Abstract
Disrupted-in-schizophrenia 1 (DISC1) has emerged as a schizophrenia-susceptibility gene affecting various neuronal functions. In this study, we characterized Mitofilin, a mitochondrial inner membrane protein, as a mediator of the mitochondrial function of DISC1. A fraction of DISC1 was localized to the inside of mitochondria and directly interacts with Mitofilin. A reduction in DISC1 function induced mitochondrial dysfunction, evidenced by decreased mitochondrial NADH dehydrogenase activities, reduced cellular ATP contents, and perturbed mitochondrial Ca2+ dynamics. In addition, deficiencies in DISC1 and Mitofilin induced a reduction in mitochondrial monoamine oxidase-A activity. The mitochondrial dysfunctions evoked by the deficiency of DISC1 were partially phenocopied by an overexpression of truncated DISC1 that is associated with schizophrenia in human. DISC1 deficiencies induced the ubiquitination of Mitofilin, suggesting that DISC1 is critical for the stability of Mitofilin. Finally, the mitochondrial dysfunction induced by DISC1 deficiency was partially reversed by coexpression of Mitofilin, confirming a functional link between DISC1 and Mitofilin for the normal mitochondrial function. According to these results, we propose that DISC1 plays essential roles for mitochondrial function in collaboration with a mitochondrial interacting partner, Mitofilin.
Keywords: IMMT, mitochondrial dysfunctions, hyperdopaminergia, calcium buffering, psychiatric disorders
Schizophrenia is a mental illness characterized by impairments in an individual's perceptions of reality, which are commonly manifested in the form of hallucinations, paranoid or bizarre delusions, and/or disorganized speech and thinking (1). The lifetime prevalence of schizophrenia is 0.55–1% of population, with up to 80% of genetic inheritance (2). Although the molecular mechanisms underlying the etiology and pathophysiology of schizophrenia have yet to be clearly defined, increased dopaminergic activities in the mesolimbic pathway have been consistently associated with several of the positive symptoms of schizophrenia (3).
Disrupted-in-schizophrenia 1 (DISC1) has emerged as a convincing candidate gene that may explain many aspects of schizophrenia and related mood disorders. Familial mutations in the DISC1 gene, including t(1;11)(q42.1;q14.3) translocation, cosegregate with symptoms related to schizophrenia, bipolar disorders, and recurrent major depression (4). DISC1 plays a significant role in the processes of neurodevelopment, including neurite outgrowth, neuronal migration, neurogenesis (5–8), intracellular cAMP signaling (9), and many other neuronal processes in collaboration with other cellular factors such as nudE nuclear distribution gene E homolog (A. nidulans)-like 1 (NDEL1) (6). Additionally, genetic evidence supporting a functional link between DISC1 and NDEL1 in schizophrenia pathogenesis has been reported (10).
Mitochondria play critical roles in fundamental cellular processes, encompassing energy production, programmed cell death, Ca2+ homeostasis, and monoamine metabolism. Intriguingly, mitochondrial dysfunction has been found to be associated with various psychiatric disorders, including schizophrenia (11). For example, Maurer et al. (12) reported a deficit in the process of oxidative phosphorylation in the postmortem brains of patients with schizophrenia. Deformations and reductions in the number of mitochondria have also been reported in postmortem studies (13). Although these observations are critical insights into the link between schizophrenia and mitochondrial dysfunctions, the underlying molecular mechanisms explaining this relationship have not been well defined.
In this study, we characterized Mitofilin, also called inner-membrane protein, mitochondrial (IMMT) (14), as a mitochondrial interacting partner of DISC1, through which DISC1 plays essential roles in mitochondria. Mitofilin has been well-characterized as an essential component for the functional integrity of mitochondria (15, 16); thus, the data shown in this study may provide a mechanistic link between DISC1 and mitochondrial function, the deregulation of which might have implications in the pathogenesis of schizophrenia.
Results
Mitofilin as an Interacting Partner of DISC1.
To elucidate DISC1 functions potentially relevant to schizophrenia, we attempted to identify components of the DISC1–NDEL1-containing protein complex by using a yeast two-hybrid screen on a human fetal brain library using NDEL1 as bait. Then, we tested whether any of the positives from this screening could interact with DISC1. As a result, we identified a cDNA clone encoding amino acid residues, 230–758, of Mitofilin, a mitochondrial inner membrane protein. We noticed that the potential interaction between DISC1 and Mitofilin has been consistently recognized in the previous reports describing various potential DISC1 interacting partners, mostly from yeast two-hybrid screens (17, 18), although it has not been further analyzed or characterized functionally.
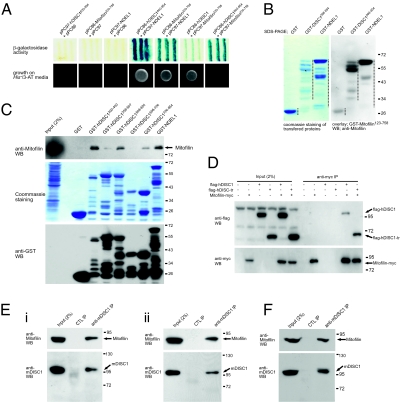
The interactions were confirmed by interaction-dependent β-galactosidase activity and growth on selective media (Fig. 1A). In the subsequent interaction analyses using yeast two-hybrid assays, regions of DISC1, amino acid residues 600–854 and 1–347, showed interaction-dependent signals with Mitofilin371–590 (Fig. S1A). The DISC1600–854 region showed strong interaction-dependent signals with Mitofilin400–590 (Fig. S1B). In addition, DISC1200–400 did not interact with Mitofilin371-590 (Fig. S1A) but showed signal with Mitofilin230–758 (Fig. S1C), suggesting that additional DISC1 interaction regions may exist in the outside of the 371–590 region in Mitofilin. Mitofilin exhibited significant interactions with DISC1 only when its N-terminal regions were deleted (Fig. 1A and Fig. S1C). It is likely because of an intrinsic limitation of the yeast two-hybrid assay; the hydrophobic transmembrane domain and the mitochondrial targeting signal located in the 1–187 residues may have hindered its localization to the nucleus of yeast cells and/or stable expression of Mitofilin fusion protein (14–16).
Fig. 1.
Interactions among DISC1, NDEL1, and Mitofilin. (A) Interactions of DISC1, NDEL1, and Mitofilin in the yeast two-hybrid assay. (Upper) Interaction-dependent β-galactosidase expression. (Lower) Growth on HIS− selective media containing 20 mM 3-amino-1,2,4-triazol (3-AT). hDISC1, human DISC1. (B) Interaction of Mitofilin with DISC1 and NDEL1 in vitro analyzed by a blot overlay assay. The PVDF membrane with the purified hDISC1 and NDEL1 protein bands transferred from an SDS/PAGE was first stained with Coomassie Blue R250 (Left) and subjected to GST–Mitofilin blot overlay followed by anti-Mitofilin Western blot analyses (Right). Dashed lines are the distribution of purified recombinant proteins with degradation products. WB, Western blotting. (C) Associations of Mitofilin with DISC1 and NDEL1 determined by GST pull-down assays. Lysates were prepared from SH-SY5Y cells that were differentiated for 36 h. (D) Coimmunoprecipitation of Mitofilin with full-length and truncated DISC1. HEK293 cells were transfected with flag-hDISC1 or flag-hDISC1-tr (amino acid residues 1–598), and Mitofilin-myc constructs as indicated. IP, immunoprecipitation. (E) Coimmunoprecipitation of endogenous DISC1 and Mitofilin from the differentiated CAD cells (i) and cultured mouse primary neurons (ii). Lysates prepared from CAD cells differentiated in the serum-free media for 36 h or mouse primary neurons cultured for 5 d in vitro (DIV5) were used. Anti-mDISC1 IP, IP with goat anti-mDISC1 antibody (N-16; Santa Cruz); CTL IP, control IP with goat anti-PhyA (Phytochrome A, Arabidopsis thaliana) antibody. (F) Coimmunoprecipitation of DISC1 and Mitofilin in the mouse brain lysate. Anti-mDISC1 IP, IP with goat anti-mDISC1 antibody (N-16; Santa Cruz); CTL IP, control IP with goat anti-human DISC1 antibody (K-20; Santa Cruz).
To further confirm the direct interactions among DISC1, Mitofilin, and NDEL1, a series of blot overlay assays were performed. When recombinant GST–hDISC1598–854 and GST–NDEL1 proteins were fractionated on the PVDF membrane and overlaid with recombinant GST–Mitofilin123–758, GST–Mitofilin123–758 was detected at the molecular weight corresponding to GST–NDEL1 and GST–hDISC1 (Fig. 1B). Other combinations of blot overlay assays consistently showed protein bands corresponding to the other binding partners, exhibiting varying affinities to degradation species (Fig. S1D, a and b). Furthermore, in the GST pull-down assay from differentiated SH-SY5Y cell lysates using purified recombinant GST–hDISC1 deletion proteins and GST–NDEL1 proteins, significant amounts of endogenous Mitofilin were consistently detected in the precipitates with GST–hDISC1200–400, GST–hDISC1598–854, and GST–NDEL1, confirming the capacity of Mitofilin to interact with DISC1 and NDEL1 in this condition (Fig. 1C). Consistently, both full-length hDISC1 and truncated hDISC1 (amino acid residues 1–598) (19) were detected in the Mitofilin immunoprecipitates from transfected HEK293 cells (Fig. 1D), indicating that multiple interaction interfaces exist between DISC1 and Mitofilin. To confirm the potential complex formation between DISC1–Mitofilin in more physiologically relevant conditions, we performed coimmunoprecipitation experiments in the lysates from differentiated central adrenergic tyrosine hydroxylase expressing (CATH) a-differentiated (CAD) (20) (Fig. 1E, i), cultured mouse neurons (Fig. 1E, ii), and a mouse brain (Fig. 1F). A significant amount of endogenous Mitofilin was consistently detected in the mDISC1 immunoprecipitates. Taken together, these results suggest that DISC1 and Mitofilin interact directly with each other to form a complex.
DISC1–Mitofilin Complex in Mitochondria.
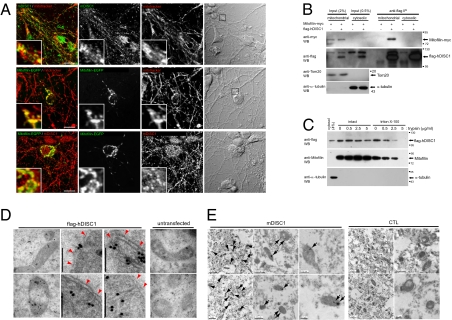
To examine if the DISC1–Mitofilin complex is associated with mitochondria, we performed immunocytochemical analyses in the cultured mouse primary neurons. For this, Mitofilin–EGFP was overexpressed because of the limitation that anti-Mitofilin antibody is not applicable to immunocytochemical experiments using mouse cells. Endogenous mDISC1 is dispersed throughout the cell (Fig. 2A Top), whereas Mitofilin is localized mainly, if not exclusively, to mitochondria, as indicated by extensive costaining with mitotracker, replicating the localization of endogenous Mitofilin in mitochondria in previous studies (Fig. 2A Middle) (14–16, 21). Significant colocalization between endogenous mDISC1 with Mitofilin–EGFP was detected in cytosolic area and processes (Fig. 2A Bottom). Consistently, colocalization of overexpressed hDISC1 and Mitofilin in mouse primary neurons was observed (Fig. S2A). Also, we performed a set of coimmunoprecipitation experiments in conjunction with mitochondrial fractionation from HEK293 cells expressing flag-hDISC1. Association of hDISC1 and Mitofilin was detected in the hDISC1 immunoprecipitate selectively from the mitochondrial fraction (Fig. 2B), further supporting the restricted association of the hDISC1–Mitofilin complex in mitochondria.
Fig. 2.
Localization of the DISC1–Mitofilin complex in mitochondria. (A) Colocalization of Mitofilin and DISC1 in mitochondria. Mouse primary neurons in culture were transfected with Mitofilin–EGFP plasmid on the DIV3 (green; Middle and Bottom), treated with mitotracker (red; Top and Middle) on the DIV4 as indicated, and applied to immunocytochemistry using goat anti-mDISC1 antibody (N-16; Santa Cruz; Top and Bottom), and Mitofilin–EGFP was detected by fluorescence from EGFP (Middle and Bottom). (Scale bar: 10 μm.) (B) Coimmunoprecipitation of Mitofilin with DISC1 from the mitochondrial fraction. The mitochondrial and cytosolic fractions from HEK293 cells transfected as indicated were subjected to immunoprecipitation followed by Western blotting. The same membrane was reprobed for Tom20, a mitochondrial marker, and α-tubulin. (C) Increased trypsin sensitivity of mitochondrial DISC1 by Triton X-100 treatment. The mitochondrial fraction was prepared from flag-hDISC1–transfected HEK293 cells en masse and divided into aliquots for the treatments of Triton X-100 (final 2%) and trypsin followed by Western blotting. (D) Localization of overexpressed DISC1 to the inside of mitochondria. HEK293 cells transfected with the flag-hDISC1 construct (Left) along with untransfected control cells (Right) were subjected to anti-flag IGEM. Mitochondrial outer membrane is indicated by red arrowheads. (Scale bar: 100 nm.) (E) Localization of endogenous DISC1 to the inside of mitochondria. Mouse brain sections were subjected to goat anti-mDISC1 IGEM (Left). The clusters of 10-nm immuno-gold particles for endogenous mDISC1 in mitochondria are indicated by arrows. CTL, IGEM with goat anti-PhyA antibody (Right).
DISC1 has been consistently observed in the mitochondrion-associated cellular fractions (9, 22). However, the nature of this association has not been clearly defined. Because Mitofilin is an integral mitochondrial inner membrane protein with most of the protein portion protruding to the intermembrane space of mitochondria (15, 16), the interaction of DISC1 with Mitofilin suggests that DISC1 can localize to the inside of mitochondria. To address this point, we examined the trypsin sensitivity of hDISC1 in parallel with that of endogenous Mitofilin on Triton X-100 treatment. The trypsin sensitivity of hDISC1 and Mitofilin in the Triton X-100–treated mitochondrial fraction was similarly increased compared with those of hDISC1 and Mitofilin in the intact mitochondrial fraction, indicating that a fraction of DISC1 is localized to the inside of mitochondria (Fig. 2C). Moreover, in the immuno-gold electron microscopy (IGEM), a significant fraction of flag-hDISC1 was consistently detected as associated with the electron-dense area in mitochondria of transfected HEK293 cells (Fig. 2D). Furthermore, in the anti-mDISC1 IGEM analyses, endogenous mDISC1 was consistently detected in the electron-dense area within mitochondria both in the differentiated CAD cells (Fig. S2B) and mouse brain sections (Fig. 2E). Collectively, these results support the idea that a fraction of DISC1 is localized to the inside of mitochondria and forms a complex with Mitofilin, implying a role for DISC1 in intrinsic mitochondrial functions.
Mitochondrial Dysfunctions Induced by DISC1 and Mitofilin Deficiencies.
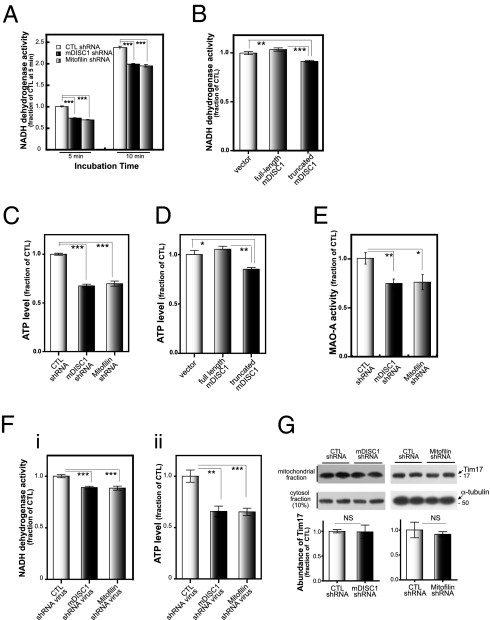
We next tested the potential impact of reduced DISC1 on various aspects of mitochondrial function. For this, we generated shRNA constructs against mouse forms of Mitofilin and DISC1 and confirmed their knockdown efficiencies (Fig. S3 A and B). We carried out a series of experiments to assess potential mitochondrial dysfunction in DISC1 and Mitofilin knockdown conditions. First, we examined the activity of mitochondrial NADH dehydrogenase, a key component in the mitochondrial electron transport chain. As shown in Fig. 3A, the mitochondrial NADH dehydrogenase activity, measured by a colorimetric assay (23), was significantly reduced in both mDISC1 and Mitofilin shRNA-transfected differentiated CAD cells. Notably, a mild but significant reduction of mitochondrial dehydrogenase activity was detected in the differentiated CAD cells overexpressing truncated mDISC1 (Fig. 3B). Consistently, ATP contents in the lysates of the cells transfected with mDISC1 shRNA, Mitofilin shRNA, or truncated mDISC1 constructs were significantly reduced (Fig. 3 C and D). A simultaneous knockdown of both DISC1 and Mitofilin did not seem to cause considerable further decrease of the NADH dehydrogenase activity or ATP levels (Fig. S4). Also, these DISC1 and Mitofilin knockdown phenotypes were partially, but effectively, rescued by coexpression of human forms of DISC1 and Mitofilin, removing the off-target effect of shRNA constructs (Fig. S4). These results indicate that the loss of DISC1 or overexpression of a pathogenic form of DISC1 can result in mitochondrial dysfunctions related to mitochondrial bioenergetics.
Fig. 3.
Compromised mitochondrial function by deficiencies in DISC1 and Mitofilin. (A) Down-regulation of mitochondrial NADH dehydrogenase activity by mDISC1 or Mitofilin knockdown. CAD cells were transfected with shRNA constructs and differentiated for an additional 5 d. Media were replaced with the assay solution, and the time-dependent conversion of soluble formazan from a tetrazolium salt was measured at OD495. Error bars are mean ± SEM. ***P < 0.001, two-tailed t test (n = 6). (B) Down-regulation of mitochondrial NADH dehydrogenase activity by an overexpression of truncated mDISC1. CAD cells were transfected with plasmids as indicated and differentiated for 72 h before NADH-dehydrogenase activity assay. Incubation time was 10 min. Error bars are mean ± SEM. **P < 0.01; ***P < 0.001, two-tailed t test (n = 6). (C) Reduction of ATP contents in the mDISC1 or Mitofilin knockdown cells. CAD cells were transfected with shRNA constructs and differentiated for an additional 5 d. ATP contents in the cell lysates were measured. Error bars are mean ± SEM. ***P < 0.001, two-tailed t test (n = 6). (D) Reduction of ATP contents by an overexpression of truncated DISC1. CAD cells were transfected with full-length mDISC1 or truncated mDISC1, and after 72 h of differentiation in serum-free media, the ATP contents in lysates were measured. Error bars are mean ± SEM. *P < 0.05; **P < 0.01, two-tailed t test (n = 6). (E) Decreased monoamine oxidase-A (MAO-A) activities in the mDISC1 or Mitofilin knockdown cells. SN4741 cells were transfected with shRNA constructs and differentiated for an additional 5 d. Lysates were subjected to an MAO-A activity assay. Error bars are mean ± SEM. *P < 0.05; **P < 0.01, two-tailed t test (n = 6). (F) Reduction of NADH dehydrogenase activity and ATP contents by DISC1 and Mitofilin knockdown in cultured primary neurons. Cultured mouse cortical neurons (DIV2) were infected with lentiviruses containing mDISC1 or Mitofilin shRNA and cultured for an additional 6 d before the NADH dehydrogenase activity assays (i) and ATP measurement (ii). Error bars are mean ± SEM. **P < 0.01; ***P < 0.001, two-tailed t test (n = 6). (G) Unaltered mitochondrial contents in mDISC1 and Mitofilin knockdown cells. CAD cells were transfected with mDISC1 and Mitofilin shRNAs and differentiated for 5 d. The Tim17 levels in mitochondrial and α-tubulin in corresponding cytosol fractions were analyzed. Ratios of Tim17/α-tubulin were subjected to statistical analysis. Error bars are mean ± SEM. NS, not significant, two-tailed t test (n = 4).
It is well-known that monoamine oxidases (MAOs), key components in the monoamine catabolic pathway, are located in mitochondria, which makes the mitochondrion a critical organelle in the maintenance of monoamine homeostasis. To determine whether DISC1 and Mitofilin deficiencies compromise MAO function, the activity of the MAO-A was examined in differentiated SN4741 cells, a rodent neuronal cell line with catecholaminergic properties (24). We confirmed expression of MAO-A (Fig. S5A) and association of DISC1 and Mitofilin in this cell line by coimmunoprecipitation experiments (Fig. S5B). As shown in Fig. 3E, MAO-A activity was markedly reduced in both mDISC1 and Mitofilin knockdown cells, implying that the monoamine homeostasis can be compromised by reduced DISC1 and Mitofilin function. We see a consistent tendency to a decrease in MAO activity with the truncated DISC1 overexpression, but the extent of the reduction did not reach a statistically significant level. Because the mitochondrial dysfunctions induced by truncated DISC1 are generally weaker than those by DISC1 knockdown, truncated DISC1 may be hypomorphic to complete DISC1 loss of function in mitochondria, and MAO-A phenotype may exemplify this notion. It is also possible that the truncated DISC1 may still retain the capacity to regulate MAO-A activity.
We also examined the DISC1 deficiency-induced mitochondrial dysfunction in the cultured mouse primary neurons using lentiviruses containing mDISC1 and Mitofilin shRNA constructs (Fig. S3C). Similar reductions of NADH dehydrogenase activities and ATP contents were observed in the virus-infected neurons (Fig. 3F), strengthening the physiological relevance of mitochondrial function of DISC1.
It has been reported that deficiency in Mitofilin can slightly increase apoptosis in proliferating HeLa cells (15). Thus, we tested if the observed down-regulation of mitochondrial function associated with mDISC1 and Mitofilin knockdown is because of a decrease in the contents of cells in samples. The levels of Tim17, a mitochondrial marker that reflects the cellular contents of mitochondria, were not altered in the shRNA-transfected differentiated CAD cells (Fig. 3G). Also, no significant decrease in the total number and mean diameter of mitochondria was detected in mDISC1 or Mitofilin shRNA-transfected CAD cells (Fig. S6). Moreover, the total cell numbers were not significantly altered in the equivalent conditions (Fig. S7A). Also, the extent of apoptotic cells in this population of cells, monitored by the TUNEL assay and generation of cleaved caspase-3, did not increase on the transfection of mDISC1 or Mitofilin shRNA constructs (Fig. S7 B and C). However, we detected a slight increase in the number of mitochondria with morphological abnormalities in the DISC1 and Mitofilin knockdown cells, which is reminiscence of morphological defects linked to Mitofilin deficiency in HeLa cells (Fig. S8) (15). Collectively, these results indicate that potential changes in cellular contents of mitochondria, cell proliferation, and/or apoptosis, if any, did not exert significant impacts on the experimental settings used in this study, leaving the possibility of involvement of morphological defect in the DISC1 and Mitofilin knockdown cells.
Perturbed Mitochondrial Ca2+ Dynamics Induced by DISC1 and Mitofilin Deficiencies.
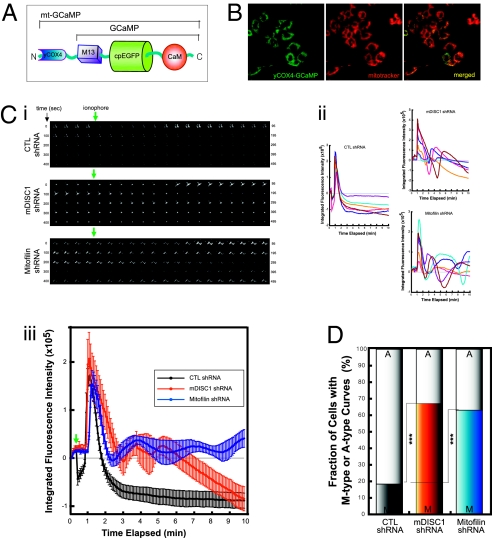
The Ca2+-buffering function of mitochondria is particularly important in the nervous system in that long neuronal processes require local Ca2+ stores in proximity to cope with transient and local changes in Ca2+ induced by various neuronal activities (11). As such, we test if loss of DISC1 function has an impact on Ca2+-buffering capacity of mitochondria. For this purpose, we generated a mitochondrion-targeted protein Ca2+ sensor (mt-GCaMP) by adding the presequence of yeast cytochrome C oxidase 4 (yCOX4) (25) to the N terminus of GCaMP (Materials and Methods) (Fig. 4A) (26). The targeting of the mt-GCaMP to mitochondria was confirmed by extensive colocalization of mt-GCaMP expression with mitotracker (Fig. 4B).
Fig. 4.
Abnormal mitochondrial Ca2+ buffering by deficiencies in DISC1 and Mitofilin. (A) A schematic diagram of mt-GCaMP, a mitochondrial Ca2+ sensor. (B) Restricted expression of mt-GCaMP in mitochondria. Mitotracker (red) and anti-GFP antibody (green) in differentiated CAD cells. (C) Time-lapse mitochondrial Ca2+ imaging. CAD cells were cotransfected with mt-GCaMP constructs in combination with shRNA constructs (mt-GCaMP:shRNA plasmid = 1:5, molar ratio), and changes in fluorescence in response to ionomycin treatment were measured by time-lapse imaging 5 d after transfection. (i) Examples of time-lapse images for 495 s. Numbers, the time elapsed in seconds; green arrows, time point of ionomycin addition. (ii) Individual mitochondrial Ca2+ in response to ionomycin; six representative measurements of each are shown. Fluorescence intensity was normalized by fluorescence at the resting condition. (iii) Mean values of the curves shown in ii. Error bars are mean ± SEM. (D) The increased fraction of the cells showing abnormal fluctuation of Ca2+ (M-type curves) in mDISC1 and Mitofilin shRNA-transfected cells. ***P < 0.001, Fisher's exact test (n = 16 for CTL shRNA, n = 23 for mDISC1 shRNA, n = 10 for Mitofilin shRNA).
A time-lapse imaging technique was used to assess mitochondrial Ca2+ concentration in response to acutely increased intracellular Ca2+ concentrations by ionomycin treatment (Fig. 4C). In CAD cells cotransfected with control shRNA and mt-GCaMP constructs, ionomycin treatment transiently increased the Ca2+-dependent fluorescence, but it decreased and stabilized to the basal level within 5 min (designated as A-type curves) (Fig. 4C, i Top and ii Left graph). In contrast, in the majority of mDISC1 or Mitofilin shRNA-transfected CAD cells, a characteristic pattern of fluorescence fluctuation was observed (designated as M-type curves) (Fig. 4C, i and ii right graphs, and iii). When quantified, over 60% of the mDISC1 or Mitofilin shRNA-transfected CAD cells showed an M-type curve pattern with abnormal fluctuation of mitochondrial Ca2+ concentrations (Fig. 4D). Collectively, although the mechanistic nature of the changes in mitochondrial Ca2+ dynamics in DISC1-deficient cells should be studied further, the data clearly show that a reduction in DISC1 and Mitofilin functions perturbs mitochondrial Ca2+-buffering activity in response to acutely elevated intracellular Ca2+ concentrations.
Potential Role for DISC1 in the Stability of Mitofilin.
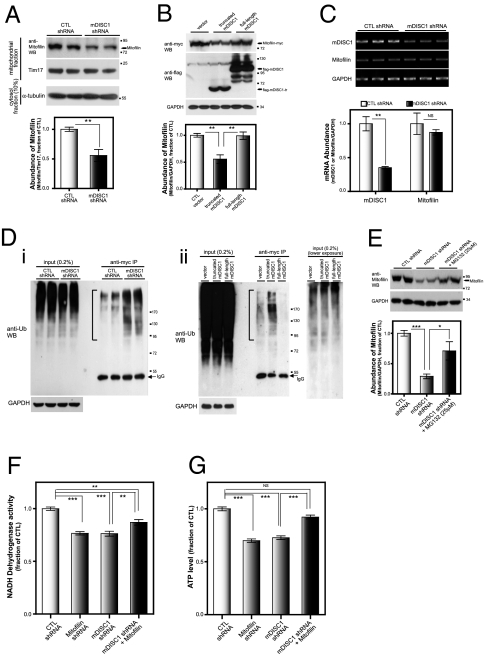
To understand the mode of which DISC1 affects Mitofilin function, we examined Mitofilin expression when DISC1 function was reduced. Notably, knockdown of mDISC1 caused a significant reduction in Mitofilin protein levels in the mitochondrial fraction from differentiated CAD cells (Fig. 5A). Interestingly, an overexpression of truncated mDISC1, which serves to mimic the pathogenic form of DISC1 found in the Scottish pedigree (19), also reduced heterologously expressed Mitofilin protein in the differentiated CAD cells (Fig. 5B). This phenomenon is not caused by transcriptional changes in Mitofilin expression, because Mitofilin mRNA levels were not significantly altered in the mDISC1 knockdown cells (Fig. 5C).
Fig. 5.
Down-regulation of Mitofilin by aberrant DISC1 function. (A) Down-regulation of Mitofilin protein levels by knockdown of DISC1. CAD cells were transfected with control or mDISC1 shRNA constructs and differentiated for 5 d. Mitochondrial fraction was isolated, and endogenous Mitofilin protein levels were analyzed by anti-Mitofilin Western blotting. Error bars are mean ± SEM. **P < 0.01, two-tailed t test (n = 6). (B) Down-regulation of Mitofilin protein levels by overexpression of truncated DISC1 protein. CAD cells were transfected with constructs of flag-mDISC1, truncated flag-mDISC1 (amino acid residues 1–598), and Mitofilin-myc and were differentiated for 72 h. Whole-cell lysates were analyzed. Ratios of Mitofilin/GAPDH were subjected to statistical analyses. Error bars are mean ± SEM. **P < 0.01, two-tailed t test (n = 4). (C) Unaffected Mitofilin mRNA level in DISC1 knockdown cells. CAD cells were transfected with shRNA constructs and differentiated for 5 d. mDISC1 and Mitofilin mRNA levels were measured by RT-PCR. Error bars are mean ± SEM. **P < 0.01, two-tailed t test (n = 4). (D) Increased ubiquitination of Mitofilin by DISC1 knockdown or overexpression of truncated DISC1. (i) CAD cells were cotransfected with shRNA, HA-Ub, and Mitofilin-myc constructs, and after 68 h of differentiation, anti-Myc immunoprecipitates from the cell lysates were analyzed by anti-Ub Western blotting. (ii) CAD cells were transfected with truncated mDISC1 or full-length mDISC1 along with Mitofilin-myc constructs, differentiated, and analyzed as in a. A lower exposure blot of input is also shown. Brackets indicate ubiquitinated Mitofilin. (E) Inhibition of down-regulation of Mitofilin by a proteasome inhibitor. CAD cells were transfected with control or mDISC1 shRNA constructs and differentiated for 5 d. MG132 (Sigma, 25 μM) was treated for 8 h ahead of the preparation of lysates. Error bars are mean ± SEM. *P < 0.05; ***P < 0.001, two-tailed t test (n = 4). (F and G) Partial rescue of the DISC1 knockdown-mediated reduction of NADH dehydrogenase activity and ATP contents by coexpression of Mitofilin. CAD cells were transfected with the constructs as indicated and differentiated for 5 d, and NADH dehydrogenase activity (F) and ATP contents (G) were measured. Error bars are mean ± SEM. **P < 0.01; ***P < 0.001, NS, not significant, two-tailed t test (n = 6).
Proteasome-mediated proteolysis is known to be a key checkpoint by which excess or misfolded mitochondrial proteins can be eliminated (27). Therefore, we attempted to assess the ubiquitination level of Mitofilin proteins in the mDISC1 knockdown or truncated mDISC1 overexpression conditions. As shown in Fig. 5D, the ubiquitinated Mitofilin was increased in these conditions. Furthermore, the down-regulation of Mitofilin was significantly blocked by MG132, a proteasome inhibitor (Fig. 5E). These results indicate that the impaired DISC1 function facilitates ubiquitination of Mitofilin and presumably, proteasome-mediated degradation of Mitofilin.
Finally, we attempted to directly test whether the reduction of Mitofilin proteins associated with DISC1 deficiency is linked to the mitochondrial DISC1 function. Remarkably, a coexpression of Mitofilin can partially reverse the reduction of mitochondrial NADH dehydrogenase activity and ATP contents caused by DISC1 knockdown (Fig. 5 F and G), whereas overexpression of Mitofilin alone did not significantly alter the mitochondrial function (Fig. S9). However, the rescue is partial, especially for the NADH dehydrogenase activity. Although it is possible that mDISC1 knockdown may have additional effects other than the ones mediated by Mitofilin, it is also likely that technical difficulties in generating absolute complementation of Mitofilin in all of the mDISC1 knockdown cells may be involved. In sum, these results strengthen the interpretation that mitochondrial function of DISC1 is tightly associated with Mitofilin.
From a mechanistic point of view, the potential roles of NDEL1 in mitochondrial function of DISC1 seemed interesting. However, our initial attempt to detect NDEL1–Mitofilin complex in cellular environments was not successful (Fig. S10A), although a small fraction of overexpressed NDEL1 was detected in the mitochondrial fraction (Fig. S10B). Also, the knockdown or overexpression of NDEL1 in the differentiated CAD cells did not significantly alter mitochondrial functionalities monitored by NADH dehydrogenase activity and cellular ATP contents (Fig. S10 C and D). Thus, it is unclear whether NDEL1 also participates in the mitochondrial function of DISC1.
Discussion
In this study, we showed the evidence that DISC1 plays roles for intrinsic mitochondrial functions through regulation of the stability of Mitofilin, a mitochondrial interacting partner. This is intriguing, because the aberrant functioning of mitochondria has been linked to the pathogenesis of schizophrenia without a clear mechanistic basis. For example, a reduction in the number of mitochondria, an abnormal morphology of mitochondria, and likely, as a consequence, defects in oxidative phosphorylation have been reported in postmortem brain studies of schizophrenia patients (12, 13). More recently, gene expression studies using the postmortem brains of schizophrenia patients also suggest a role of mitochondrial dysfunction in the development of schizophrenia (28). Furthermore, the Mitofilin gene is located in a genomic region, in which chromosomal abnormalities have been linked to schizophrenia (29, 30). Thus, the role of DISC1 in normal mitochondrial function, in conjunction with Mitofilin, may provide a molecular insight into the mitochondrial hypothesis of schizophrenia.
Mechanistic links between the reduced function of DISC1–Mitofilin complex and a wide range of mitochondrial defects remain to be further understood. Intriguingly, it has recently been shown that Mitofilin is associated with the general protein complex involved in the import of mitochondrial proteins such as sorting and assembly machinery component 50 homolog (SAM50), metaxin1, and metaxin2 (31). Mitofilin has actually been implicated in the mitochondrial localization of poly-ADP ribose polymerase-1 (PARP-1) (32). Thus, we can speculate a possibility that the function of DISC1 in mitochondria is linked to the mitochondrial protein import machinery through Mitofilin.
Although correlations between mesolimbic hyperdopaminergia and the positive symptoms of schizophrenia are relatively well-established, the causative cellular basis for this association is not clearly defined (33). In this regard, it is noteworthy that the knockdown of DISC1 can cause reductions in mitochondrial MAO-A activity. MAO-A has been considered to be involved in human psychopathologies, including schizophrenia, because of its activity in the catabolic pathway of biogenic amines, including dopamine (34). Down-regulation of MAO-A by compromised functioning of the DISC1–Mitofilin complex, which is likely to cause up-regulated monoamine contents in monoaminergic neurons, may help explain the cellular basis underlying schizophrenia-associated neurochemical disturbances. Thus, a direct link between abnormalities of DISC1 and dopamine homeostasis should be of immediate interest.
Materials and Methods
SI Materials and Methods has descriptions of the following experimental procedures and reagents, including yeast two-hybrid screening and interaction analyses, antibodies, GST pull-down assays, blot overlay assays, immunoprecipitation, immunocytochemistry, IGEM, trypsin sensitivity assay, NADH dehydrogenase activity assay, ATP measurement, MAO-A activity assay, mitochondrial Ca2+ imaging, and ubiquitination analysis.
Supplementary Material
Acknowledgments
We thank Dr. Li-Huei Tsai (Massachusetts Institute of Technology) for kind comments. We thank Dr. Chin Ha Chung (Seoul National University, Seoul, Korea) for providing critical reagents and technical advice. This work was supported by Brain Research Center of the 21st Century Frontier Research Program Grant 2010K000812 and in part by Mid-Career Researcher Program Grant 3-200900000001605 funded by the Korean Ministry of Education, Science and Technology (MEST). This work was also funded by MEST Grants 331-2007-1-C00213 and 20090076351. S.K.P. was a recipient of the 2004 and 2006 National Alliance for Research on Schizophrenia and Depression Young Investigator Awards. M.D.N. holds a Career Development Award from HFSPO and a New Investigator Award from the CIHR.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004361107/-/DCSupplemental.
References
- 1.American Psychiatric Association—Task Force on DSM-IV . Diagnostic and Statistical Manual of Mental Disorders: DSM-IV-TR. 4th Ed. Washington, DC: American Psychiatric Association; 2000. p. xxxvii. [Google Scholar]
- 2.Cardno AG, Gottesman II. Twin studies of schizophrenia: From bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–17. [PubMed] [Google Scholar]
- 3.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: On the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 4.Mackie S, Millar JK, Porteous DJ. Role of DISC1 in neural development and schizophrenia. Curr Opin Neurobiol. 2007;17:95–102. doi: 10.1016/j.conb.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Kamiya A, et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 6.Ozeki Y, et al. Disrupted-in-Schizophrenia-1 (DISC-1): Mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci USA. 2003;100:289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan X, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao Y, et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millar JK, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 10.Burdick KE, et al. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: Evidence of epistasis and competitive binding. Hum Mol Genet. 2008;17:2462–2473. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben-Shachar D, Laifenfeld D. Mitochondria, synaptic plasticity, and schizophrenia. Int Rev Neurobiol. 2004;59:273–296. doi: 10.1016/S0074-7742(04)59011-6. [DOI] [PubMed] [Google Scholar]
- 12.Maurer I, Zierz S, Möller H. Evidence for a mitochondrial oxidative phosphorylation defect in brains from patients with schizophrenia. Schizophr Res. 2001;48:125–136. doi: 10.1016/s0920-9964(00)00075-x. [DOI] [PubMed] [Google Scholar]
- 13.Kung L, Roberts RC. Mitochondrial pathology in human schizophrenic striatum: A postmortem ultrastructural study. Synapse. 1999;31:67–75. doi: 10.1002/(SICI)1098-2396(199901)31:1<67::AID-SYN9>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 14.Gieffers C, Korioth F, Heimann P, Ungermann C, Frey J. Mitofilin is a transmembrane protein of the inner mitochondrial membrane expressed as two isoforms. Exp Cell Res. 1997;232:395–399. doi: 10.1006/excr.1997.3539. [DOI] [PubMed] [Google Scholar]
- 15.John GB, et al. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell. 2005;16:1543–1554. doi: 10.1091/mbc.E04-08-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odgren PR, Toukatly G, Bangs PL, Gilmore R, Fey EG. Molecular characterization of mitofilin (HMP), a mitochondria-associated protein with predicted coiled coil and intermembrane space targeting domains. J Cell Sci. 1996;109:2253–2264. doi: 10.1242/jcs.109.9.2253. [DOI] [PubMed] [Google Scholar]
- 17.Millar JK, Christie S, Porteous DJ. Yeast two-hybrid screens implicate DISC1 in brain development and function. Biochem Biophys Res Commun. 2003;311:1019–1025. doi: 10.1016/j.bbrc.2003.10.101. [DOI] [PubMed] [Google Scholar]
- 18.Camargo LM, et al. Disrupted in Schizophrenia 1 Interactome: Evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 19.Millar JK, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 20.Qi Y, Wang JK, McMillian M, Chikaraishi DM. Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J Neurosci. 1997;17:1217–1225. doi: 10.1523/JNEUROSCI.17-04-01217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Icho T, et al. A novel human gene that is preferentially transcribed in heart muscle. Gene. 1994;144:301–306. doi: 10.1016/0378-1119(94)90394-8. [DOI] [PubMed] [Google Scholar]
- 22.Brandon NJ, et al. Subcellular targeting of DISC1 is dependent on a domain independent from the Nudel binding site. Mol Cell Neurosci. 2005;28:613–624. doi: 10.1016/j.mcn.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 24.Son JH, et al. Neuroprotection and neuronal differentiation studies using substantia nigra dopaminergic cells derived from transgenic mouse embryos. J Neurosci. 1999;19:10–20. doi: 10.1523/JNEUROSCI.19-01-00010.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurt EC, Pesold-Hurt B, Suda K, Oppliger W, Schatz G. The first twelve amino acids (less than half of the pre-sequence) of an imported mitochondrial protein can direct mouse cytosolic dihydrofolate reductase into the yeast mitochondrial matrix. EMBO J. 1985;4:2061–2068. doi: 10.1002/j.1460-2075.1985.tb03892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 27.Radke S, et al. Mitochondrial protein quality control by the proteasome involves ubiquitination and the protease Omi. J Biol Chem. 2008;283:12681–12685. doi: 10.1074/jbc.C800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prabakaran S, et al. Mitochondrial dysfunction in schizophrenia: Evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- 29.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavlidis P, Noble WS. Matrix2png: A utility for visualizing matrix data. Bioinformatics. 2003;19:295–296. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
- 31.Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007;581:3545–3549. doi: 10.1016/j.febslet.2007.06.052. [DOI] [PubMed] [Google Scholar]
- 32.Rossi MN, et al. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J Biol Chem. 2009;284:31616–31624. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owen F, Simpson M. The neurochemistry of schizophrenia. Mol Cell Biol Hum Dis Ser. 1994;4:133–159. doi: 10.1007/978-94-011-0709-9_6. [DOI] [PubMed] [Google Scholar]
- 34.Maas JW, et al. Studies of catecholamine metabolism in schizophrenia/psychosis—II. Neuropsychopharmacology. 1993;8:111–116. doi: 10.1038/npp.1993.12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.