Abstract
IκBζ, encoded by Nfibiz, is a nuclear IκB-like protein harboring ankyrin repeats. IκBζ has been shown to regulate IL-6 production in macrophages and Th17 development in T cells. However, the role of IκBζ in natural killer (NK) cells has not be understood. In the present study, we found that the expression of IκBζ was rapidly induced in response to IL-18 in NK cells, but not in T cells. Analysis of Nfkbiz−/− mice revealed that IκBζ was essential for the production of IFN-γ production and cytotoxic activity in NK cells in response to IL-12 and/or IL-18 stimulation. IL-12/IL-18–mediated gene induction was profoundly impaired in Nfkbiz−/− NK cells. Whereas the phosphorylation of STAT4 was normally induced by IL-12 stimulation, STAT4 was not recruited to the Ifng gene regions in Nfkbiz−/− NK cells. Acetylation of histone 3 K9 on Ifng regions was also abrogated in Nfkbiz−/− NK cells. IκBζ was recruited on the proximal promoter region of the Ifng gene, and overexpression of IκBζ together with IL-12 activated the Ifng promoter. Furthermore, Nfkbiz−/− mice were highly susceptible to mouse MCMV infection. Taken together, these results demonstrate that IκBζ is essential for the activation of NK cells and antiviral host defense responses.
Keywords: IFN-γ, transcription, virus infection, STAT4
Antiviral host defense is mediated by coordinated action of multiple cell types, such as innate immune cells, T cells, and natural killer (NK) cells (1, 2). In response to virus infection, NK cells induce apoptosis of infected cells by expressing perforins and granzymes and produce high amounts of IFN-γ (3–5). Various cytokines are important for activating NK cells to produce IFN-γ and increasing their cytotoxic activity. Type I IFNs produced from innate immune cells as well as cytokines such as IL-2, IL-12, IL-15, IL-18, and IL-21 are critical for NK-cell activation. Furthermore, NK-cell receptors harboring immunoreceptor tyrosine-based activation motifs (ITAMs) or those interacting with ITAM-containing adaptor proteins induce cytolytic activity and IFN-γ production (4). For instance, antibody-coated target cells are recognized by FcgRIII, which mediates antibody-dependent cellular cytotoxicity and IFN-γ production.
The expression of the Ifng gene is one of the hallmarks of NK-cell activation. Because Ifng is also highly expressed in T helper 1 (Th1) cells, the mechanisms of the Ifng promoter activation has been extensively studied in Th1 cells and NK cells (6, 7). Stimulation of NK cells with IL-12 and IL-18 leads to production of IFN-γ as well as elevation of cytotoxic activity (8, 9). It has been shown that sequence elements conserved among species (also known as conserved noncoding sequences, CNSs) present up to 100 kb from the transcription start site (TSS) of Ifng are involved in controlling the expression of IFN-γ (7, 10–12). Various transcription factors are recruited to the CNS of Ifng. IL-12 induces phosphorylation and nuclear translocation of STAT4, which is recruited to the cis-regulatory region of Ifng (7, 13, 14). IFN-γ–inducible T-box transcription factor (T-bet) is critical for inducing Ifng by controlling histone acetylation in Th1 cells (11, 15, 16). T-bet is also required for the development of NK cells (17). In T cells, T-bet–dependent chromatin remodeling of the Ifng locus induces recruitment of the NF-κB p65 subunit to Ifng cis-elements (12).
IL-18 is structurally related to IL-1β and known to work together with IL-12 to activate Th1 cells to produce IFN-γ (18). However, IL-18 does not induce Th1 development by itself, unlike IL-12. Recent reports show that IL-18 is involved in the differentiation of Th2 cells and Th17 cells in murine experimental models, in addition to Th1 (18, 19). Although IL-18 is reported to activate AP-1 via MAP kinases to express Ifng (20), the mechanism of how IL-18 potentiates IFN-γ production in NK cells is yet to be clarified.
IκBζ, also known as INAP or MAIL, is a nuclear factor belonging to the Bcl-3 family, which contains a nuclear localization domain in the N terminus and C-terminal ankylin repeats (21). IκBζ is encoded by the Nfkbiz gene, and the expression of Nfkbiz is rapidly induced in response to various Toll-like receptor (TLR)/IL-1 receptor (IL–1R) stimuli in macrophages (22). The expressed IκBζ interacts with NF-κB p50 subunit and positively regulates expression of a set of genes including Il6, Il12b, Csf2, Defb2, Lcn2, and so on (22, 23). In addition to NF-κB binding sequences, flanking C/EBP-binding sites are also important for IκBζ-mediated control of transcription (23). Furthermore, a recent study shows that IκBζ is required for chromatin remodelling downstream of TLR stimuli (24). Recently, it was shown that T cells lacking IκBζ showed severe defects in development of Th17 cells, and IκBζ regulates IL-17 expression by cooperating with Rorgt (25). On the other hand, a report showed that overexpression of IκBζ induced Ifng in a cell line, although the mechanism was not understood (26). Nevertheless, it is unclear whether IκBζ plays any role in the activation of NK cells.
In the present study, we found that IκBζ was required for the activation of NK cells in response to IL-12 and IL-18. IL-12/IL-18–mediated gene expression including Ifng was profoundly impaired in Nfkbiz−/− NK cells. IκBζ was recruited to the proximal promoter region of Ifng and able to transactivate the Ifng together with IL-12. Furthermore, Nfkbiz−/− mice were highly susceptible to infection with mouse CMV (MCMV), a model virus known to be cleared by NK cells. Taken together, IκBζ plays an essential role in the activation of NK cells by triggering dynamic transcription factor recruitment and chromatin remodeling.
Results
IκBζ Expression Is Induced in Response to IL-18 in NK Cells.
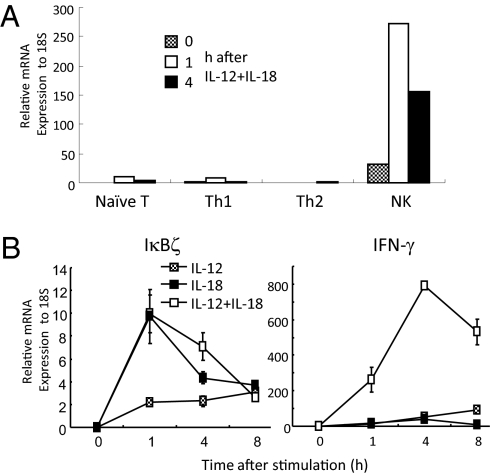
To investigate whether IκBζ plays a role in controlling cells other than macrophages, we first examined the expression levels of IκBζ in NK cells and T cells. We prepared splenic NK, and naïve T cells. Then, T cells were cultured in the presence of either IL-12, anti-IL-4 Ab, and anti-CD3 to develop Th1 cells or of IL-4, anti-IL-12, and anti-CD3 to develop Th2 cells. Quantitative-PCR (Q-PCR) analysis using RNAs prepared from these cells revealed that NK cells expressed the IκBζ gene higher than naïve T cells (Fig. 1A). Polarization to Th1 or Th2 cells did not increase the expression of Nfkbiz. The expression of IκBζ was highly induced in response to IL-12 and IL-18 stimulation in NK cells (Fig. 1A). Next, we examined kinetics of the IκBζ expression in NK cells in response to IL-12, IL-18, or both. IκBζ expression was induced at 1 h after stimulation with IL-18 alone or after costimulation with IL-12 and IL-18, and gradually decreased by 8 h after stimulation (Fig. 1B). In contrast, IL-12 alone slowly increased Nfkbiz about twofold at 8 h after stimulation. On the other hand, Ifng expression was increased in response to both IL-12 and IL-18, but not to either cytokine alone, and the expression peaked at 4 h after stimulation in NK cells (Fig. 1B). These results demonstrate that IκBζ expression is higher in NK cells than in T cells, and that this is an early induced gene in response to IL-18 stimulation in NK cells.
Fig. 1.
Expression of IκBζ in NK cells in response to IL-12 and IL-18 stimulation. (A) The expression of IκBζ in naïve T, Th1, Th2 cells, and NK cells in response to IL-12 and IL-18. (B) NK cells were stimulated with IL-12 and/or IL-18 for the indicated periods, and the expression levels of IκBζ and IFN-γ were determined by Q-PCR analysis. Data represent means ± SD of triplicates. Similar results were obtained in three independent experiments.
IκBζ Is Critical for Activation of NK Cells in Response to IL-12 and IL-18.
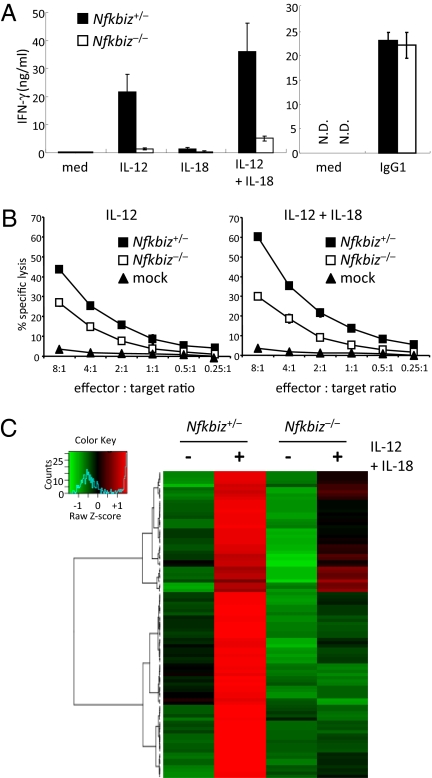
The expression of IκBζ in NK cells prompted us to investigate a functional role of IκBζ in NK cells by using IκBζ-deficient (Nfkbiz−/−) mice. Stimulation of Nfkbiz+/− splenic NK cells with IL-12, or costimulation with IL-12 and IL-18, induced production of IFN-γ (Fig. 2A). In contract, IFN-γ production was severely impaired in Nfkbiz−/− NK cells in response to IL-12, and IL-12 together with IL-18. These results indicated that IκBζ is indispensable for IFN-γ production in response to IL-12 and IL-18. Production of IFN-γ in response to culture with immobilized IgG1 was comparable in wild-type NK cells (Fig. 2A), indicating that signaling pathways from FcgRIII to induce Ifng expression is not affected in the absence of IκBζ. We then analyzed cytotoxic activity of NK cells to IL-12 and IL-18 stimulation by a standard 51Cr release assay against YAC1 target cells. Cytotoxic activity of NK cells stimulated with IL-12 alone or costimulated with IL-12 and IL-18 was reduced in Nfkbiz−/− mice (Fig. 2B). IL-18 alone failed to increase cytotoxic activity even in wild-type NK cells (data not shown). These results demonstrate that IκBζ is critical for activation of NK cells in response to IL-12 and IL-18.
Fig. 2.
Role of IκBζ in the activation of NK cells in response to IL-12 and IL-18 stimulation. (A) Splenic NK cells from Nfkbiz+/− and Nfkbiz−/− mice were stimulated with 1 ng/mL IL-12, 10 ng/mL IL-18, and a combination of IL-12 and IL-18 or the cells were cultured on the plates coated with IgG1 for 24 h. The concentrations of IFN-γ in the culture supernatant were measured by ELISA. Data represent means ± SD. Similar results were obtained in three independent experiments. (B) IL-2–induced splenic NK cells were cultivated with IL-12 together with or without IL-18 for 4 h. NK-cell cytotoxicities against YAC-1 cells were measured by a 51Cr release assay at different E:T ratios. The percentages of specific cell lysis are indicated. Data represent the mean ± SD of duplicate measurements. The data shown are representative of two independent experiments. (C) Splenic NK cells from Nfkbiz+/− and Nfkbiz−/− mice were stimulated with IL-12 and IL-18 for 24 h, and total RNA was subjected to microarray analysis using Affymetrix mouse genome 430 2.0 microarray chips. The data were normalized by robust multichip analysis (RMA), and genes up-regulated more than twofold in wild-type NK cells after stimulation were selected. The genes were hierarchically clustered as follows: The expression values for each gene were standardized as average = 0 and SD = 1. Complete linkage method was then applied using absolute value of Pearson correlation as distance. The resulted heatmap, dendrogram, and a color key are shown. A blue line in the color key indicates the frequency distribution colors in the heatmap. For the gene lists shown in this figure, see Dataset S1.
IκBζ Is Dispensable for NK-Cell Development.
We next analyzed the differentiation of splenic NK cells in Nfkbiz−/− mice. It is known that NK-cell progenitors initially express CD122 (IL15R), CD127 (IL7R), and c-kit (27). Upon maturation, NK cells down-regulate CD127 and c-kit. The DX5+CD3− NK-cell population in spleen was comparable between Nfkbiz+/− and Nfkbiz−/− mice (Fig. S1). Furthermore, CD122 was highly expressed, and c-kit and CD127 expressions were comparably suppressed both in wild-type and Nfkbiz−/− splenic NK cells (Fig. S1). Collectively, IκBζ is dispensable for controlling differentiation of NK cells.
Profound Impairment in Gene Expression in Nfkbiz−/− NK Cells in Response to IL-12 and IL-18 Stimulation.
Next we examined the role of IκBζ in IL-12 and IL-18–mediated gene expression in NK cells. Wild-type and Nfkbiz−/− NK cells were stimulated with IL-12 and IL-18 for 24 h, and gene expression profile was assessed by microarray analysis. In response to stimulation with IL-12 and IL-18, 96 genes were up-regulated more than twofold in wild-type NK cells. We then hierarchically clustered genes induced by IL-12 and IL-18 and found that they are roughly classified into those completely (58 genes) or partially (38 genes) dependent on IκBζ (Fig. 2C) (Dataset S1). Genes completely dependent on IκBζ include Ifng, Il22, Ccr5, and Il18r1. These data demonstrate that IκBζ is critical for IL-12 and IL-18–mediated gene expression in NK cells.
IκBζ Promotes STAT4 Accessibility in IFN-γ Genes in Response to IL-12 and IL-18 in NK Cells.
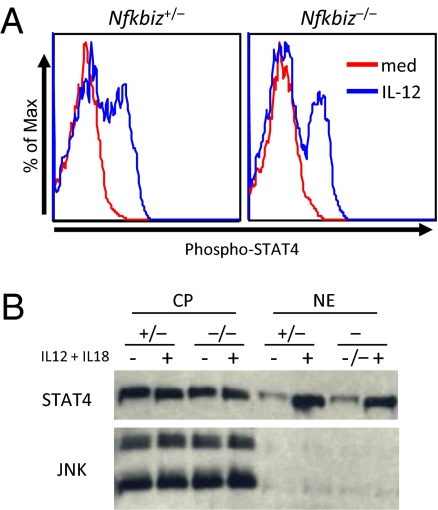
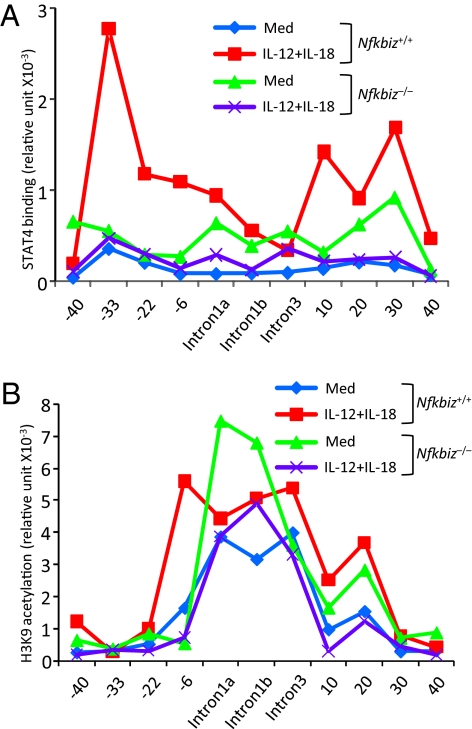
IL-12 activates Janus kinases (JAKs) that phosphorylate STAT4 (14). STAT4 is localized in the cytoplasm in the resting cells, and phosphorylated STAT4 translocates to the nucleus for the transcriptional activation. Treatment with IL-12 greatly increased NK cells stained with anti-phospho-STAT4 (p-STAT4), and the levels of p-STAT4 positive cells were comparable between Nfkbiz+/− and Nfkbiz−/− mice (Fig. 3A). Furthermore, nuclear translocation of STAT4 was comparably induced in Nfkbiz+/− and Nfkbiz−/− NK cells in response to stimulation with IL-12 and IL-18 (Fig. 3B), indicating that IκBζ was dispensable for phosphorylation and nuclear translocation of STAT4. We then examined recruitment of STAT4 to Ifng CNSs by chromatin immunoprecipitation (ChIP) coupled with Q-PCR (ChIP-Q-PCR) analysis. We found that STAT4 was widely recruited to CNSs (−33 kb, −22 kb, −6 kb, intron 1a, +10 kb, +20 kb, and +30 kb from the TSS) of Ifng in response to IL-12 and IL-18 stimulation in wild-type NK cells (Fig. 4A). In contrast, STAT4 recruitment to the Ifng conserved elements was severely impaired in Nfkbiz−/− NK cells through the CNS (Fig. 4A). These results suggested that IκBζ promotes STAT4 accessibility to the CNS of Ifng gene in stimulated NK cells.
Fig. 3.
Nuclear translocation of STAT4 in NK cells in the absence of IκBζ. (A) Splenic NK cells were stimulated with IL-12 for 30 min. Phosophorylation of STAT4 was then determined by intracellular staining of cells with anti-phospho-STAT4 Ab followed by flow cytometry. (B) NK cells were stimulated with IL-12 and IL-18 for 1 h. Cytoplasmic (CP) and nuclear (NE) proteins were prepared and immunoblotted using anti STAT4 Ab and anti-JNK as a control.
Fig. 4.
Requirement of IκBζ in the recruitment of STAT4 and acetylation of H3K9 in NK cells in response to IL-12 and IL-18. Chromatin was isolated from wild-type and Nfkbiz−/− NK cells stimulated with IL-12 and IL-18 for 4 h; recruitment of STAT4 (A) and acetylation of H3K9 (B) to indicated Ifng gene regions was determined by ChIP-QPCR analysis. The data are representative of two independent experiments.
IκBζ Is Required for Change in Histone 3 Lysine 9 Acetylation in Response to IL-12 and IL-18 in NK Cells.
It has been shown that histones of the Ifng loci were hyperacetylated even in the absence of stimulation in NK cells, compared with T cells (10). We performed ChIP analysis with anti-acetyl histone 3 lysine 9 (H3K9) antibody to assess H3K9 acetylation in Ifng CNS. The analysis revealed that intron regions of Ifng were hyperacetylated even without stimulation in wild-type and Nfkbiz−/− NK cells (Fig. 4B). Whereas H3K9 acetylation levels at the −6-kb region of Ifng were up-regulated in response to IL-12 and IL-18 in wild-type NK cells, Nfkbiz−/− NK cells failed to induce H3K9 acetylation (Fig. 4B). These results suggest that IκBζ is required for change in H3K9 acetylation on Ifng loci in response to IL-12 and IL-18 stimulation.
IκBζ Is Recruited to the Ifng Proximal Promoter Region.
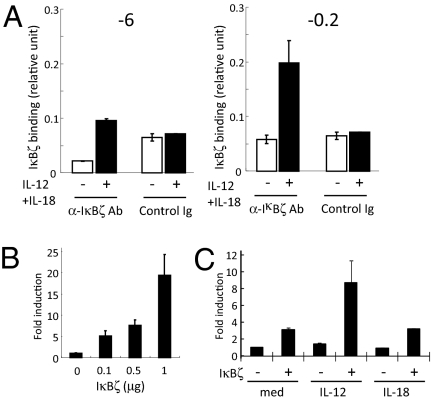
To examine the recruitment of IκBζ to the Ifng promoter, we examined ChIP analysis using anti-IκBζ antibody. In contrast to STAT4 recruitment or H3K9 acetylation, IκBζ was not recruited to the −6-kb region of Ifng locus in NK cells in response to IL-12 and IL-18 (Fig. 5A). In contrast, IκBζ was recruited to the proximal promoter region (−0.2 kb from the TSS) of Ifng (Fig. 5A). IκBζ failed to associate with other CNSs such as −22 kb and intron 1a (data not shown). On the other hand, STAT4 was not recruited to the proximal Ifng promoter (data not shown). To investigate whether IκBζ directly regulates Ifng through binding to the proximal promoter region, we expressed a reporter construct with the human Ifng promoter region (−3.6 kb to +70 k) linked to the luciferase gene, together with IκBζ in EL4 cells. As shown in Fig. 5B, overexpression of IκBζ enhanced luciferase reporter activity in a dose-dependent manner. Furthermore, stimulation with IL-12, but not IL-18, synergistically activated the Ifng promoter with overexpression of IκBζ (Fig. 5C), suggesting that IκBζ expression facilitates IL-12–mediated Ifng promoter activation. These observations suggest that the recruitment of IκBζ to the proximal promoter region is responsible for the transcriptional activation of Ifng.
Fig. 5.
Regulation of Ifng proximal promoter region by IκBζ in NK cells. (A) Chromatin was isolated from splenic NK cells stimulated with IL-12 and IL-18 for 4 h, recruitment of IκBζ to indicated Ifng gene regions was determined by ChIP-QPCR analysis. (B) A 3.6-kb fragment of the human Ifng promoter luciferase reporter construct was transfected to EL4 cells with increasing amounts of IκBζ construct. The luciferase activity was measured 18 h after transfection. (C) EL4 cells were cotransfected with the Ifng promoter reporter construct and IκBζ, followed by stimulation with IL-12 or IL-18. The luciferase activity was measured 18 h after stimulation.
Essential Role of IκBζ in Host Defense Against MCMV Infection.
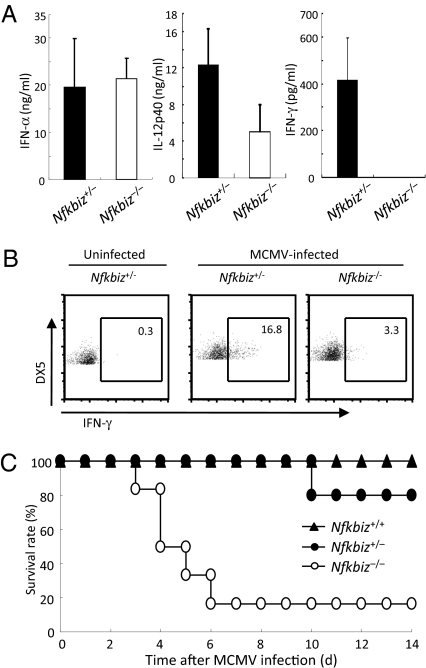
It is known that NK cells play an important role in host defense against MCMV infection (1, 5, 28). MCMV-induced activation of innate immune cells such as dendritic cells (DCs) induces the production of type I IFNs as well as IL-12, leading to activation of NK cells to produce IFN-γ. To examine the role of IκBζ in activating NK cells in vivo in response to virus infection, we intraperitoneally inoculated MCMV into wild-type and Nfkbiz−/− mice. MCMV-induced production of IL-12p40, but not IFN-α, in the sera was partially impaired in Nfkbiz−/− mice compared with wild-type mice (Fig. 6A), suggesting that IκBζ plays a role in expressing IL-12p40 in innate immune cells in response to MCMV. In contrast, the production of IFN-γ in the sera was completely abrogated in Nfkbiz−/− mice (Fig. 6A). Furthermore, flow cytometric analysis revealed that the expression of IFN-γ in splenic NK cells was severely impaired in MCMV-infected Nfkbiz−/− mice (Fig. 6B), although NK-cell cytotoxicity was not altered in Nfkbiz−/− NK cells (data not shown). Although all wild-type mice and 80% of the Nfkbiz+/− mice survived, Nfkbiz−/− mice were highly susceptible and more than 80% of the Nfkbiz−/− mice died within 6 d after MCMV infection (Fig. 6C). Taken together, IκBζ is essential for host defense against MCMV infection by inducing IFN-γ production from NK cells.
Fig. 6.
Essential role of IκBζ in host defense against MCMV infection. (A) Wild-type and Nfkbiz−/− mice (n = 3–5) were intraperitoneally infected with 1 × 105 pfu MCMV, and the concentrations of IFN-α, IFN-γ, and IL-12p40 in the sera 36 h after infection were measured by ELISA. (B) The expression of IFN-γ in splenic NK cells prepared from Nfkbiz+/− and Nfkbiz−/− mice 36 h after MCMV infection was determined by intracellular staining of cells with anti-IFN-γ Ab followed by flow cytometry. (C) Nfkbiz+/+ (n = 5), Nfkbiz+/− (n = 5), and Nfkbiz−/− (n = 6) mice were intraperitoneally infected with MCMV, and the survival of mice was monitored daily. The data are representative of two independent infection experiments.
Discussion
In the present study, we examined the role of IκBζ in NK cells and found that this nuclear protein is essential for inducing a set of genes including Ifng in response to IL-12 and IL-18. IκBζ was rapidly induced in response to IL-18 in NK cells, although the expression level was kept low in Th1 cells. IL-18 is a cytokine that is able to activate NK cells together with IL-12, although the precise mechanisms of the action of IL-18 has not been understood well (18). IL-18R triggers intracellular signaling pathways leading to the activation of NF-κB and MAP kinases (29). We have previously shown that IκBζ is a TLR/IL-1R–inducible gene in macrophages (22). Given that IL-18R and TLR/IL-1R share MyD88 as an adaptor molecule for the signaling, it is not surprising that IL-18 also induces IκBζ expression. IL-18 is known to augment IFN-γ production, when costimulated with IL-12.
IL-12 induces phosphorylation and nuclear translocation of STAT4, which is recruited to cis-regulatory regions of several genes including Ifng (6, 7). It has been shown that sequence elements conserved among species present up to 100 kb from the transcription start site of Ifng are involved in controlling the expression of Ifng (12). These distal elements may loop toward the Ifng gene for regulating its transcription (6, 7). Whereas the H3K9 acetylation as well as recruitment of STAT4 to distal conserved regions of Ifng was abrogated in Nfkbiz−/− NK cells, IκBζ was recruited to a proximal promoter region of Ifng, but not to distal sequences. These results imply that initial recruitment of IκBζ to the proximal region is required for further chromatin remodelling and efficient STAT4 binding. A report showed that IκBζ inhibits STAT3-mediated signaling by direct interaction (30). However, given that STAT4 and IκBζ are recruited to different conserved elements on the Ifng promoter, it seems reasonable to assume that IκBζ and STAT4 do not control the Ifng expression by physically interacting with each other.
IκBζ interacts with NF-κBp50 in macrophages, and the ankyrin repeats of this protein were required for synergistically increasing transcriptional activity (22). Furthermore, IκBζ is reported to be involved in histone modifications on the Lcn2 promoter in LPS-stimulated macrophages (24). A previous report shows that NF-κBp50–deficient mice showed increased, but not decreased, IFN-γ production in response to IL-12 and IL-18 stimulation in NK cells (31). Further, no clear NF-κB binding sequence was found in the proximal promoter region of Ifng. Thus, it is unlikely that NF-κBp50–IκBζ interaction plays a pivotal role in controlling IFN-γ production in NK cells. In Th17 cells, IκBζ was suggested to directly control IL-17 expression by recruiting to the Il17 promoter regions (25). However, clear IκBζ responsive elements were not found in the proximal region of Ifng. Thus, it is possible that IκBζ is directly recruited to Ifng promoter, although further studies are required to address the mechanism of Ifng transactivation by IκBζ.
In contrast to the critical role of IκBζ in NK-cell activation in response to IL-12 and IL-18, Nfkbiz−/− Th1 cells developed in the presence of IL-12 and IL-18 did not show a defect in producing IFN-γ in response to antigen-receptor stimulation (25). In contrast, IκBζ is involved in the development of Th-17 cells (25). We found that IL-18 stimulation failed to up-regulate IκBζ expression in T cells. Although NK cells produced IFN-γ by stimulation with IL-12 and IL-18, Th1 cells require T-cell receptor (TCR) stimulation in addition to culture with these cytokines. In addition to the difference in IκBζ expression levels in NK cells and T cells, differential requirement of antigen-receptor signaling in producing IFN-γ may explain the role of IκBζ in NK cells but not in Th1 cells.
Although IκBζ was essential for IFN-γ production in response to IL-12 and IL-18 stimulation, Nfkbiz−/− NK cells produced IFN-γ normally in response to culture with IgG1, which is known to activate NK cells via FcgRIIIa. In contrast to IL-12 and IL-18, FcgRIII activates the intracellular signaling via recruiting ITAM-motif containing adaptors activating Syk tyrosine kinase and ZAP-70 (32). Subsequently, phospholipase C (PLC)-γ is activated to induce Ca2+ influx, MAP kinase activation, and NF-κB activation. The FcgRIII and TCR largely share the signaling molecules, and future studies are required for identifying the mechanisms of how activating FcgRIII and TCR induce production of IFN-γ independent of IκBζ.
NK cells are important for the elimination of MCMV (1, 5). Type I IFNs and IL-12 produced from DC subsets such as plasmacytoid DC and CD8α+ DC are important for the NK-cell cytotoxic activity and production of IFN-γ, respectively (3, 4). Whereas MCMV-mediated production of IFN-α in the sera was normal, IL-12p40 levels were decreased in Nfkbiz−/− mice. Because IκBζ is required for the production of IL-6 and IL-12p40 in response to TLR ligands in macrophages (22), impaired production of IL-12p40 in Nfkbiz−/− mice is likely due to the defect in TLR-mediated MCMV recognition. Although type I IFNs are known to activate NK cells in response to virus infection, it has been shown that IFN-γ production to MCMV infection was not decreased even in the absence of type I IFN production or IFNR signaling. On the other hand, we did not find a defect in MCMV-induced NK-cell cytotoxicity in Nfkbiz−/− mice. Because NK-cell cytotoxicity to MCMV infection was reported to be IL-12–independent, normal activation of NK-cell cytotoxicity to MCMV infection in Nfkbiz−/− mice suggests that IκBζ is not involved in the type I IFN signaling.
In summary, our data demonstrate that IκBζ is essential for NK-cell activation in response to IL-12 and IL-18 stimulation by inducing various genes including IFN-γ. Furthermore, induction of IκBζ is one of the important host defense mechanisms against virus infection via NK cells. Given that IκBζ looks to control different set of genes in a cell-type specific manner, further studies will clarify the mechanisms of IκBζ in controlling gene expression.
Materials and Methods
Mice, Cells, and Reagents.
Nfkbiz−/− (22) and littermate control mice were used at 6–16 wk of age. Mouse experiments were conducted in accordance with institutional guidelines of animal care and use committees. Splenic NK cells were enriched via MACS separation with DX5 MicroBeads (Miltenyi Biotec). YAC-1 cells were as described previously (33). EL4 cells were obtained from ATCC (TIB-39). IL-12 and IL-2 were purchased from R&D Systems. Biotinylated rabbit anti-mouse IgG1 was purchased from Zymed. ELISA kits for IFN-γ and IL-12p40 were purchased from R&D Systems. IL-18 and ELISA kit for IFNα were purchased from PBL.
Q-PCR.
Total RNA was isolated using TRIzol (Invitrogen) and reverse transcription was performed with ReverTra Ace (Toyobo) according to the manufacturer's instructions. For Q-PCR, cDNA fragments were amplified by real-time PCR Master Mix (Toyobo), Fluorescence from the taqman probe for each cytokine was detected by a 7500 real-time PCR system (Applied Biosystems). To determine the relative induction of cytokine mRNA in response to various stimuli, the mRNA expression level of each gene was normalized to the expression level of 18S rRNA.
Cytotoxicity Assay.
Splenic NK cells were cultured with IL-2 (2,000 U/mL) for 7 d. IL-2–induced NK cells were stimulated with IL-12 (0.1 ng/mL) and IL-18 (1 ng/mL) for 4 h. The 51Cr-labeled YAC-1 cells (1 × 104 cells/well) were cultured with indicated numbers of NK cells. After 4-h incubation, the supernatant was harvested and the 51Cr radioactivity of supernatants were measured as described previously (33).
Intracellular Staining of STAT4.
Antibodies for flow cytometry were purchased from BD or eBioscience. Splenocytes were stimulated with IL-12 (2 ng/mL) for 30 min and were fixed immediately with PhosFlow Lyse/Fix buffer (BD) for 10 min. Cells were washed once with PBS and stained with DX5-PE and CD3e-APC. Cells were then permeabilized by BD PhosFlow Perm Buffer III and incubated for 25 min on ice. Cells were washed with BD Pharmingen Stain Buffer and stained with Phospho-STAT4-FITC at room temperature for 30 min in the dark. Cells were washed and acquired on a FACS Calibur flow cytometer (BD) and analyzed using FlowJo (Tree Star).
Immunoblot Analysis.
NK cells were lysed in a buffer containing 0.1% Nonidet-P40, 10 mM KCl, 10 mM Hepes-KOH (pH 7.8), 0.1 mM EDTA and a protease inhibitor mixture (Roche). Cell lysates were centrifuged at 5,000 rpm for 5 min, and the supernatant was harvested as cytoplasmic proteins. Pellets were lysed in a buffer containing 5 mM MgCl2, 420 mM KCl, 50 mM Hepes-KOH (pH 7.8), 0.1 mM EDTA and 20% glycerol for 30 min on ice. The lysates were centrifuged at 18,500 × g for 15 min, and the supernatant was harvested as nuclear proteins. Cell lysates were dissolved by SDS/PAGE and transferred onto a polyvinylidene difluoride membrane. The membrane was blotted with the specific antibodies to indicated proteins.
Chromatin Immunoprecipitation (ChIP).
ChIP was performed by using a chip assay kit (Millipore). NK cells were stimulated with IL-12 (20 ng/mL) and IL-18 (100 ng/mL) for 4 h and were fixed with 1% formaldehyde for 10 min or 10 mM DMA for 20 min. Cells were then washed and resuspended in SDS buffer. Lysates were sonicated to obtain DNA fragments with a peak in size between 200 and 1,000 bp. Lysates precleared with protein A agarose/salmon sperm DNA were incubated with antibodies against STAT4 (Santa Cruz) or H3K9 (Abcam) and IκBζ (kindly provided by Drs. Kayama and Takeda, Osaka University, Suita, Japan), and immunoprecipitated at 4 °C overnight. The immune complexes were absorbed with beads. Beads were washed with low salt buffer, high salt buffer, LiCl wash buffer, and in TE buffer. Immune complexes were extracted with elution buffer (1% SDS 100 mM NaHCO3, 10 mM DTT) and cross links were reversed by incubation overnight at 65 °C. After proteinase K treatment for 1 h, DNA was then purified via ethanol precipitation. The purified DNA was used in qPCR to assess the presence of target sequences. Quantitative RT-PCR was performed with Thunderbird qPCR Mix (Toyobo) in an Applied Biosystem 7300. Primers used for amplifying Ifng loci were shown previously (10, 34).
Reporter Assay.
EL4 cells were transfected with either an empty vector or the IκBζ vector, human Ifng reporter plasmid containing the firefly luciferase gene (1 μg) (Addgene plasmid 17598) (35) and 50 ng of pRL-TK (Promega) by using DMRIE-C reagent (Invitrogen). EL4 cells were incubated in the presence of IL-12 (2 ng/mL) and IL-18 (10 ng/mL) for 18 h after transfection. Luciferase activities were measured with a Dual-Luciferase reporter assay system (Promega).
Microarray.
Splenic NK cells were stimulated with IL-12 (1 ng/mL) and IL-18 (10 ng/mL) for 24 h. Then, total RNA was extracted with TRIzol (Invitrogen Life Technologies) and further purified using an RNeasy kit (Qiagen). Biotin-labeled cDNA was synthesized from 100 ng of total RNA using the Ovation Biotin RNA amplification and labeling system (Nugen) according to the manufacturer's protocol. Hybridization, staining, washing, and scanning of Affymetrix mouse Genome 430 2.0 microarray chips were conducted according to the manufacturer's instructions. Data analysis was performed using R.
Supplementary Material
Acknowledgments
We thank Dr. M. Ueda at Kyoto Prefectural University of Medicine (Kyoto, Japan) for providing Nfkbiz−/− mice, Dr. M. Dalod at Université de la Méditerranée (Marseille, France) for providing MCMV, and Drs. H. Kayama and K. Takeda at Osaka University (Suita, Japan) for providing anti-IκBζ antibody. We thank all of our colleagues in our laboratory, E. Kamada for secretarial assistance, and Y. Fujiwara, M. Kumagai, and R. Abe for technical assistance. This research was supported by Funding Program for World-Leading Innovative R&D on Science and Technology of the Japan Society for the Promotion of Science, the Special Coordination Funds of the Japanese Ministry of Education, Culture, Sports, Science and Technology, grants from the Ministry of Health, Labor and Welfare in Japan, the Global Center of Excellence Program, and the National Institutes of Health Grant P01 AI070167.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012977107/-/DCSupplemental.
References
- 1.Beutler B, et al. Genetic analysis of resistance to viral infection. Nat Rev Immunol. 2007;7:753–766. doi: 10.1038/nri2174. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 4.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 5.Lanier LL. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 7.Aune TM, Collins PL, Chang S. Epigenetics and T helper 1 differentiation. Immunology. 2009;126:299–305. doi: 10.1111/j.1365-2567.2008.03026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamura H, Kashiwamura S, Tsutsui H, Yoshimoto T, Nakanishi K. Regulation of interferon-gamma production by IL-12 and IL-18. Curr Opin Immunol. 1998;10:259–264. doi: 10.1016/s0952-7915(98)80163-5. [DOI] [PubMed] [Google Scholar]
- 9.Takeda K, et al. Defective NK cell activity and Th1 response in IL-18-deficient mice. Immunity. 1998;8:383–390. doi: 10.1016/s1074-7613(00)80543-9. [DOI] [PubMed] [Google Scholar]
- 10.Chang S, Aune TM. Histone hyperacetylated domains across the Ifng gene region in natural killer cells and T cells. Proc Natl Acad Sci USA. 2005;102:17095–17100. doi: 10.1073/pnas.0502129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatton RD, et al. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Balasubramani A, et al. Modular utilization of distal cis-regulatory elements controls Ifng gene expression in T cells activated by distinct stimuli. Immunity. 2010;33:35–47. doi: 10.1016/j.immuni.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thierfelder WE, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 14.Levy DE, Darnell JE., Jr Stats: Transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 15.Szabo SJ, et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 16.Avni O, et al. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 17.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 18.Nakanishi K, Yoshimoto T, Tsutsui H, Okamura H. Interleukin-18 regulates both Th1 and Th2 responses. Annu Rev Immunol. 2001;19:423–474. doi: 10.1146/annurev.immunol.19.1.423. [DOI] [PubMed] [Google Scholar]
- 19.Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: An effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Nakahira M, et al. Synergy of IL-12 and IL-18 for IFN-gamma gene expression: IL-12-induced STAT4 contributes to IFN-gamma promoter activation by up-regulating the binding activity of IL-18-induced activator protein 1. J Immunol. 2002;168:1146–1153. doi: 10.4049/jimmunol.168.3.1146. [DOI] [PubMed] [Google Scholar]
- 21.Yamazaki S, Muta T, Takeshige K. A novel IkappaB protein, IkappaB-zeta, induced by proinflammatory stimuli, negatively regulates nuclear factor-kappaB in the nuclei. J Biol Chem. 2001;276:27657–27662. doi: 10.1074/jbc.M103426200. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto M, et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature. 2004;430:218–222. doi: 10.1038/nature02738. [DOI] [PubMed] [Google Scholar]
- 23.Matsuo S, Yamazaki S, Takeshige K, Muta T. Crucial roles of binding sites for NF-kappaB and C/EBPs in IkappaB-zeta-mediated transcriptional activation. Biochem J. 2007;405:605–615. doi: 10.1042/BJ20061797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kayama H, et al. Class-specific regulation of pro-inflammatory genes by MyD88 pathways and IkappaBzeta. J Biol Chem. 2008;283:12468–12477. doi: 10.1074/jbc.M709965200. [DOI] [PubMed] [Google Scholar]
- 25.Okamoto K, et al. IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature. 2010;464:1381–1385. doi: 10.1038/nature08922. [DOI] [PubMed] [Google Scholar]
- 26.Raices RM, et al. A novel role for IkappaBzeta in the regulation of IFNgamma production. PLoS ONE. 2009;4:e6776. doi: 10.1371/journal.pone.0006776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Santo JP. Natural killer cell developmental pathways: A question of balance. Annu Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 28.Vidal SM, Lanier LL. NK cell recognition of mouse cytomegalovirus-infected cells. Curr Top Microbiol Immunol. 2006;298:183–206. doi: 10.1007/3-540-27743-9_10. [DOI] [PubMed] [Google Scholar]
- 29.Hoshino K, et al. Cutting edge: Generation of IL-18 receptor-deficient mice: Evidence for IL-1 receptor-related protein as an essential IL-18 binding receptor. J Immunol. 1999;162:5041–5044. [PubMed] [Google Scholar]
- 30.Wu Z, et al. Nuclear protein IkappaB-zeta inhibits the activity of STAT3. Biochem Biophys Res Commun. 2009;387:348–352. doi: 10.1016/j.bbrc.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 31.Tato CM, et al. Opposing roles of NF-kappaB family members in the regulation of NK cell proliferation and production of IFN-gamma. Int Immunol. 2006;18:505–513. doi: 10.1093/intimm/dxh391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 33.Miyake T, et al. Poly I:C-induced activation of NK cells by CD8 alpha+ dendritic cells via the IPS-1 and TRIF-dependent pathways. J Immunol. 2009;183:2522–2528. doi: 10.4049/jimmunol.0901500. [DOI] [PubMed] [Google Scholar]
- 34.Bandyopadhyay S, Qui HZ, Adler AJ. In vitro and in vivo differentiated effector CD8 T cells display divergent histone acetylation patterns within the Ifng locus. Immunol Lett. 2009;122:214–218. doi: 10.1016/j.imlet.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonsky R, et al. Mucosa-specific targets for regulation of IFN-gamma expression: Lamina propria T cells use different cis-elements than peripheral blood T cells to regulate transactivation of IFN-gamma expression. J Immunol. 2000;164:1399–1407. doi: 10.4049/jimmunol.164.3.1399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.