Abstract
Angelman syndrome (AS) and Prader–Willi syndrome (PWS) are neurodevelopmental disorders of genomic imprinting. AS results from loss of function of the ubiquitin protein ligase E3A (UBE3A) gene, whereas the genetic defect in PWS is unknown. Although induced pluripotent stem cells (iPSCs) provide invaluable models of human disease, nuclear reprogramming could limit the usefulness of iPSCs from patients who have AS and PWS should the genomic imprint marks be disturbed by the epigenetic reprogramming process. Our iPSCs derived from patients with AS and PWS show no evidence of DNA methylation imprint erasure at the cis-acting PSW imprinting center. Importantly, we find that, as in normal brain, imprinting of UBE3A is established during neuronal differentiation of AS iPSCs, with the paternal UBE3A allele repressed concomitant with up-regulation of the UBE3A antisense transcript. These iPSC models of genomic imprinting disorders will facilitate investigation of the AS and PWS disease processes and allow study of the developmental timing and mechanism of UBE3A repression in human neurons.
Keywords: antisense transcript, epigenetic, neuronal differentiation
Angelman syndrome (AS) is a neurogenetic disorder characterized by profound intellectual disability, absent speech, frequent seizures, motor dysfunction, and a characteristic happy demeanor (1, 2). Prader–Willi syndrome (PWS) is characterized hyperphagia/obesity; small stature, hands, and feet; and behavioral problems that are likened to obsessive compulsive disorder (3). AS is caused by loss of function of the maternally inherited allele of the E3 ubiquitin ligase UBE3A. UBE3A is subject to tissue-specific genomic imprinting; although both alleles are expressed in most tissues, the paternally inherited allele is repressed in the brain (4–6). Imprinted expression of UBE3A is thought to occur as a result of reciprocal expression of a long noncoding antisense transcript, UBE3A-ATS, which is part of a >600-kb transcript initiating at the differentially methylated PWS imprinting center (IC) located in exon 1 of the SNURF-SNRPN gene (7–9). PWS is associated with the loss of several species of small nucleolar RNAs (snoRNAs) (10); however, its genetic basis is currently unknown, and there is no mouse model that recapitulates all features of PWS.
Mouse models of AS have proved significant in studying important aspects of the AS disease mechanism. There are, however, differences in the tissue specificity of the transcript that harbors UBE3A-ATS between humans and mice (11), indicating that the timing and mechanism of UBE3A repression may diverge between these species. The ability to study the developmental timing and mechanism of brain-specific repression of the paternal UBE3A allele in a model of human development is critical for better understanding the AS disease process and for using live neurons from patients with AS to discover previously undescribed therapeutic interventions. Here, we have developed such a model via human induced pluripotent stem cell (iPSC) technology.
Results
Human iPSCs Generated from AS and PWS Fibroblasts.
iPSC lines were established using fibroblasts from two different patients with AS who carried maternally inherited deletions of chromosome 15q11-q13 and from one patient with PWS who carried a paternal deletion of chromosome 15q11-q13. iPSCs were generated using retroviral vectors encoding OCT4, SOX2, KLF4, MYC, and LIN28 (12, 13). Reprogrammed colonies were initially identified morphologically and were subsequently validated using immunocytochemistry to verify the expression of the pluripotency markers NANOG, SSEA4, TRA1-60, and TRA1-81 (Fig. 1 and Fig. S1). AS iPSCs were shown to have the expected karyotypes of 46, XX or XY, del(15) (pter > q11::q13 > qter) and to express genes associated with pluripotency at levels comparable to the H9 (WA09) human ES cell (hESC) line (Fig. 1 and Fig. S2). Human stem cell pluripotency arrays were used for quantitative RT-PCR (qRT-PCR) for gene expression analysis of AS iPSC-derived embryoid bodies (EBs) after 14 d of spontaneous differentiation. These data demonstrate that AS iPSCs are capable of multilineage differentiation, because early lineage markers representing each of the three embryonic germ layers as well as the trophectoderm layer are expressed (Fig. S3).
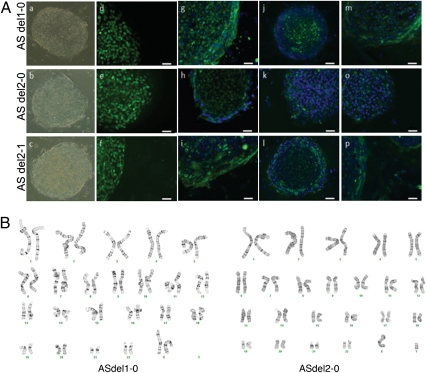
Fig. 1.
AS iPSCs express markers of pluripotency and have expected karyotypes. (A) Phase-contrast (a–c) images of AS iPSCs show hESC-like morphology. Immunocytochemistry for pluripotency markers on representative iPSC lines shows expression of NANOG (d–f), SSEA4 (g–i), TRA1-60 (j–l), and TRA1-81 (m–p). (B) Karyotype analysis on G-banded chromosomes was performed by Cell Line Genetics. Representative G-banded chromosomes from two AS iPSC lines show normal chromosome counts of 46 XX (ASdel1-0) and 46 XY (ASdel2-0).
AS and PWS iPSCs Maintain the Appropriate Methylation Imprint Following Reprogramming.
Because nuclear reprogramming of somatic cells into pluripotent stem cells may involve global erasure of epigenetic marks, it has been argued that the differential methylation that marks the PWS-IC could also be erased during reprogramming, making it difficult to generate an iPSC model of AS (14). In this regard, aberrant DNA methylation at the imprinted H19 and KCNQOT1 genes in iPSCs was recently reported (15). We assessed the methylation imprint in normal, AS, and PWS iPSCs by methylation-specific PCR (16, 17). iPSCs from normal individuals and from persons with AS and PWS showed the same methylation patterns as the fibroblast lines from which they were derived. A methylated maternal allele and an unmethylated paternal allele were both present in normal iPSCs, whereas only an unmethylated paternal allele was observed in AS iPSCs and only a methylated maternal allele was observed in PWS iPSCs (Fig. 2 and Fig. S4A). These results were confirmed using a methylation-sensitive restriction endonuclease quantitative PCR assay (Fig. S4B).
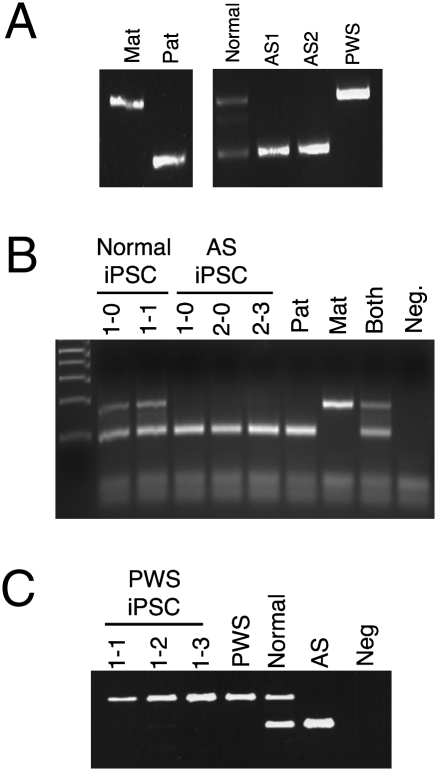
Fig. 2.
Methylation imprint at the PWS-IC is maintained during reprogramming. (A) Methylation-specific PCR analysis of genomic DNA from normal, AS, and PWS fibroblasts. Primers specific for the methylated allele amplify a band that is 174 bp, whereas primers specific for the unmethylated allele amplify a 100-bp product. Mat, maternal; Pat, paternal. (B) Methylation-specific PCR using genomic DNA from three different AS iPSC lines and two different normal iPSC lines. Neg, negative. (C) Methylation-specific PCR using genomic DNA from three different PWS iPSC lines.
AS iPSCs Can Be Differentiated into Functional Neurons.
Because AS is a neurogenetic disorder, we sought to generate functional neurons from AS iPSCs. AS and normal iPSCs were differentiated into neurons using an EB intermediate to approximate normal human development (18, 19) (Figs. S5 and S6). Consistent with a recent report (20), considerable variability in differentiation efficiency was observed among different iPSC lines. Therefore, we selected one normal line and one AS iPSC line that displayed efficient neuronal lineage differentiation for further studies.
The developmental timing of neural differentiation for both AS and normal iPSCs closely followed that previously reported for hESCs (19). Following 4 wk of differentiation, MAP2- and TUJ1-positive neurons could be identified (Fig. 3 A and B and Fig. S6 B–D). Astrocytes were detected after 6 wk of differentiation by staining for S100β (Fig. 3C and Fig. S6B). Concomitant with the appearance of astrocytes, the development of synapses was evident by the appearance of Synapsin I-positive cells (Figs. S5D and S6C). After 10 wk of development, immunostaining with a sodium channel antibody (PanNav) indicated that sodium channels localized to the axon initial segment in some but not all neurons (Figs. S5 E and F and S6E).
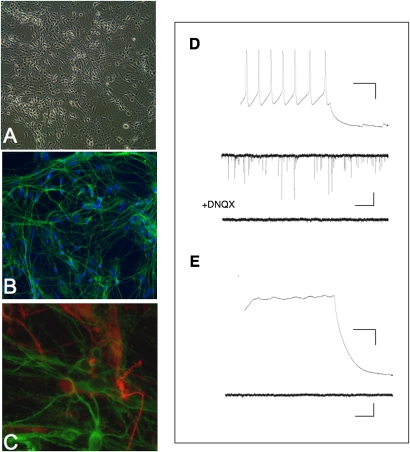
Fig. 3.
Functional neurons can be generated from AS iPSCs. (A) Phase-contrast image of 10-wk-old in vitro differentiated neurons generated from AS iPSCs. (B) iPSC-derived neurons stain positively for βIII-TUBULIN (TUJ1, green). (C) S100β-positive astrocytes spontaneously arise in AS iPSC-derived neuron cultures after 6 wk of differentiation. (D) Electrophysiological recordings from a mature AS iPSC-derived neuron after 10 wk of differentiation. (Upper) Train of action potentials evoked by depolarizing current injection (50 pA). (Calibration bars: 20 mV, 0.1 s.) (Lower) Example sweeps of spontaneous EPSCs recorded from the same neuron in the absence or presence of the AMPA receptor antagonist DNQX (10 μM). (Calibration bars: 10 pA, 1 s.) (E) Electrophysiological recordings from an immature AS iPSC-derived neuron after 10 wk of differentiation. (Upper) Response to a 60-pA depolarizing current injection. (Lower) Lack of spontaneous synaptic activity. Calibration bars are the same as in D.
These data suggested that the iPSC-derived neurons were maturing in culture and that some but not all neurons were functionally mature. In agreement with immunocytochemistry data, 10-wk differentiated AS iPSC cultures contained neurons that exhibited trains of action potentials and excitatory postsynaptic currents (EPSCs) mediated by AMPA receptor activation (example in Fig. 3D). Neurons that did not have repetitive trains of action potentials and/or robust EPSCs could also be identified (Fig. 3E). Normal iPSC-derived neurons exhibited a similar diversity after 10 wk of development (example in Fig. S7). Although mature functional neurons with synaptic activity can be generated from AS iPSCs by in vitro development, 10-wk cultures are heterogeneous in their maturity. This is not surprising because only 14% of human cortical plate neurons sampled at 16 wk of gestation fire repetitive action potentials (21).
Paternal UBE3A Is Repressed During in Vitro Neurogenesis of AS iPSCs.
In addition to faithful maintenance of the PWS-IC methylation imprint, the other key epigenetic characteristic of an appropriate model of AS is repression of the paternal UBE3A allele on neural differentiation. Although both UBE3A alleles are expressed in most tissues, the paternally inherited allele is repressed in the brain (4–6). Imprinted expression of UBE3A is thought to occur as a result of reciprocal expression of a long noncoding antisense transcript, UBE3A-ATS, which is part of a >600-kb transcript initiating at the differentially methylated PWS-IC located in exon 1 of the SNURF-SNRPN gene (7–9).
The ability to derive mixed populations of mature neurons from AS and normal iPSCs allowed us to test whether the epigenetic reprogramming process had affected the tissue-specific regulation of UBE3A imprinting. UBE3A expression was assayed by qRT-PCR in iPSCs and iPSC-derived neuron cultures from normal patients and patients with AS. Fig. 4A shows qRT-PCR for UBE3A expression in AS and normal iPSCs and iPSC-derived neurons. Reduced levels of UBE3A expression were observed in both AS iPSCs and iPSC-derived neuron cultures relative to their normal counterparts. Furthermore, UBE3A expression in AS iPSC-derived neurons is decreased relative to AS iPSCs, demonstrating its epigenetic repression on neuronal differentiation. Conversely, UBE3A expression levels do not change between normal iPSCs and iPSC-derived neurons, despite the apparent repression of the paternal UBE3A allele.
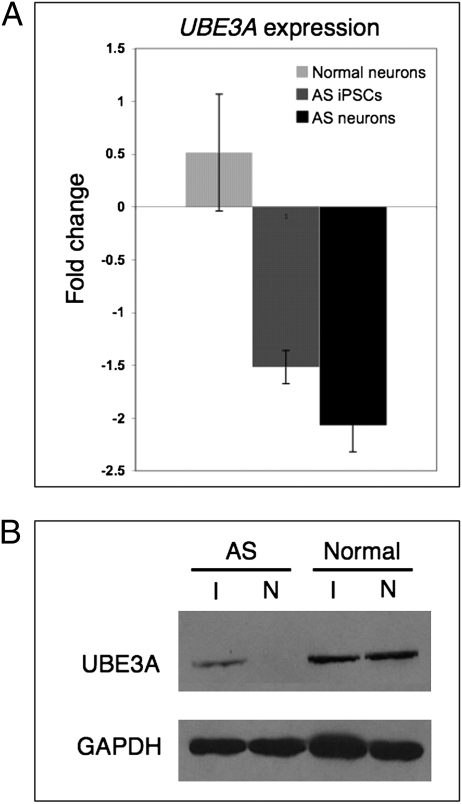
Fig. 4.
Paternal UBE3A is repressed in AS iPSC-derived neurons. (A) qRT-PCR analysis of UBE3A expression in AS and normal iPSCs and iPSC-derived neurons. Gene expression is normalized to GAPDH and is presented as the fold change relative to UBE3A expression levels in normal iPSCs. Error bars indicate SD for three independent cultures. (B) Western blot analysis of normal and AS iPSCs (I) and 10-wk-old iPSC-derived neurons (N).
To determine whether the repressed paternal UBE3A allele leads to reduced UBE3A protein levels in our in vitro model of AS, Western blot analysis was performed on AS and normal iPSCs and iPSC-derived neuron cultures (Fig. 4B). Similar to qRT-PCR results, UBE3A protein is markedly reduced in AS compared with normal iPSCs. Consistent with tissue-specific imprinting, UBE3A protein levels were further reduced in AS iPSC-derived neuronal cultures compared with AS iPSCs. UBE3A protein levels were similar between normal iPSCs and iPSC-derived neuron cultures. In our iPSC model system, the paternal UBE3A allele is significantly repressed in neuronal cells derived from AS iPSCs following 10 wk of differentiation, recapitulating the main epigenetic characteristic of the disease in vitro.
Neuron-Specific Up-Regulation of UBE3A-ATS Correlates with UBE3A Repression.
Although the mechanism of UBE3A repression in the brain has not been entirely elucidated, it does not result from tissue-specific differential DNA methylation of the UBE3A promoter because the CpG-rich promoter of UBE3A is unmethylated in all human and mouse tissues that have been examined, including human postmortem brain tissue (1). We confirmed that the paternal UBE3A promoter is unmethylated in iPSC-derived neurons as well as in fibroblasts and iPSCs (Fig. S8). We conclude that the silencing of the paternal UBE3A allele in iPSC-derived neurons is not the result of spurious methylation of the UBE3A promoter during the reprogramming process but, rather, that it reflects the normal mechanism of UBE3A imprinting in the brain.
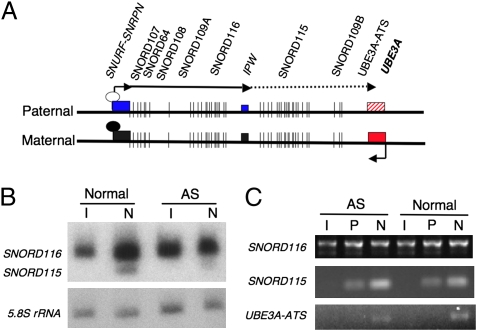
There is evidence to suggest that brain-specific UBE3A repression is mediated by a long noncoding antisense RNA (7–9) (Fig. 5A). To determine whether the UBE3A antisense transcript UBE3A-ATS displays the expected tissue specificity of expression, the downstream noncoding exons of SNURF-SNRPN were analyzed. Northern blot hybridization was used to assess expression of the processed snoRNAs, SNORD116 (also called HBII-85) and SNORD115 (also called HBII-52), demonstrating that SNORD116 is expressed in both iPSCs and neurons, whereas SNORD115 expression is restricted to iPSC-derived neurons (Fig. 5B), consistent with other reports that it is specifically expressed in the brain (11). RT-PCR using primers to recognize spliced exons in the SNORD116 cluster, the SNORD115 cluster, and UBE3A-ATS (9) confirmed this finding (Fig. 5C). These data further indicate that although the coding and proximal noncoding SNURF-SNRPN exons are expressed and processed in human pluripotent stem cells, more distal exons become increasingly expressed and processed as in vitro neural development progresses. Because the UBE3A-ATS transcript, which overlaps UBE3A, is only detected in the neuron cultures, we conclude that the neuron-specific repression of UBE3A may occur relatively late during neurogenesis, coincident with up-regulation of SNORD116, SNORD115, and UBE3A-ATS.
Fig. 5.
UBE3A-ATS is only expressed in neuron cultures and correlates with paternal UBE3A repression. (A) Map of the SNURF-SNRPN and UBE3A transcripts. White and black oval-shaped circles indicate the unmethylated and methylated PWS-IC, respectively. The solid line with arrows indicates transcripts expressed in iPSCs, whereas the dotted line indicates neural-specific transcripts. AS iPSCs and their neural derivatives lack the entire maternally inherited allele of this region. (B) Northern blot using total RNA from AS and normal iPSCs (I) and iPSC-derived 10-wk-old neurons (N) shows expression of the snoRNAs SNORD116 and SNORD115 during in vitro neural development. (C) RT-PCR using primers spanning multiple exons of SNORD116, SNORD115, and UBE3A-ATS in iPSCs (I), 3-wk-old iPSC-derived neural precursor cells (P), and 10-wk-old iPSC-derived neuron cultures (N).
Discussion
iPSCs may provide robust models of complex human genetic disorders (22–24). One impediment to the use of iPSCs to model human disorders involving genomic imprinting is the possibility that the imprint would either be erased during reprogramming or not maintained during culture of iPSCs. Our data (Fig. 2 and Fig. S4), indicating that the methylation imprint at the PWS-IC is not erased or reset during the nuclear reprogramming process in both male and female iPSC lines, supports the widely accepted model that the PWS-IC methylation imprint is erased in the germline, established either in the germline or during the early stages of preimplantation development, and then maintained throughout development (25–28). These findings suggest that the PWS-IC methylation imprint is refractory to the global erasure of epigenetic marks induced by the reprogramming process. Interestingly, this imprint is also stably maintained during long-term culture of both human and murine ES cells (29–31).
Although we observed variability in the neural differentiation potential of our AS iPSCs, we were able to demonstrate that we could produce live functional AS neurons in culture. Importantly, these neurons showed AMPA receptor-mediated EPSCs. Recently, Greer et al. (32) demonstrated that excitatory synapses in murine neurons with reduced UBE3A have fewer AMPA receptors and reduced AMPA receptor function. Further electrophysiological and immunocytochemical studies of mature iPSC-derived neurons are underway to determine whether AMPA receptor localization and function differ between AS and normal cells in our human neuron cultures.
One of the most striking findings of our study is that the expression of the downstream noncoding SNURF-SNRPN exons becomes more neuron-specific in a proximal-to-distal fashion. This is different from mouse models, where all the downstream noncoding exons of Snurf-Snrpn are brain-specific (11). Evidence suggests that the regulation of this transcript and, more specifically, the UBE3A-ATS portion of the transcript plays an important role in the repression of paternal UBE3A (7–9). The coordinated expression of paternal UBE3A-ATS and repression of paternal UBE3A in the AS iPSCs during in vitro neurogenesis allow us to use this model to study the regulation of UBE3A-ATS processing and its effects on the chromatin structure of the paternal UBE3A promoter during human neural development.
Our human cell culture model of AS recapitulates the tissue-specific pattern of UBE3A imprinting, and thus provides an important tool to address the timing and mechanism of epigenetic UBE3A silencing during human neural development. The in vitro AS model system described here will also be useful to characterize the physiological abnormalities of the disease at a cellular level, including electrophysiological analyses of AS iPSC-derived neurons. The in vitro development of AMPA receptor-mediated spontaneous synaptic activity indicates that these iPSC-derived neurons can be used to study synaptic structure and development in AS neurons. Our iPSC model of PWS will be useful in further investigating the genetic basis of PWS, including how loss of the several species of snoRNAs contributes to the disorder. We also plan to use iPSC technology to model chromosome 15q11-q13 duplication-associated autism. The cell culture models of these and other human neurogenetic disorders will provide important tools to advance the understanding of disease mechanisms and to develop unique tools for drug discovery.
Materials and Methods
Patient-Specific Fibroblasts.
Fibroblasts from patient ASdel1 isolated and provided by Bruce Korf (Boston Children's Hospital, Boston, MA) in 1995 were subsequently stored as anonymized tissue in liquid nitrogen. FISH was performed to confirm the 15q11-q13 deletion before reprogramming. ASdel2, PWSdel1, and normal fibroblasts were obtained from the Repository for Mutant Human Cell Strains at McGill University Health Centre/Montreal Children's Hospital (Montreal, QC, Canada) and are cell line numbers WG1631, WG1534, and MCH065, respectively.
Cell Culture.
Human fibroblasts, mouse embryonic fibroblasts (MEFs), and 293T/17 cells were maintained in DMEM (Invitrogen) supplemented with 10% (vol/vol) FCS. iPSCs were cultured in conventional hESC medium consisting of DMEM-F12 (Invitrogen) supplemented with 20% (vol/vol) knock-out serum replacer, nonessential amino acids, 1 mM L-glutamine, 100 mM β-mercaptoethanol, and 4 ng/mL basic FGF. Cells were maintained in a humid incubator at 37 °C with 5% (vol/vol) CO2. iPSCs were passaged approximately once a week by manually cutting colonies with a needle.
Retrovirus Production.
pMXs retroviral vectors carrying the inserts for OCT4 (plasmid 17217), SOX2 (plasmid 17218), KLF4 (plasmid 17219), and C-MYC (plasmid 17220) were obtained from Addgene. A pBABE-GFP retroviral vector carrying LIN28 was generated by subcloning PCR-amplified LIN28 cDNA between the SnaBI and EcoRI sites. Clones were verified by restriction digestion and sequenced.
The retroviral packaging construct, pMDL.Gagpol, was a gift from Alexander Lichtler (University of Connecticut Health Center, Farmington, CT), and the envelope plasmid, pMD2.G, was obtained from Addgene (plasmid 12259). Retroviruses were produced in 293T/17 cells by transfection using Lipofectamine 2000 (Invitrogen). Virus-containing supernatants were collected at 24 and 48 h posttransfection and used directly for reprogramming or frozen for later use.
iPSC Generation.
iPSCs were generated following previously published protocols (12). Specifically, two serial transductions using equal volumes of the five retrovirus-containing supernatants were performed over 48 h. Six days after the first transduction, 105 transduced fibroblasts were plated to an irradiated MEF feeder layer on a 10-cm2 dish. Transduced fibroblasts were fed with hESC medium every other day. Colonies with hESC morphology (very tight and very round) were picked to freshen irradiated MEF feeder layers after ≈4 wk.
Neuronal Differentiation.
Neuronal differentiation was carried out according to published protocols (18, 19) with minor changes. Specifically, iPSCs were manually cut and lifted from the mouse fibroblast feeder layer rather than being dissociated with Dispase (Invitrogen).
The normal neurons used in our study were generated from passage 20 iPSCs and were at least 10 wk old; the AS neurons were cultured from passage 15–20 iPSCs and were also at least 10 wk old for all experiments shown. Neural precursor cells were taken after 3 wk of culture, before the second plating on laminin-coated dishes.
Methylation-Specific PCR.
To perform methylation-specific PCR, genomic DNAs were subjected to bisulfite conversion using the EZ DNA Methylation-Gold Kit (Zymo Research) according to the manufacturer's instructions. Bisulfite-converted DNA was used to seed a multiplex PCR using the following primers: 15maternalF, 5′-TAATAAGTACGTTTGCGCGGTC-3′; 15maternalR, 5′-AACCTTACCCGCTCCATCGCG-3′; 15paternalF, 5′-GTAGGTTGGTGTGTATGTTTAGGT-3′; and 15paternalR, 5′-ACATCAAACATCTCCAACAACCA-3′. Maternal and paternal controls were performed by using maternal or paternal primer sets alone on previously amplified templates. The maternal fragment size is 174 bp, and the paternal fragment size is 100 bp.
Immunocytochemistry.
Immunocytochemistry was performed on iPSCs and neurons according to previously published methods (33), with minor changes. Specifically, coverslips destined for staining with nonnuclear markers were not permeablized with cytoskeletal buffer (CSK) before fixation, and coverslips to be stained with cell surface markers were not permeabilized. Coverslips stained with GAD65/67 and TUJ1 were fixed in methanol for 10 min at −20 °C and then rehydrated for 5 min in PBS instead of paraformaldehyde fixation. Stained slides were visualized by fluorescence microscopy and imaged using a Zeiss Axiovision microscope at magnifications of 5×, 20×, and 63×.
Electrophysiology.
Cells in culture grown on individual coverslips were transferred to the recording chamber (room temperature) fixed to the stage of an Olympus BX51WI upright microscope fitted with a 40× water-immersion objective lens. During recordings, coverslips were continuously perfused at 2 mL/min with bath solution. The pH was equilibrated by continuous bubbling with 95% (vol/vol) O2–5% (vol/vol) CO2. Whole-cell voltage-clamp [Vhold (holding voltage) = −70 mV] and current-clamp recordings were obtained from the cells in culture using previously established techniques (34). All electrical events were filtered at 2.9 kHz and digitized at ≥6 kHz using a HEKA ITC-16 digitizer built into an EPC9/2 amplifier (Heka Elektronic). Series resistance was compensated 70% at a lag of 10–100 μs. Input resistance was monitored with 5-mV (50-ms) hyperpolarizing voltage steps. Off-line analysis was carried out using Clampfit (Axon Instruments).
Supplementary Material
Acknowledgments
We thank Dr. David S. Rosenblatt and Gail Dunbar for assisting with identification of the appropriate normal, AS del2, and PWS del1 fibroblast samples from the Canadian Repository for Mutant Human Cell Strains and Dr. Bruce R. Korf (University of Alabama at Birmingham) for generously providing AS del1. We thank Dr. Ren-He Xu and the staff of the University of Connecticut Stem Cell Core for advice and support. We thank Dr. Peter Benn, Judy Delach, Karen Ronski, Dounia Djeghloul, Nicholas Jannetty, and Heather Glatt-Deeley for their assistance. This study was supported by an endowment to S.J.C. from the Raymond and Beverly Sackler foundation and by the State of Connecticut under the Connecticut Stem Cell Research Grants Program (Grant 09SCAUCHC14 to S.J.C.). The work is also supported by a grant from the Foundation for Prader–Willi Research (to M.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004487107/-/DCSupplemental.
References
- 1.Lossie AC, et al. Distinct phenotypes distinguish the molecular classes of Angelman syndrome. J Med Genet. 2001;38:834–845. doi: 10.1136/jmg.38.12.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams CA, et al. Angelman syndrome 2005: Updated consensus for diagnostic criteria. Am J Med Genet A. 2006;140:413–418. doi: 10.1002/ajmg.a.31074. [DOI] [PubMed] [Google Scholar]
- 3.Cassidy SB, Driscoll DJ. Prader-Willi syndrome. Eur J Hum Genet. 2009;17:3–13. doi: 10.1038/ejhg.2008.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- 5.Vu TH, Hoffman AR. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat Genet. 1997;17:12–13. doi: 10.1038/ng0997-12. [DOI] [PubMed] [Google Scholar]
- 6.Albrecht U, et al. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- 7.Rougeulle C, Cardoso C, Fontés M, Colleaux L, Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 8.Chamberlain SJ, Brannan CI. The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics. 2001;73:316–322. doi: 10.1006/geno.2001.6543. [DOI] [PubMed] [Google Scholar]
- 9.Runte M, et al. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 10.de Smith AJ, et al. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum Mol Genet. 2009;18:3257–3265. doi: 10.1093/hmg/ddp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavaillé J, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 14.Colman A, Dreesen O. Pluripotent stem cells and disease modeling. Cell Stem Cell. 2009;5:244–247. doi: 10.1016/j.stem.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Pick M, et al. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–2690. doi: 10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- 16.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosaki K, McGinniss MJ, Veraksa AN, McGinnis WJ, Jones KL. Prader-Willi and Angelman syndromes: Diagnosis with a bisulfite-treated methylation-specific PCR method. Am J Med Genet. 1997;73:308–313. [PubMed] [Google Scholar]
- 18.Pankratz MT, et al. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25:1511–1520. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson MA, Weick JP, Pearce RA, Zhang SC. Functional neural development from human embryonic stem cells: Accelerated synaptic activity via astrocyte coculture. J Neurosci. 2007;27:3069–3077. doi: 10.1523/JNEUROSCI.4562-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu BY, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore AR, et al. Electrical excitability of early neurons in the human cerebral cortex during the second trimester of gestation. Cereb Cortex. 2009;19:1795–1805. doi: 10.1093/cercor/bhn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee G, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moretti A, et al. Patient-Specific Induced Pluripotent Stem-Cell Models for Long-QT Syndrome. N Engl J Med. 2010;6:407–411. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 24.Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obata Y, Kono T. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J Biol Chem. 2002;277:5285–5289. doi: 10.1074/jbc.M108586200. [DOI] [PubMed] [Google Scholar]
- 26.Kimura Y, Tateno H, Handel MA, Yanagimachi R. Factors affecting meiotic and developmental competence of primary spermatocyte nuclei injected into mouse oocytes. Biol Reprod. 1998;59:871–877. doi: 10.1095/biolreprod59.4.871. [DOI] [PubMed] [Google Scholar]
- 27.Brannan CI, Bartolomei MS. Mechanisms of genomic imprinting. Curr Opin Genet Dev. 1999;9:164–170. doi: 10.1016/S0959-437X(99)80025-2. [DOI] [PubMed] [Google Scholar]
- 28.El-Maarri O, et al. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet. 2001;27:341–344. doi: 10.1038/85927. [DOI] [PubMed] [Google Scholar]
- 29.Schumacher A, Doerfler W. Influence of in vitro manipulation on the stability of methylation patterns in the Snurf/Snrpn-imprinting region in mouse embryonic stem cells. Nucleic Acids Res. 2004;32:1566–1576. doi: 10.1093/nar/gkh322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Hum Mol Genet. 2007;16(Spec No. 2):R243–R251. doi: 10.1093/hmg/ddm245. [DOI] [PubMed] [Google Scholar]
- 31.Kim KP, et al. Gene-specific vulnerability to imprinting variability in human embryonic stem cell lines. Genome Res. 2007;17:1731–1742. doi: 10.1101/gr.6609207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greer PL, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fortin DA, Levine ES. Differential effects of endocannabinoids on glutamatergic and GABAergic inputs to layer 5 pyramidal neurons. Cereb Cortex. 2007;17:163–174. doi: 10.1093/cercor/bhj133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.