cIN, the handedness being just a consequence of its value. Indeed, it is expected that in various situations the N∗ would require a large concentration: (i) in shorter sequences—because it is necessary to aggregate more monomers; (ii) when the terminal coupling is weaker—because the mean aggregate length grows with concentration and coupling strength (18); and (iii) when duplexes are bent or have tilted bases—because the N∗ ordering requires straight aggregates thus setting in a competing condition between ordering and aggregation.
is the average value of the interval of pitches observed for each given sequence. Even though the experimental points are scattered, it is quite clear that the handedness depends on concentration, the RH ordering being preferred at large cIN. In Fig. 3 we also plot points referring to the achiral AAC (black dot), the inversion point for allCG2 (blue open dot), the long DNA (red open dot). It is thus tempting to speculate that the factors introduced and discussed so far—length, terminals, helical conformation—all combine to determine cIN, the handedness being just a consequence of its value. Indeed, it is expected that in various situations the N∗ would require a large concentration: (i) in shorter sequences—because it is necessary to aggregate more monomers; (ii) when the terminal coupling is weaker—because the mean aggregate length grows with concentration and coupling strength (18); and (iii) when duplexes are bent or have tilted bases—because the N∗ ordering requires straight aggregates thus setting in a competing condition between ordering and aggregation.
According to this scenario, as the concentration is increased beyond a critical distance between two neighboring helices or stacks of helices, a new regime sets in. For some reason, the interhelical coupling is inverted and the favored symmetry is RH, in some cases dominating over the effect of approaching the N∗-COL boundary, expected to favor unwinding. The mean interaxial distance dH corresponding to the inversion concentration of allCG2 (see Fig. 3) is dINV ≈ 3.2 nm (†), an intriguing result because it matches the most fundamental characteristic length of the chiral structure of the double-helix; i.e. the helical pitch of B-DNA duplexes.
Conclusions
The LC chiral nematic phase of concentrated solutions of ultrashort DNA duplexes displays a much richer behavior than N∗ phase of long DNA double strands. Minor changes in oligomer length or sequence, or modifications in the mode of end-to-end aggregation, strongly affect the properties of the nDNA solutions giving rise to a previously unexpected, unobserved, and unaccounted variety of behavior. Particularly interesting observations are: (i) the existence of a RH N∗ phase for a number of oligomer sequences; (ii) the opposite N∗ handedness between DD and sDD, identical in their sequence pattern along the chains and only differing by the mode of end-to-end aggregation, BE vs. SD; (iii) the opposite handedness for two oligomers differing by only 4 nucleotides (ACC and AAT) and the existence of a nonchiral nematic phase for the intermediate sequence AAC; (iv) the occurrence of N∗ handedness inversion, from LH to RH, at increasing concentration in one of the sequences (allCG2); and (v) the opposite temperature dependence of the pitch for DD and pDD, only differing by a terminal phosphate group (see SI Text).
Analysis of the data reveals that the propagation of chirality from the nDNA helix to the N∗ phase depends on various independent factors. A scenario compatible with the data is that all factors here explored affect the chirality of the system mainly by determining the range of concentrations for the N∗ phase and that the mean interhelical distance is the single parameter controlling the handedness of the system.
Materials and Methods
Synthetic oligonucleotide sequences, purified by HPLC, were purchased from Primm s.r.l. as liophylized powders. Concentrated solutions (200–1000 mg/mL) were prepared in milliQ water. The solutions were loaded between parallel plate glass plates with gaps in the range 5–30 μm. In the cells aimed at quantitative analysis, the solution drop was sealed with a fluorinated oil and generally left at 60 °C for some hours to let the DNA concentration homogenize.
Microscopic observations were made either on a Nikon TE200 or on a Nikon Optiphot2 microscope. Images were acquired on a Nikon DS-5M camera. To measure the handedness of the N∗ phase, spectra were acquired with a S200 UV-VIS spectrometer (Ocean Optics) connected to the TE200 microscope. Further details are given in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank A. Ferrarini for discussions. Our work was supported by Cariplo Foundation Grant 2008-2413 (to G.Z., F.G., M.B., R.C., and T.B) and National Science Foundation (NSF) Grant DMR 0606528 and NSF Materials Research Science and Engineering Center Grant DMR 0820579 (to N.A.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011199107/-/DCSupplemental.
†This estimate follows from assuming the DNA helixes density ∼1800 mg/mL, as reported in literature.
References
- 1.Timsit Y, Várnai P. Helical chirality: A link between local interactions and global topology in DNA. PLoS One. 2010;5:e9326. doi: 10.1371/journal.pone.0009326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris AB, Kamien RD, Lubensky TC. Molecular chirality and chiral parameters. Rev Mod Phys. 1999;71:1745–1757. [Google Scholar]
- 3.Coates D, Gray GW. The synthesis of a cholesterogen with hydrogen-deuterium asymmetry. Mol Cryst Liq Cryst. 1973;24:163–177. [Google Scholar]
- 4.Cherstvy AG. DNA cholesteric phases: The role of DNA molecular chirality and DNA–DNA electrostatic interactions. J Phys Chem B. 2008;112:12585–12595. doi: 10.1021/jp801220p. [DOI] [PubMed] [Google Scholar]
- 5.Bonazzi S, et al. Four-stranded aggregates of oligodeoxyguanylates forming lyotropic liquid crystals: A study by circular cichroism, optical microscopy, and X-ray diffraction. J Am Chem Soc. 1991;113:5809–5816. [Google Scholar]
- 6.Livolant F, Maestre MF. Circular cichroism microscopy of compact forms of DNA and chromatin in vivo and in vitro: Cholesteric liquid-crystalline phases of DNA and single dinoflagellate nuclei. Biochemistry. 1988;27:3056–3068. doi: 10.1021/bi00408a058. [DOI] [PubMed] [Google Scholar]
- 7.Spada GP, Brigidi P, Gottarelli G. The determination of the handedness of cholesteric superhelices formed by DNA fragments. J Chem Soc Chem Commun. 1988:953–954. [Google Scholar]
- 8.Tombolato F, Ferrarini A, Grelet E. Chiral nematic phase of suspensions of rodlike viruses: Left-handed phase helicity from a right-handed molecular helix. Phys Rev Lett. 2006;96:258302. doi: 10.1103/PhysRevLett.96.258302. [DOI] [PubMed] [Google Scholar]
- 9.Tomar S, Green MM, Day LA. DNA–protein interactions as the source of large-length-scale chirality evident in the liquid crystal behavior of filamentous bacteriophages. J Am Chem Soc. 2007;129:3367–3375. doi: 10.1021/ja068498d. [DOI] [PubMed] [Google Scholar]
- 10.Barry E, Beller D, Dogic Z. A model liquid crystalline system based on rodlike viruses with variable chirality and persistence length. Soft Matter. 2009;5:2563–2570. [Google Scholar]
- 11.Straley JP. Theory of piezoelectricity in nematic liquid crystals, and of the cholesteric ordering. Phys Rev A. 1976;14:1835–1841. [Google Scholar]
- 12.Kornyshev AA, Lee DJ, Leikin S, Wynvenn A. Structure and interactions of biological helices. Rev Mod Phys. 2007;79:943–996. [Google Scholar]
- 13.Tombolato F, Ferrarini A. From the double-stranded helix to the chiral nematic phase of B-DNA: A molecular model. J Chem Phys. 2005;122:054908. doi: 10.1063/1.1839859. [DOI] [PubMed] [Google Scholar]
- 14.Kornyshev AA, Leikin S. Electrostatic interaction between long, rigid helical macromolecules at all interaxial angles. Phys Rev E. 2000;62:2576–2596. doi: 10.1103/physreve.62.2576. [DOI] [PubMed] [Google Scholar]
- 15.Stanley CB, Hong H, Strey HH. DNA cholesteric pitch as a function of density and ionic strength. Biophys J. 2005;89:2552–2557. doi: 10.1529/biophysj.105.064550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rutgers University. Atlas of Nucleic Acid Structures. Available at ndbserver.rutgers.edu/atlas.
- 17.Nakata M, et al. End-to-end stacking and liquid crystal condensation of 6– to 20–base pair DNA duplexes. Science. 2007;318:1276–1279. doi: 10.1126/science.1143826. [DOI] [PubMed] [Google Scholar]
- 18.Zanchetta G, Nakata M, Buscaglia M, Clark NA, Bellini T. Liquid crystal ordering of DNA and RNA oligomers with partially overlapping sequences. J Phys-Condens Mat. 2008;20:494214. [Google Scholar]
- 19.Chandrasekhar S. Liquid Crystals. 2nd Ed. Cambridge, UK: Cambridge University Press; 1992. [Google Scholar]
- 20.Makow DM. Peak reflectance and color gamut of superimposed left and right-handed cholesteric liquid crystals. Appl Opt. 1980;19:1274–1277. doi: 10.1364/AO.19.001274. [DOI] [PubMed] [Google Scholar]
- 21.Maddalena P, Arnone G, Abbate G, Marrucci L, Santamato E. Optically induced reorientation in nematics doped by chiral agents. Mol Cryst Liq Cryst. 1995;261:113–122. [Google Scholar]
- 22.Morii N, Kido G, Suzuki H, Nimori S, Morii H. Molecular chain orientation of DNA films induced by both the magnetic field and the interfacial effect. Biomacromolecules. 2004;5:2297–2307. doi: 10.1021/bm0496460. [DOI] [PubMed] [Google Scholar]
- 23.Livolant F, Leforestier A. Condensed phases of DNA: Structures and phase transitions. Prog Polym Sci. 1996;21:1115–1164. [Google Scholar]
- 24.Leforestier A, Livolant F. Supramolecular ordering of DNA in the cholesteric liquid crystalline phase: An ultrastructural study. Biophys J. 1993;65:56–72. doi: 10.1016/S0006-3495(93)81063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grelet E, Fraden S. What is the origin of chirality in the cholesteric phase of virus suspensions? Phys Rev Lett. 2003;90:198302. doi: 10.1103/PhysRevLett.90.198302. [DOI] [PubMed] [Google Scholar]
- 26.Berman HM. Crystal studies of B-DNA: The answers and the questions. Biopolymers. 1997;44:23–44. doi: 10.1002/(SICI)1097-0282(1997)44:1<23::AID-BIP3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 27.Packer MJ, Dauncey MP, Hunter CA. Sequence-dependent DNA structure: Dinucleotide conformational maps. J Mol Biol. 2000;295:71–83. doi: 10.1006/jmbi.1999.3236. [DOI] [PubMed] [Google Scholar]
- 28.Pérez A, Lankas F, Luque FJ, Orozco M. Towards a molecular dynamics consensus view of B-DNA flexibility. Nucleic Acids Res. 2008;36:2379–2394. doi: 10.1093/nar/gkn082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campos L, et al. Overview of the structure of all-AT oligonucleotides: Organization in helices and packing interactions. Biophys J. 2006;91:892–903. doi: 10.1529/biophysj.106.084210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hizver J, Rozenberg H, Frolow F, Rabinovich D, Shakked Z. DNA bending by an adenine–thymine tract and its role in gene regulation. Proc Natl Acad Sci USA. 2001;98:8490–8495. doi: 10.1073/pnas.151247298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zanchetta G, Nakata M, Bellini T, Clark NA. Physical polymerization and liquid crystallization of RNA oligomers. J Am Chem Soc. 2008;130:12864–12865. doi: 10.1021/ja804718c. [DOI] [PubMed] [Google Scholar]
- 32.Zanchetta G, Nakata M, Buscaglia M, Bellini T, Clark NA. Phase separation and liquid crystallization of complementary sequences in mixtures of nanoDNA oligomers. Proc Natl Acad Sci USA. 2008;105:1111–1117. doi: 10.1073/pnas.0711319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.