Abstract
Regulation of the storage of glycogen, one of the major energy reserves, is of utmost metabolic importance. In eukaryotes, this regulation is accomplished through glucose-6-phosphate levels and protein phosphorylation. Glycogen synthase homologs in bacteria and archaea lack regulation, while the eukaryotic enzymes are inhibited by protein kinase mediated phosphorylation and activated by protein phosphatases and glucose-6-phosphate binding. We determined the crystal structures corresponding to the basal activity state and glucose-6-phosphate activated state of yeast glycogen synthase-2. The enzyme is assembled into an unusual tetramer by an insertion unique to the eukaryotic enzymes, and this subunit interface is rearranged by the binding of glucose-6-phosphate, which frees the active site cleft and facilitates catalysis. Using both mutagenesis and intein-mediated phospho-peptide ligation experiments, we demonstrate that the enzyme’s response to glucose-6-phosphate is controlled by Arg583 and Arg587, while four additional arginine residues present within the same regulatory helix regulate the response to phosphorylation.
Keywords: allosteric activation, glycosyltransferase
Glycogen is a major energy repository in eukaryotes, and the biosynthetic pathway of glycogen synthesis is highly conserved from yeast to humans. The structure of glycogen is characterized by linear chains of glucose linked by α-1,4 glycosidic bonds and branch points occurring every 10 to 13 residues through introduction of α-1,6 linkages. In eukaryotes, glycogen synthase catalyzes the linear polymerization of glucose by transferring glucose residues from UDP-glucose to the 4’-hydroxyl end of a growing glycogen chain and is rate-limiting for synthesis under most circumstances (1).
As the first known intracellular target of insulin action (2), the regulation of glycogen synthase (GS) has been a subject of intense investigation for over 50 years. However, only within the past seven years has structural information on any GS enzyme become available. To date, structures for three members of the GS family have been determined—monomeric Escherichia coli enzyme (3), dimeric Agrobacterium enzyme (4), and trimeric Pyrococcus enzyme (5). GS enzymes are members of the GTB fold of glycosyl transferases, which are further divided into two families, GT3 and GT5, based on sequence identity and their responses to posttranslational regulation (6, 7). The eukaryotic GT3 family of enzymes is activated by glucose-6-phosphate, is inhibited by protein phosphorylation, and shares less than 15% pair-wise sequence identity to the GT5 family enzymes (1).
Similar to many higher eukaryotes, Saccharomyces cerevisiae has two distinct genes encoding glycogen synthases, GSY1 (708 residues*) and GSY2 (705 residues), of which GSY2 is nutritionally regulated and is the more important isoenzyme for glycogen accumulation (8). In higher eukaryotes the two genes (GYS1 and GYS2) encode distinct isozymes of 738 and 704 residues, respectively, that differ in their tissue expression patterns and the precise nature of signaling inputs, though the common themes of glucose-6-phosphate activation and inhibition by phosphorylation are retained (1). The action of Ser/Thr protein kinases on residues outside the conserved central catalytic core of the GT3 enzymes inhibits enzyme activity (1). In yeast Gsy2p, the sites of phosphorylation occur at positions 651, 655, and 668, while in mammalian muscle glycogen synthase (GYS1) the known sites of phosphoryation occur at positions 8, 11, 641, 645, 649, 653, 656, 698, and 710. The mammalian liver enzymes (GYS2) lack the phosphorylation sites at positions corresponding to 698 and 710. In all cases, inhibition can be overcome by the allosteric activator glucose-6-phosphate or reversed by the action of Ser/Thr phosphatases (1). Thus, the ratio of the activity of glycogen synthase in the absence to that in the presence of glucose-6-phosphate (the activity ratio) serves as an index of its activity state. A group of six conserved arginines were identified in the C-terminal region of the eukaryotic GS enzymes that mediate the enzymes’ sensitivity to glucose-6-phosphate and phosphorylation (9). In Gsy2p, simultaneous mutation of the first three arginines in a conserved cluster (Gsy2p-R580A/R581A/R583A or Gsy2p-580A3) created an enzyme with an intermediate activity level that could neither be activated nor inhibited (9). Simultaneous mutation of the second set of three arginines (Gsy2p-R587A/R589A/R592A or Gsy2p-587A3) created an enzyme that could be inhibited by phosphorylation but could not be activated by glucose-6-phosphate (9).
Based on prior work (10–12) and the kinetic studies of these arginine mutants, a three-state model for regulation of Gsy2p activity was proposed (9). The dephosphorylated enzyme exists in an intermediate, or basal, activity state that can be converted to a low activity state by phosphorylation. Binding of glucose-6-phosphate converts the enzyme to the high activity state. Since mutations of the conserved arginines interfere with both activation and inhibition of the enzyme, the arginines are believed to be intimately involved in the transition between the different activity states. Because the GT5 members lack these conserved arginines and are not subject to regulatory action, the available structural information does not provide insight into the mechanism by which the GT3 enzymes transition between different activity states.
We sought to investigate the structural properties that underlie the ability of eukaryotic GS to respond to regulatory control. Toward this goal, we determined the crystal structure of the GT3 family member yeast Gsy2p in its basal activity state and in its glucose-6-phosphate activated form. The binding of glucose-6-phosphate to Arg583 and Arg587 anchors the activator such that the interaction of the glucose moiety with residues 280–284 triggers an extensive rotation and translation of the subunits, which ultimately frees the active site cleft from structural constraints and affords better substrate access. In addition, we combined a new series of mutational studies within the arginine cluster of Gsy2p, as well as intein-mediated phospho-peptide ligation (13) studies, to delineate the regulatory roles of the arginine cluster in mediating the responses to covalent phosphorylation and glucose-6-phosphate activation. These studies demonstrate that the regulatory arginines, through their interactions with the phosphate of glucose-6-phosphate or phosphorylated Thr668, act as sensors which directly control the transitions between conformational states in Gsy2p.
Results and Discussion
Overall Structure and Oligomeric Arrangement.
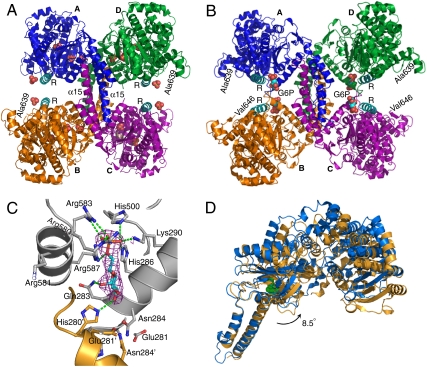
The structures for the basal state and glucose-6-phosphate activated state of Gsy2p have been solved to 3.0 Å and 2.4 Å resolution, respectively, in two distinct crystal forms (Table 1). In all cases, the subunits lack interpretable electron density C-terminal to residue 646, and both activity states of yeast glycogen synthase are tetramers (Fig. 1). The binding of glucose-6-phosphate induces extensive translations and rotations among the subunits that alter the nature of the subunit interface (Fig. 1B). The Gsy2p monomer folds into two structural domains, each dominated by its individual Rossmann folds (Figs. S1 and S2). In addition to the canonical GTB fold, the N-terminal Rossmann domain has two inserts, one following strand β2 and the other following strand β7. The C-terminal Rossmann domain has a unique insert found in all eukaryotic synthases following strand β11 (Figs. S2 and S3). The major structural feature of the C-terminal domain insert is a pair of long helices (α15–16) that extend away from the Rossmann fold, forming the majority of the intersubunit interface in both activity states (Fig. 1). The cluster of conserved arginine residues present in eukaryotic glycogen synthases is located in the first of the two interdomain helices, which we will refer to as the regulatory or R helix (α22, Figs. S1 and S2), in light of the regulation conferred by these arginine residues. The yeast enzyme is representative of all eukaryotic enzymes because, aside from the regulatory N- and C-terminal phosphorylation sequences, only the lengths of two loops connecting conserved elements of secondary structure differ across species (Figs. S2 and S3).
Table 1.
Data collection, phasing, and refinement statistics
| Basal | TaBr1 | TaBr2 | UDP Complex | G-6-P Activated | |
| Data Collection | |||||
| Space group | P21 | P21 | P21 | P21 | I222 |
| Cell dimensions | |||||
| a, b, c (Å) | 96.5, 166.1, 121.0 | 97.3, 167.6, 122.2 | 96.5, 167.3, 121.6 | 96.6, 167.2, 121.2 | 192.7, 206.9, 205.8 |
| α, β, γ (°) | 90.0, 103.4, 90.0 | 90.0, 103.3, 90.0 | 90.0, 103.1, 90.0 | 90.0, 102.7, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50-3.0 (3.1-3.0) | 50.0-4.5 (4.6-4.5) | 50.0-4.5 (4.6-4.5) | 50.0-3.5 (3.57-3.5) | 50.0-2.4 (2.44-2.40) |
| Rmerge (%) | 8.2 (47.5) | 10.9 (46.9) | 12.5 (37.8) | 11.0 (44) | 8.7 (55) |
| I/σI | 13.5 (2.1) | 14.7 (1.7) | 7.5 (1.9) | 9.9 (2.2) | 13.9 (2.0) |
| Completeness (%) | 99.6 (96.8) | 92.8 (94.7) | 82.7 (64.1) | 99.0 (92.7) | 98.5 (79) |
| Redundancy | 3.7 (2.6) | 2.9 (2.6) | 3.3 (3.4) | 3.6 (2.7) | 4.8 (3.1) |
| Refinement | |||||
| Resolution (Å) | 50.0-3.0 | 50.0-3.5 | 50.0-2.4 | ||
| No. reflections | 70557 | 46330 | 148274 | ||
| Rwork/Rfree (%) | 20.3/25.4 | 21.9/24.9 | 20.3/24.3 | ||
| No. of atoms | |||||
| Protein | 19770 | 19692 | 20818 | ||
| Ligand/ion | 80 | 155 | 124 | ||
| Water | 580 | ||||
| B-factors | |||||
| Protein | 64.37 | 104.4 | 57.41 | ||
| Ligand/ion | 90.05 | 127.7 | 47.11 | ||
| Water | 48.63 | ||||
| R.m.s deviations | |||||
| Bond lengths (Å) | 0.008 | 0.009 | 0.009 | ||
| Bond angles (°) | 1.07 | 1.11 | 1.15 |
Fig. 1.
Structures of the basal and activated states of Gsy2p. (A) Ribbon diagram of the basal state conformation in which the individual subunits are labeled (A–D) and colored separately. The regulatory helices (α22) are colored cyan and labeled R, while the intersubunit helices are labeled α15 as is the last ordered residue at the C terminus of subunits B and D. Ordered sulfate ions in this structure are represented using space filling atoms. (B) Ribbon diagram of the activated state of Gsy2p. The color scheme, subunit, and regulatory helix labeling is identical to panel A. The bound glucose-6-phosphate molecules at the interface are represented using space filling atoms and labeled G6P. (C) Glucose-6-phosphate binding site in Gsy2p. Glucose-6-phosphate and the surrounding amino acid residues are represented using ball-and-stick models and displayed using atom type coloring. Elements from distinct subunits are labeled and denoted with separate ribbon and carbon atom coloring. The electron density map displayed is the original 2Fo-Fc map (contoured at 1 standard deviation) for glucose-6-phosphate prior to its inclusion in the structural model. The local hydrogen bonding pattern is represented using dashed green lines. (D) Ribbon representation of the superposed basal state and activated state monomers. The glucose-6-phosphate molecule bound to the activated state structure (blue) is represented using green space filling atoms. The alignment was generated by superposing helices α15 and α16 (residues 365–399 and 416–434) using the program superpose in the CCP4 suite (21). [Produced using Pymol (23) for Windows.]
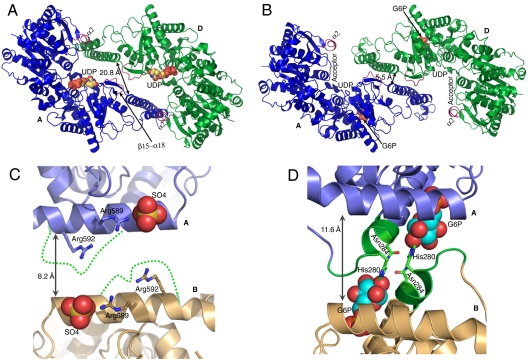
While it is not clear what catalytic advantage is conferred by oligomerization of the GT5 enzymes, the oligomeric assembly of Gsy2p, and GT3 members in general, provides a means for integrating allosteric ligand binding and covalent phosphorylation in the regulation of activity. Consistent with this hypothesis, the most striking feature of our structural analyses of Gsy2p is the rearrangement of the subunits upon glucose-6-phosphate binding (Figs. 1 and 2). In both structures, the primary subunit interface is formed by a coiled-coil helical arrangement of helices α15 and α16 (residues 365–431). A 12-residue loop between these two helices (residues 401–412) is disordered in the basal state but becomes ordered as the interface changes in response to glucose-6-phosphate binding. In addition to an 8.5 ° rotation of the C- and N-terminal Rossmann domains away from the subunit interface (Fig. 1D), there is also a rotation and translation of the individual subunits toward the center of the tetrameric interface. This motion can be described as a 14.2 Å translation and 7 ° rotation of the B/D dimer pair toward the A/C dimer pair when the helical domains of subunits A/C are used to align the two conformational states (Figs. 1 A and B and 2 A and B). In particular, when viewed from the A/D subunit interface, residues 482–487 (loop β15–α18) lie across the molecular two-fold from each other in the basal state but form new interactions with helix α15 in the activated state (Fig. 2 A and B).
Fig. 2.
Comparison of the basal and activated state conformations. (A) Ribbon representation of the AD dimer pair in the basal state with bound UDP represented using space filling atoms and ribbon coloring identical to Fig. 1A. The loop region β15-α18 and helix α2 are highlighted in purple and labeled. The distance between equivalent positions of the α15 helices is provided. (B) Ribbon representation of the AD dimer pair in the activated state using the same coloring scheme as in panel A. The glucose-6-phosphate molecules are represented using space filling atoms and labeled, as are the relative positions of UDP and structural elements contributing to glycogen acceptor binding. (C) Ribbon representation of the regulatory interface between subunits A and B in the basal state. The individual subunits are indicated with labels and distinct ribbon colors. The bound sulfate molecule near the N terminus of each regulatory helix is displayed using space filling atoms. The disordered region between residues 277–285 is represented as a dashed green line solely to indicate the connectivity. (D) Ribbon representation of the regulatory helix interface between subunits A and B in the activated state of the R589A/R592A mutant. The coloring scheme is identical to that in panel C and the bound glucose-6-phosphate is displayed using space filling atoms. The residues interacting with glucose-6-phosphate from the 277–285 loop region are represented using green coloring. [Produced using Pymol (23) for Windows.]
Glucose-6-Phosphate Binding.
As might be expected for an allosteric ligand that induces such large conformational changes, the interactions between the enzyme and glucose-6-phosphate are extensive and involve residues from more than one subunit. Glucose-6-phosphate binds in a surface pocket adjacent to the N terminus of the regulatory helix (Figs. 1C and 2D). The 6-phosphate is completely sequestered in a binding pocket comprising five residues (His286, Lys290, His500, Arg583, and Arg587) that are conserved across eukaryotes. The last two residues are the third and fourth arginine residues in the regulatory arginine cluster.
The C1’ and C2’ hydroxyl groups of the glucose moiety form hydrogen bonds to Gln283 and His280, respectively. The C2’ hydroxyl is also an average of 3.7 Å from the side chain of Asn284, which together with His280 form critical intersubunit contacts at the regulatory interface (Figs. 1C and 2D). Unlike the phosphate-binding site, the glucose binding site is poorly formed prior to binding. In particular, residues 278 to 284 are disordered in the basal state and adopt a stable conformation only after the activator is bound (Fig. 2 C and D). Thus, most of the conformational changes are a consequence of interactions contributed by this loop within its own subunit and across the regulatory interface. In this context, the phosphoryl group serves as the anchor upon which to build the necessary interactions to drive the conformational changes.
Based on the changes in local structure, it would appear that the trigger for the conformational changes that underlie activation is the interaction contributed by His280, Gln283, and Asn284, the last of which hydrogen bonds with Asn284 from the adjacent subunit. Although residues His286 and Lys290 also contribute hydrogen bonds to the bound glucose-6-phosphate molecule, their relative positions change very little upon glucose-6-phosphate binding. Rather, the ordering of the loop between 278 and 284 and drawing of this loop into interactions across the regulatory interface stabilize the activated conformation by inducing rigid body rotations and translations in order to achieve optimal interactions for His280, Gln283, and Asn284.
Conformational Changes and Enzyme Activation.
The conformational changes induced by the binding of glucose-6-phosphate are initiated by the interactions at the A/B and C/D subunit interfaces (Fig. 2D), but the global effects on enzyme activity are best illustrated by examining the A/D or B/C subunit interfaces since these interfaces contain the nucleotide-donor and acceptor substrate binding sites (Fig. 2 A and B). We gained insight into the structural basis for substrate binding by solving the basal state structure of Gsy2p in a complex with UDP (Table 1 and Fig. S4). Similar to the bacterial and archeal enzymes (3–5), the nucleotide sugar donor binds to the C-terminal Rossmann domain (Fig. 2A) and the glycogen acceptor substrate is thought to bind along the surface of the N-terminal Rossmann domain (3, 14) (Fig. 2B). Both kinetic and structural data show that glucose-6-phosphate has only minor effects on UDP-glucose utilization as UDP binds to our basal state structure, and the Km for UDP-glucose in the nonphosphorylated enzyme is unaffected by the presence of glucose-6-phosphate (9). In contrast, the Km for glycogen is reduced by 50% upon glucose-6-phosphate activation (9). The glucose-6-phosphate-induced reorganization of the subunit interfaces likely leads to increased catalytic efficiency because acceptor substrate access and domain closure is now unimpeded by constraining interactions between the N-terminal Rossmann domains at the periphery of the tetramer and the α16 helices (Fig. 2 A and B). Structural studies with glycogen synthase from E. coli have demonstrated that the transition between the resting open and catalytically poised closed conformations of the enzyme involves a 15° interdomain rotation (3). The relative domain orientation of Gsy2p in our basal state structure is most similar to the open-form of the E. coli enzyme and domain closure similar to the E. coli enzyme is hindered by the interactions between helix α2 and strand β3 with helix α16 (Fig. 2A). The extensive rotation and translation of the subunits induced by activator binding releases these constraints such that the enzyme can now more easily adopt the opened and closed domain conformations required for efficient donor and acceptor binding and release. Consistent with this hypothesis, when compared to the basal state structure, the N-terminal domains of the A, C, and D subunits are rotated an additional 4.6° away from the interface in the activated structure (Fig. S5A). Interestingly, the B subunit is 12.8° more closed than in the basal state structure. This domain closure appears to be precipitated by binding of an additional glucose-6-phosphate molecule within the active site that triggers the domain closure (Fig. S5B). No such variation in domain orientation was observed in the basal state structure which is consistent with our contention that the subunit positioning in the activated state facilitates catalytically important conformational changes.
Roles of the regulatory arginines in activation and inhibition.
To delineate the role(s) of the conserved arginines in mediating glucose-6-phosphate activation and inhibition by phosphorylation, we combined site-directed mutagenesis and expressed protein ligation experiments. Consistent with their interactions with the 6-phosphate of glucose-6-phosphate in our structure, detailed mutagenesis of the regulatory arginines demonstrates that the presence of Arg583 and Arg587 are necessary and sufficient for conferring glucose-6-activation (Table 2). However, mutations of the first two or last two arginines had disparate effects on the enzyme’s activity state. Mutation of Arg580 and Arg581 had little effect on basal activity levels or activation by glucose-6-phosphate but clearly impacted the response to phosphorylation (Table 2). In contrast, the R589A/R592A double mutant, whose glucose-6-phosphate activated structure is reported here, exhibited a specific activity and activity ratio similar to the phosphorylated enzymes and could be completely activated by the presence of glucose-6-phosphate (Table 2). Thus, the two N-terminal and two C-terminal arginine residues in this cluster establish the conformational set points from which the effects of phosphorylation or glucose-6-phosphate activation are further manifested.
Table 2.
Specific activity and activity ratio of Yeast Gsy2p
| Synthase Specific activity (mmol· min-1) | Activity Ratio | ||
| Enzyme | No G6P | 7.2 mM G6P | −G6P/+G6P |
| Wild type | 1.03 ± 0.03 | 1.73 ± 0.03 | 0.60 ± 0.01 |
| R580A | 0.99 ± 0.01 | 1.55 ± 0.03 | 0.64 ± 0.01 |
| R580/581A2 | 1.07 ± 0.05 | 1.59 ± 0.05 | 0.67 ± 0.02 |
| R580/581/583A3 | 0.55 ± 0.02 | 0.51 ± 0.03 | 1.09 ± 0.02 |
| R587/589/592A3 | 0.66 ± 0.01 | 0.58 ± 0.02 | 1.14 ± 0.05 |
| R589/592A2 | 0.17 ± 0.01 | 1.56 ± 0.07 | 0.11 ± 0.01 |
| Wild type 640 truncated | 1.25 ± 0.01 | 1.95 ± 0.05 | 0.64 ± 0.01 |
| Wild type 35mer Nonphospho-peptide ligated | 1.12 ± 0.02 | 1.77 ± 0.04 | 0.63 ± 0.01 |
| Wild type 35mer Phospho-peptide ligated | 0.27 ± 0.01 | 1.96 ± 0.05 | 0.14 ± 0.01 |
| R580/581/583A3 35mer Phospho-peptide ligated | 0.44 ± 0.01 | 0.44 ± 0.02 | 1.05 ± 0.01 |
| Wild type 49mer Nonphospho-peptide ligated | 1.08 ± 0.02 | 1.78 ± 0.02 | 0.61 ± 0.02 |
| Wild type 49mer Phospho-peptide ligated | 0.10 ± 0.01 | 1.92 ± 0.04 | 0.05 ± 0.01 |
| R580A 49mer Phospho-peptide ligated | 0.34 ± 0.01 | 1.74 ± 0.10 | 0.20 ± 0.02 |
| R580/581A2 49mer Phospho-peptide ligated | 0.58 ± 0.01 | 1.50 ± 0.01 | 0.39 ± 0.01 |
| R580/581/583A3 49mer Phospho-peptide ligated | 0.43 ± 0.02 | 0.40 ± 0.01 | 1.04 ± 0.04 |
Prior work showed that phosphorylation of Thr668 had the greatest impact on activity (10). Consequently, the effects of phosphorylation of this residue on enzyme activity was examined by fusing C-terminal phosphopeptides to a recombinant Gsy2p core, either with native sequence for residues 1–641 or with mutations within the regulatory helix. The recombinant wild-type enzyme, the 641 residue construct and the control, nonphosphorylated peptide-ligated enzymes exhibit similar activities and activity ratios (Table 2). In contrast, introduction of a single phosphothreonine at position 668 in the peptide-ligated construct inhibits the enzyme in a manner comparable to the in vitro phosphorylated wild-type enzyme (10). Ligation of the 35-residue phospho-peptide to the wild-type sequence produces an enzyme truncated at position 676 with basal activity decreased by 70% and an activity ratio about fourfold lower (Table 2). Ligation of the 49-residue phospho-peptide produces an enzyme truncated at residue 690 whose basal activity is decreased by 90% with an activity ratio similar to the wild-type enzyme phosphorylated by Pho85p/Pcl10p (9).
Our group had previously demonstrated that the Gsy2p-580A3 triple mutant enzyme is resistant to inhibition by Pho85p/Pcl10p phosphorylation whereas the Gsy2p-587A3 mutant showed moderate inhibition (9), suggesting that response to phosphorylation is mediated by residues 580 and 581. Here we show that the activity ratios of the peptide-ligated constructs were inversely correlated with the number of arginine residues in this first cluster, with the 580/581A2 mutant having the highest activity ratio and the wild-type sequence the lowest activity ratio (Table 2). Despite the fact that Arg580 forms an interaction with the 6-phosphate of glucose-6-phosphate (Fig. 1C), it would not appear to be directly competitive with phosphorylated residue binding, because mutation of 580 or 581 affected only inhibition by phosphorylation and not the ability to be activated by glucose-6-phosphate. Thus, residues 580 and 581 appear critical for stabilizing the inhibited state. We propose that when Thr668 is phosphorylated, Arg580, and Arg581 play a role in stabilizing the inhibited conformation by directly interacting with the phosphate group at or near the N terminus of the regulatory helix, possibly near one of the sulfate binding sites in our basal state structure (Fig. 2C). Even though residues 580/581/583 were mutated to alanine in this structure, the sulfate is positioned near the N terminus of the regulatory helix, residue 581, and the side chain of Arg589.
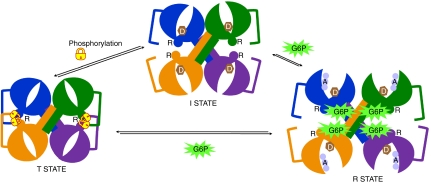
In contrast to the mutations at positions 580 and 581, which did not affect the basal state activity, mutation of Arg589 and Arg592 dramatically lowered the activity of the enzyme in the absence of glucose-6-phosphate, but this mutant could be fully activated to wild-type levels with glucose-6-phosphate. Consequently, the activity ratio of this mutant resembled that of the phosphorylated forms (Table 2). We hypothesize that the role of these last two arginine residues is to prevent the collapse of the regulatory helices toward the interface and that charge neutralization—either through mutation to alanine or through interactions with phosphorylated residues—results in an inhibited conformation, which can be transformed upon glucose-6-phosphate binding. Thus, rather than directly triggering the conformational changes upon activator binding, the roles of these six arginine residues are to provide anchoring interactions for phosphate groups—583 and 587 toward the 6-phosphate of glucose-6-phosphate in the activated R-state and the remaining residues toward establishing and stabilizing the I-state and inhibited T-state (Fig. 3).
Fig. 3.
Schematic representation of the conformational states underlying regulation of Gsy2p activity. The individual subunits are colored according to the coloring scheme in Fig. 1. The regulatory helices containing the arginines are labeled R and the approximate positions of nucleotide-donor sugar (D) and glycogen acceptor (A) are shown for the I- and R-states, respectively. Phosphorylation of Thr668 is shown as “locking” the enzyme in the T-state conformation through intersubunit interactions across the regulatory interface. Glucose-6-phosphate binding frees these constraining interactions to fully activate the enzyme in the R-state conformation.
It is clear that either an intra- or intersubunit interaction with phospho-Thr668 would fulfill the requirement for charge neutralization of the regulatory arginines. However, an intersubunit interaction would add an additional conformational constraint in the form of a locking strap running from the N-terminal domain of the opposing subunit, where the electron density terminates in the basal state structure at residue 639 approximately 25–30 Å from the regulatory interface (Figs. 1A and 3), across the interface to the regulatory helix located on the opposing C-terminal domain. Indeed the additional residues observed at the C terminus of the glucose-6-phosphate activated structure of the R589A/R592A mutant are directed precisely toward this regulatory interface (Fig. 1B). The basal activity of the R589A/R592A double mutant supports an intermolecular locking mechanism as its lower activity ratio does not fully approach the activity ratio of the phosphorylated wild-type enzyme and an intermolecular mechanism is also supported by the stronger inhibition of the longer phospho-peptide construct (Table 2). Binding of glucose-6-phosphate presumably disrupts these intersubunit locking interactions by changing the conformational relationships surrounding the regulatory interface and throughout the tetramer (Fig. 3).
In summary, the structure of yeast Gsy2p establishes the paradigm for regulatory control of the eukaryotic glycogen synthases. In particular, the unusual tetrameric interface and its reorganization upon activator binding provide a molecular basis for the enzyme to exist in distinct activity states (Fig. 3). Our new mutational and intein-mediated phosphoenzyme data support the idea that the six conserved arginines in the regulatory helix act as a finely tuned sensor and response interface, with each residue contributing either positively or negatively toward each of the possible conformational states. The basal I-state likely represents a dynamic ensemble of conformations between the T- and R-states but based on kinetic data appears to be closer to the activated state, with phosphorylation or glucose-6-phosphate binding altering the equilibrium toward each of these two end states. The binding of glucose-6-phosphate triggers the conformational transition to a more open active site cleft permitting easier substrate access and eliminates the constraints to domain motion during catalysis. The triggering event in this transition is the recruitment of the disordered interdomain connecting loop (residues 280–284) into forming stable interactions within the glucose binding site of the neighboring subunit. These interactions are made possible by the anchoring of the phosphate group through extensive interactions with basic residues near the N terminus of the regulatory helix (Fig. 3). Phosphorylation of the intermediate state locks the enzyme in the T-state through the interaction of Arg580 and 581, as well as possibly Arg589 and 592, with the phosphorylated residue(s). The additional phosphorylation sites present in the higher eukaryotic enzymes provide additional points of contact with the regulatory interface, permitting potentially more complex regulation of activity that can integrate multiple signaling inputs to different phosphorylations. The molecular insight provided by these structures will permit the design of unique activator molecules that could bypass cellular signaling mechanisms and constraints on cellular glucose-6-phosphate concentrations to reactivate glycogen synthase under conditions where it is inappropriately inhibited by phosphorylation, such as in diabetes.
Methods
Expression and Purification and Activity Measurement of Gsy2p.
His-tagged Gsy2 proteins were expressed in E.coli BL21(DE3) cells and purified by a two step procedure that included affinity chromatography on Ni2+-nitrilotriacetic acid-agarose (9) and ion exchange purification on Q-sepharose. Gsy2p bound to the Q-sepharose column was eluted with a linear gradient of 0–1 M NaCl in a buffer containing 50 mM Tris-HCl, pH 8.0 and 1 mM 2-mercaptoethanol. Eluted fractions containing Gsy2p were pooled, dialyzed against 20 mM Tris-HCl pH 8.0 and 1 mM 2-mercaptoethanol. The PCR-based site-directed mutagenesis approach was used to generate mutants of Gsy2p.
The Gsy2p cDNA fragment encoding amino acids 1-641 was subcloned into the NdeI and SapI sites of pTXB1 vector (13, 15), and expression was driven in the E.coli ER2566 strain. Details on the production and purification of the peptide-ligated Gsy2p constructs, including the peptide design, can be found in SI Text.
GS activity was determined by the method of Thomas et al. (16) with reaction at 30 °C for 15 min with 6.7 mg/mL glycogen, 4.4 mM UDP-glucose and in the absence or presence of 7.2 mM glucose-6-phosphate.
Crystal Growth and Data Collection.
Crystals of the R580/581/583A3 Gsy2p were grown by hanging drop vapor diffusion after combining 2 μL of protein at 3 mg/mL with 2 μL of reservoir solution containing 100 mM Tris-HCl, pH 8.0-8.5, 200 mM Li2SO4 and 18–22% PEG 3400 and were cryoprotected through slow introduction of a cryogenic solution containing 20% glycerol in the mother liquor. Heavy atom derivative complexes were prepared by soaking the crystals in mother liquor containing 1 mM Ta6Br12 for 6–12 h. The UDP complex was prepared by soaking the crystals in mother liquor containing 20 mM UDP for 10 h. The R589/592A2 mutant was cocrystallized with 25 mM glucose-6-phosphate by combining 2 μL of protein at 3 mg/mL with 2 μL of reservoir solution containing 100 mM Bis-Tris, pH 6.2-6.5 and 22-25% PEG 300 in the hanging drop geometry and cryoprotected by coating the crystals with immersion oil.
The basal activity dataset and the tantalum derivative datasets were collected at beamline 19-ID operated by the Structural Biology Center Collaborative Access Team at the Argonne National Laboratory. Fluorescence scans were performed on the tantalum derivatives and data collection was initiated at the peak wavelength for these derivatives, 1.25 Å. The basal state UDP complex and the activated state datasets were collected at beamline 23-ID operated by the General Medicine and Cancer Institutes Collaborative Access Team. The data collected were indexed, integrated, and scaled using the HKL2000 program suite.
Structure Solution, Model Building and Refinement.
The basal state diffraction data were phased by multiple isomorphous replacement using the diffraction data from the tantalum derivatives between 25 and 5.5 Å, followed by molecular averaging and density modification to extend the phases to 3.0 Å. Methods used to phase and solve the basal activity structure can be found in supporting information. Refinement of the model to 3.0 Å was performed with PHENIX (17, 18) with visual map inspection and manual model modification performed using COOT (19). The activated state was solved by molecular replacement using the program package AMoRE (20) as implemented in CCP4 (21) and the basal state monomer structure as the search model and refined in PHENIX to 2.4 Å. All refinement protocols utilized NCS restraints and either tightly restrained individual temperature factor refinement (basal state and G-6-P activated state) or single refined group isotropic temperature factor for each subunit or ligand (UDP complex). All final models displayed good stereochemistry as analyzed by the program PROCHECK (22).
Supplementary Material
Acknowledgments.
We wish to thank Drs. Millie M. Georgiadis and Samantha Perez-Miller for valuable guidance and discussions, Dr. Lou Messerle for providing the hexatantalum tetradecabromide compound, and Dr. Thomas C. Terwilliger for help with the phasing scripts for PHENIX. We wish to acknowledge the assistance of the staff at the 19-ID beamline facility, Marianne Cuff, Norma Duke, and Steven Ginell. Results shown in this report are derived from the work performed at the Structural Biology Center and the General Medicine and Cancer Institutes Collaborative Access Team at the Advanced Photon Source located at the Argonne National Laboratory. Argonne is operated by U Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357. This work is supported by National Institutes of Health Grant R37-DK027221.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank, http://www.rcsb.org/pdb/ (RCSB ID codes 3naz, 3nch, 3nb0, and 3o3c).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006340107/-/DCSupplemental.
*This manuscript will utilize the new open reading frame numbering scheme in which residue numbering begins at the initiator methionine. Consequently, all residue numbers in this publication are incremented by 1 when compared to prior literature on glycogen synthase isoenzymes.
References
- 1.Roach PJ. Glycogen and its metabolism. Curr Mol Med. 2002;2(2):101–120. doi: 10.2174/1566524024605761. [DOI] [PubMed] [Google Scholar]
- 2.Villar-Palasi C, Larner J. Insulin-mediated effect on the activity of UDPG-glycogen transglucosylase of muscle. Biochim Biophys Acta. 1960;39:171–173. doi: 10.1016/0006-3002(60)90142-6. [DOI] [PubMed] [Google Scholar]
- 3.Sheng F, Jia X, Yep A, Preiss J, Geiger JH. The crystal structures of the open and catalytically competent closed conformation of Escherichia coli glycogen synthase. J Biol Chem. 2009;284(26):17796–17807. doi: 10.1074/jbc.M809804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buschiazzo A, et al. Crystal structure of glycogen synthase: Homologous enzymes catalyze glycogen synthesis and degradation. EMBO J. 2004;23(16):3196–3205. doi: 10.1038/sj.emboj.7600324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horcajada C, Guinovart JJ, Fita I, Ferrer JC. Crystal structure of an archaeal glycogen synthase: Insights into oligomerization and substrate binding of eukaryotic glycogen synthases. J Biol Chem. 2006;281(5):2923–2931. doi: 10.1074/jbc.M507394200. [DOI] [PubMed] [Google Scholar]
- 6.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328(2):307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 7.Cid E, Geremia RA, Guinovart JJ, Ferrer JC. Glycogen synthase: Towards a minimum catalytic unit? FEBS Lett. 2002;528(1-3):5–11. doi: 10.1016/s0014-5793(02)03313-6. [DOI] [PubMed] [Google Scholar]
- 8.Farkas I, Hardy TA, Goebl MG, Roach PJ. Two glycogen synthase isoforms in Saccharomyces cerevisiae are coded by distinct genes that are differentially controlled. J Biol Chem. 1991;266(24):15602–15607. [PubMed] [Google Scholar]
- 9.Pederson BA, Cheng C, Wilson WA, Roach PJ. Regulation of glycogen synthase. Identification of residues involved in regulation by the allosteric ligand glucose-6-P and by phosphorylation. J Biol Chem. 2000;275(36):27753–27761. doi: 10.1074/jbc.M003342200. [DOI] [PubMed] [Google Scholar]
- 10.Hardy TA, Roach PJ. Control of yeast glycogen synthase-2 by COOH-terminal phosphorylation. J Biol Chem. 1993;268(32):23799–23805. [PubMed] [Google Scholar]
- 11.Francois J, Villanueva ME, Hers HG. The control of glycogen metabolism in yeast. 1. Interconversion in vivo of glycogen synthase and glycogen phosphorylase induced by glucose, a nitrogen source or uncouplers. Eur J Biochem. 1988;174(3):551–559. doi: 10.1111/j.1432-1033.1988.tb14134.x. [DOI] [PubMed] [Google Scholar]
- 12.Rothman-Denes LB, Cabib E. Glucose 6-phosphate dependent and independent forms of yeast glycogen synthetase. Their properties and interconversions. Biochemistry. 1971;10(7):1236–1242. doi: 10.1021/bi00783a021. [DOI] [PubMed] [Google Scholar]
- 13.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: A general method for protein engineering. Proc Natl Acad Sci USA. 1998;95(12):6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheng F, Yep A, Feng L, Preiss J, Geiger JH. Oligosaccharide binding in Escherichia coli glycogen synthase. Biochemistry. 2009;48(42):10089–10097. doi: 10.1021/bi900916t. [DOI] [PubMed] [Google Scholar]
- 15.David R, Richter MP, Beck-Sickinger AG. Expressed protein ligation. Method and applications. Eur J Biochem. 2004;271(4):663–677. doi: 10.1111/j.1432-1033.2004.03978.x. [DOI] [PubMed] [Google Scholar]
- 16.Thomas JA, Schlender KK, Larner J. A rapid filter paper assay for UDPglucose-glycogen glucosyltransferase, including an improved biosynthesis of UDP-14C-glucose. Anal Biochem. 1968;25(1):486–499. doi: 10.1016/0003-2697(68)90127-9. [DOI] [PubMed] [Google Scholar]
- 17.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 18.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D. 2008;64(Pt 1):61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 20.Navaza J. A MoRe: An automated package for molecular replacement. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 21.Anonymous. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: A unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D. 1999;55(Pt 1):191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 23.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.