Abstract
The complement system is a major target of immune evasion by Staphylococcus aureus. Although many evasion proteins have been described, little is known about their molecular mechanisms of action. Here we demonstrate that the extracellular fibrinogen-binding protein (Efb) from S. aureus acts as an allosteric inhibitor by inducing conformational changes in complement fragment C3b that propagate across several domains and influence functional regions far distant from the Efb binding site. Most notably, the inhibitor impaired the interaction of C3b with complement factor B and, consequently, formation of the active C3 convertase. As this enzyme complex is critical for both activation and amplification of the complement response, its allosteric inhibition likely represents a fundamental contribution to the overall immune evasion strategy of S. aureus.
Keywords: allosteric modulation, complement amplification, hydrogen-deuterium exchange mass spectrometry, small angle X-ray scattering, surface plasmon resonance
The ability to evade an attack of the immune system is a hallmark of human pathogens, and a plethora of evasion strategies have been described in recent years. With some 50 factors that interfere at various stages of our immune and inflammatory system, Staphylococcus aureus appears to be particularly elaborate in this respect (1). This versatility may well contribute to the virulence of this bacterium, which is the etiologic agent for an array of conditions that vary in both clinical presentation and severity (2). Owing to its early and central role in the detection and elimination of microbial intruders and in the stimulation of subsequent immune responses, the complement system represents an ideal target for immune evasion (3). Recent discoveries revealed a multitude of complement-directed evasion proteins that influence nearly every level of the complement cascade from its initiation and amplification to its regulation and signaling (3). Because these potent inhibitors may not only be determinants for the virulence of S. aureus but may also offer structural templates for the development of therapeutics in complement-mediated diseases (3, 4), knowledge about their molecular evasion mechanisms is essential.
In the battle between complement attack and evasion, C3 convertases play a particularly prominent role. These enzyme complexes transform the abundant, yet rather inert complement component C3 into the reactive opsonin, C3b. It is this fragment that becomes covalently attached to protein and carbohydrate patches on bacterial surfaces and triggers subsequent immune responses by interacting with various ligands. Deposited C3b also forms additional surface-bound convertase complexes by interaction with factor B (fB), thereby amplifying the complement response and inducing the generation of proinflammatory anaphylatoxins, which attract immune cells to the site of infection. Binding of C3b and its degradation product iC3b to complement receptors expressed on the surface of phagocytic cells leads to the uptake of opsonized material. Further degradation of C3b/iC3b (∼175 kDa) generates the fragments C3c (∼135 kDa) and C3d (∼40 kDa), the latter of which remains covalently linked to the antigen surface as is known to substantially lower the threshold for B cell activation. Because convertase inhibition simultaneously impairs all activation, amplification, and effector pathways, it represents a powerful strategy for complement evasion. Indeed, S. aureus is known to secrete at least two families of evasion molecules that directly target convertases. Whereas the staphylococcal complement inhibitor family (SCIN) is known to bind assembled convertase complexes and trap them in an inactive state (5–7), the mechanism of complement inhibition by the extracellular fibrinogen-binding protein (Efb) and its homolog Ehp has not been fully defined (8–11).
We have previously shown that Efb forms high-affinity complexes with C3d or C3 fragments that include the corresponding thioester-containing domain (TED) via its C-terminal three-helix bundle domain of ∼9 kDa (Efb-C) (9). Whereas the interaction of Efb and Ehp with the isolated C3d fragment may affect the stimulation of adaptive immune responses (12), their potent inhibition of complement activation and amplification is primarily mediated by targeting surface-bound convertases that contain the C3b fragment (8, 9, 13). Despite detailed structural information about the C3 convertase complex (7, 14), the strong convertase-directed effects of Efb-C could not be explained, because the proposed binding site of the inhibitor on C3b is clearly distant from areas involved in convertase activity (7, 9). A detailed characterization of the binding mode and inhibitory mechanism has been further hampered by the fact that crystallization of the C3b/Efb-C complex has not yet been accomplished. Nevertheless, our previous studies indicated that formation of this complex would require structural adaptations within C3b to allow Efb-C binding (9): First, the kinetic on-rate of Efb-C was significantly lower for its interaction with C3b than with C3d (ka = 2 vs.30 × 104 M-1 s-1), and second, Efb-C binding to C3b altered its reactivity toward an antibody whose epitope is far distant from TED (9, 15). On this basis, we predicted that Efb-C binding induces conformational changes in C3b, which thereby affect the functional integrity of this versatile complement fragment.
By using an integrated biophysical and computational approach (Fig. S1), we demonstrate here that Efb-C indeed acts as an allosteric complement inhibitor, which locks C3b in a conformation where the TED domain is held apart from the remainder of the rigid core that is mainly formed by macroglobulin (MG) domains 1–5 (hereafter referred to as “MG core”). Propagation of dynamic changes throughout parts of the α-chain into functionally important recognition sites in C3b affects binding of essential ligands, such as fB, and thereby impairs assembly of the surface-bound C3 convertase. These findings not only provide important details on the inhibitory mechanism of Efb, but also represent a fascinating model on how a small bacterial inhibitor can influence the ligand-binding pattern of large plasma proteins via induction of conformational changes. Finally, our study underscores the importance of dynamic structural changes that reside at the base of many immunological processes and presents a general approach for monitoring such events.
Results
Efb-C Induces Conformational Changes Primarily in the α-Chain of C3b.
Owing to its ability to monitor solution structural dynamics of multidomain proteins such as C3b (Fig. 1 A and B) (16–21), we employed hydrogen-deuterium exchange mass spectrometry (HDX-MS) to study the conformation of C3b in the presence of Efb-C. As a negative control, we compared the exchange rates of C3b in the presence of a nonbinding, nonfunctional mutant of Efb-C (R131A+N138A), hereafter denoted RANA (9). A set of 245 and 265 peptic fragments was identified by analysis of C3b/Efb-C and C3b/RANA, respectively. Among this cohort, 190 individual peptides that spanned 81% of the C3b sequence were observed in both samples (Fig. 1D). Levels of deuteration generally increased during the seven measured HDX time points (10–10,000 s; Fig. S2). For the majority of covered peptides, exchange rates developed similarly for both complexes. However, 18 peptides in C3b/Efb-C that spanned 9 of the 14 domains in C3b (Fig. 1 C–F) displayed a significant HDX difference relative to C3b/RANA (Table 1 and Figs. S2 and S3). Whereas some affected domains were localized in the C3b β-chain, domains of the α-chain generally experienced a higher degree of HDX difference (Table 1 and Fig. 1C).
Fig. 1.
Changes in HDX levels of C3b in presence of Efb-C. (A and B) Domain organization of C3b as sequence (A) and arrangement representation (B). The ANA domain, which does not exist in C3b, is marked in gray. (C) Relative distribution of HDX changes in the C3b domains as represented by the number of altered peptides per domain (colored according to legend). (D) Sequence coverage of each domain of C3b in the presence of wild-type Efb-C (Lower) or its RANA mutant (Upper). Segments identified by HDX-MS are marked in green, of which peptides showing significantly altered HDX are highlighted in blue (≤ -10%) and orange (≥10%); unidentified sequences are in gray. (E and F) Localization of HDX peptides on the crystal structure of unbound C3b (23), visualized as cartoons (E) and surface-accessible surface (F) with color coding as in D.
Table 1.
Peptide segments possessing significant difference in HDX levels between the C3b/RANA and C3b/Efb-C complex
| C3b peptide (range, sequence) | Domain | ΔHDX, % | |
| 13–19 | RLESEET | MG1 | 11 ± 0.7 |
| 110–124 | IQTDKTIYTPGSTVL | MG2 | −10 ± 0.6 |
| 200–211 | FEVKEYVLPSFE | MG2 | −12 ± 1.1 |
| 221–230 | YYIYNEKGLE | MG3 | 13 ± 0.7 |
| 238–248 | LYGKKVEGTAF | MG3 | −11 ± 0.6 |
| 582–596 | VLNKKNKLTQSKIWD | LNK | −10 ± 0.5 |
| 773–783 | LKDSITTWEIL | MG6α | −12 ± 0.6 |
| 808–824 | FIDLRLPYSVVRNEQVE | MG7 | −12 ± 0.5 |
| 841–850 | RVELLHNPAF | MG7 | −14 ± 0.9 |
| 957–968 | RILLQGTPVAQM | CUBf-TED | 22 ± 3.0 |
| 1002–1011 | AVHYLDETEQ | TED | −10 ± 0.3 |
| 1012–1024 | WEKFGLEKRQGAL | TED | −17 ± 0.5 |
| 1127–1138 | ISLQEAKDICEE | TED | −11 ± 0.4 |
| 1258–1272 | AQYQKDAPDHQELNL | CUBg | 13 ± 0.7 |
| 1323–1323 | VTMYHAKAKD | CUBg | 11 ± 0.8 |
| 1333–1341 | QLTCNKFDL | MG8 | 12 ± 0.4 |
| 1341–1359 | LKVTIKPAPETEKRPQDAK | MG8 | 10 ± 0.5 |
| 1365–1378 | EICTRYRGDQDATM | MG8 | −16 ± 1.3 |
Our previous study has demonstrated that the C3b/Efb-C interaction is both high affinity (KD = 9 nM) and kinetically stable (t1/2 ∼ 60 min) (9). Therefore, we expected that any peptides found at the immediate Efb-C contact interface on TED would be engaged in intermolecular interactions and thus display lower HDX rates (17). Indeed, three peptide segments within TED (1002–1011, 1012–1024, 1127–1138) showed significantly decreased HDX levels in C3b/Efb-C relative to the control (Table 1, Fig. 1 C–F, and Fig. S4A). Most importantly, five of the seven TED residues that form well-characterized contacts with Efb-C (9, 22) (H1004, D1007, R1020, D1134, E1138) were likewise classified as protected by HDX-MS (Fig. S4B). The other two residues (N1069, S1075) lay in regions absent from all HDX coverage maps of C3b. Together, these observations not only confirmed the Efb-C binding site on C3b in this setting, but also validated HDX-MS as a solution-based approach for detecting changes in C3b that arise from Efb-C binding.
Besides alterations in TED that could be attributed to direct interaction with Efb-C, we also observed HDX changes in domains that are not part of the immediate binding site. The majority of these domains are part of the α-chain of C3b, with the most extensive HDX changes found in the complement C1r/C1s, Uegf, Bmp1 (CUB) domain that extends both N- and C-terminally from TED (Table 1 and Fig. 1 C–F). A nearly 30-residue span (1333–1359) of increased HDX was comprised of two adjacent peptides within MG8, whereas several protected peptides were found in MG6 and MG7 of the α-chain of C3b (Table 1 and Fig. 1 C–F). The CUB as well as the MG6-8 region have previously been identified as functionally important domains, as they undergo significant rearrangements during the activation of C3 to C3b and participate in the binding of fB and various complement regulators (14, 23–25). In addition, MG7/8 harbors the binding epitope of the anti-C3b mAb C3-9 (15), for which we previously observed changes in affinity in the presence of Efb-C (9).
Other changes affected MG1, which is in direct proximity to Efb-C, and its adjacent (MG2, MG3) or aligned (LNK) domains (Table 1 and Fig. 1 C–F). Due to the spatial arrangement and highly positive overall charge of Efb-C (9), changes in those β-chain domains may be induced by the altered steric and electrostatic environment rather than direct propagation alone. Most importantly, the HDX-MS data confirm the binding site of Efb-C on C3b and indicate a propagation of conformational changes from the TED interface via CUB to more distant domains in the α-chain of C3b.
Efb-C Separates CUB-TED from the MG Core of C3b.
To generate a conformational model of the complex, we analyzed the solution structures of C3b both free and bound to Efb-C by small angle X-ray scattering (SAXS) combined with sample repurification using size exclusion chromatography (SEC) (26, 27). SAXS is sensitive to changes in protein conformations that often accompany functional transitions (26, 28) and has previously been used in the context of complement proteins (29). Linear Guinier plots (30) of scattering profiles of collected peak fractions and its dilutions (1.2–0.5 mg/mL; Fig. S5) indicated that the protein samples were free of nonspecific aggregation and revealed radii of gyration (RG) of 46.8 ± 0.3 Å for C3b and 47.6 ± 0.3 Å for C3b/Efb-C (Fig. 2A and Fig. S5A). Separately, the derived pair-distribution functions, P(r), and the corresponding Kratky plots for interference-free SAXS profiles also indicated significant conformational changes between free C3b and the Efb-C bound state (Fig. 2B). Importantly, no further changes in the scattering curves or P(r) functions were observed when the concentration of Efb-C was increased to a fivefold molar excess relative to C3b. These results confirmed previous reports of only a single binding site between these two proteins (9), and demonstrated that changes in C3b/Efb-C conformation relative to C3b alone could be successfully detected and analyzed by SAXS.
Fig. 2.
Efb-C-induced changes in C3b as revealed by SAXS. (A) Experimental scattering curves for free C3b (black) and C3b/Efb-C (blue) were fit to either a single (red) or minimal ensemble search model (MES; green). Guinier plots (Inset) for the SAXS profile of the highest protein concentration with linear fit (red) in the range q × RG < 1.6. (B) Pair-distribution functions [P(r)] indicate conformational changes between C3b (black) and C3b/Efb-C (blue), where broadening of P(r) for C3b/Efb-C is consistent with reorientation of CUB-TED. P(r) from the atomic MES models are shown as a green-dashed line. Individual SAXS profiles are shown in the Kratky plot (Inset), wherein significant differences are highlighted by red arrows. (C) Comparison of RG for the two predominant MES conformers of either C3b (black) or C3b/Efb-C (blue) as obtained by molecular dynamics conformational sampling with their maximal dimensions (Dmax). Dot sizes represent the fraction ratio of the two conformers in each group. Rigid body modeling-derived C3b conformers are shown in gray with Efb-C highlighted in red. (D and E) Superposition of the SAXS/BILBOMD-derived conformers of free C3b (D, magenta and green) and C3b/Efb-C (E, blue/red) with the crystal structure of C3b (gray). The inset shows a schematic representation of the proposed domain rearrangements.
We further analyzed our SAXS data using a computational approach (BILBOMD) (31, 32), in which rigid body modeling is combined with simplified molecular dynamics to arrive at minimal ensemble of plausible conformers that best describe experimental data (29, 31). As the large size of C3b restricts computational simulation of the entire protein, we used a facilitated model wherein the MG core, CUB, and TED were treated as separate rigid bodies connected by flexible linkers. From 10,000 distinct conformations that were generated by constrained molecular dynamics modeling, subsets of two conformations were selected using a minimal ensemble search (31) for each sample. These subsets were analyzed for their ability to describe the experimental scattering curves by calculation of a discrepancy parameter (χ2). In the case of free C3b, a set of two distinct conformers with a volume ratio of 45% and 55% (Fig. 2C) accurately described the experimental scattering curve (χ2 of 1.1). Although both conformers generally resembled the orientation of the CUB-TED region observed in the crystal structure of C3b, with TED in contact to the MG core, the analysis indicated significant rotations and translation of the TED domain in between the selected conformers (Fig. 2D). A similar pattern with even slightly higher separation of TED from the MG core was observed in buffers that contained 5% glycerol (initially added to avoid potential aggregation; Fig. S6), thereby supporting that C3b exists in a dynamic equilibrium in solution. In the case of C3b/Efb-C, however, the ensemble was dominated by a single conformer with a volume ratio of 79% (Fig. 2C), and a two-conformation model led to minor improvements (χ2 of 1.78 and 1.67, respectively). In the major conformer, the previously reported steric clash between Efb-C and MG1 of C3b (9) was notably absent, and Efb-C appeared to act as a wedge holding CUB-TED in an “open” state (Fig. 2E).
Given its role during activation of C3 and in ligand binding, this dislocation and stabilization of CUB-TED is likely to affect the overall structure and function of C3b. In previous studies, the structure of C3b has been described using the metaphor of a puppeteer, who holds his puppet (TED) via his arm (CUB) beside his body (MG core) (Fig. 2E, Inset) (23). Any physical movement of the puppet is not only expected to move the arm but also to induce twists in the shoulders (MG7/8). Indeed, our HDX data support this hypothesis: Aside from changes in TED and CUB, we found altered peptides within this shoulder region between the α-chain part of MG6, MG7, and MG8, which may be directly propagated by the movement of CUB. The proposed binding model of Efb-C is further supported by our previous observation concerning the slow rate of complex formation for C3b/Efb-C (9), because Efb-C can only bind to this open-like conformer if it is to avoid major steric clashes with MG1. The SAXS/BILBOMD results clearly suggest that the Efb-C-mediated allosteric changes in C3b involve separation of the CUB-TED region from the MG core, and thereby support the current HDX data and our previous findings concerning the slower binding of Efb-C to C3b and the changed reactivity of mAb C3-9 (9).
Efb-C Affects Ligand-Binding Patterns of C3b.
Conformational changes during the activation of C3 to C3b expose various ligand-binding sites and render C3b among the most versatile interaction partners in circulation (21, 23, 24, 33). As binding of Efb-C results in structural changes in regions similar to those exposed during activation of C3 (e.g., MG7/8, CUB, TED), we predicted that these changes might perturb the ligand recognition pattern of C3b. We tested this hypothesis by employing surface plasmon resonance (SPR)-based binding assays, for which we selected a panel of C3b-specific ligands based on their coverage of distinct binding areas and the availability of cocrystal structures (Fig. 3 A–B). This set of ligands included fB and its fragment Ba (14), the N-terminal four domains of the complement regulator factor H [i.e., fH(1–4)] (25), the complement receptor of the Ig superfamily (CRIg) (24), the peptidic C3 inhibitor compstatin (34), and SCIN (7). For each ligand, we compared the binding activity for either free C3b or a saturated C3b/Efb-C complex (Fig. 3 C–F). In parallel, we determined the contact residues for each complex in silico (Table S1) and localized them on C3b along with the HDX peptides (Fig. 3 G–L). In case of fB, the structural comparison was performed on its cocrystal with the structural C3b/C3c-homolog cobra venom factor (CVF) (14) and extrapolated to the corresponding areas of C3b (Fig. S7).
Fig. 3.
Effect of Efb-C on the ligand-binding pattern of C3b as assessed by SPR. (A and B) The cocrystal structures of (A) C3b/fH1-4 (25), C3b/CRIg (24), and C3c/Compstatin (analog 4W9A, ref. 34), and (B) C3b/Bb/SCIN (7), and CVF/fB (14) were superimposed with the crystal structure of free C3b (23). (C–L) Compstatin (C), CRIg (D), SCIN (E), and fH1-4 (F) were injected to either immobilized C3b (blue) or C3b/Efb-C (red). (G–L) Contact residues for each ligand (magenta) were determined from the corresponding cocrystal structures shown in A and B using LIGPLOT (42) and mapped on the crystal structure of C3b. The binding area of each ligand is marked by dashed circles. In the case of SCIN, both binding sites on C3b were analyzed. A more detailed binding site analysis can be found in Fig. S8 A–E. Differential HDX is marked using the color code described in Fig. 1.
No significant activity changes between free and Efb-C-bound C3b were detected for CRIg, compstatin, and SCIN (Fig. 3 C–E). In good agreement, analysis of the compstatin site revealed that none of the contact residues on C3b were situated in areas that experienced changes in HDX levels (Fig. 3G and Fig. S8A). Despite the fact that both binding interfaces for SCIN reside at the shoulder area of C3b that contains five peptides of altered HDX, detailed binding site analysis showed that none of the contact residues for SCIN are located within peptides of significant HDX change (Fig. 3 I–K and Fig. S8 C–D). Though the contact information was derived from the SCIN-stabilized, dimeric C3 convertase complex (7), recent binding studies demonstrated that SCIN interacts with free C3b at a highly similar site (5). In the case of CRIg, which occupies a large site spanning the MG3-6 and LNK domains (24), a single contact residue (Lys242) was part of an 11-residue peptide (238–248) that showed protected exchange following Efb-C binding, whereas the other six residues were in regions of no significant change (Fig. 3H and Fig. S8B). Closer analysis revealed that this 11-residue peptide is part of a loop in MG3 that has previously been found susceptible to dynamic changes (21). Furthermore, MG3 is not directly connected to TED-CUB and the decreased HDX of the peptide may be influenced by the spatial arrangement of the nearby MG7/8 (Fig. S9). As a consequence, it is plausible that Efb-C influences exposure of this peptide without disturbing the CRIg interface.
In contrast to these three ligands, the binding of fH(1–4) was completely blocked in the presence of Efb-C (Fig. 3F). Indeed, the large interaction interface for this regulator spans across several domains, in which either significantly increased or decreased HDX was observed upon binding of Efb-C (Fig. 3L). Three contact residues (K774, I1135, E1138) were localized directly within peptides of altered HDX and several others were found nearby (Fig. S8E). In particular, one segment on MG6 (773–783) was found to be adjacent to the binding region of the complement control protein (CCP) domain 2 of fH(1–4) (25). Finally, recent cocrystal data regarding this complex showed that TED participates to binding and function of the regulator via interaction with fH domain CCP4 (25). Because this contact region directly overlaps with the Efb-C binding site on TED (Fig. S10), bound Efb-C will very likely disrupt this interaction, either by direct steric clash or by dislocating this site within the wedged “open-like” conformation of C3b. Therefore, all the steric hindrance, dislocation, and conformational changes may simultaneously contribute to the complete inhibition of this N-terminal fH fragment. Whereas the strong effect on fH(1–4) directly supports our structural hypothesis, the functional consequences have to be put into the context of full-length fH. Interestingly, Efb-C significantly increased the binding of both fH and its C-terminal two-domain fragment (CCP 19–20) (Fig. S11), which is not directly involved in the regulatory functions but contributes a second binding site for C3b (35). Whether this enhanced binding partially compensates for the impaired interaction of the fH(1–4) fragment or even leads to a recruitment of this potent convertase regulator to the surface of S. aureus has to be further investigated.
Efb-C Impairs Binding of fB to C3b and Formation of the C3 Convertase.
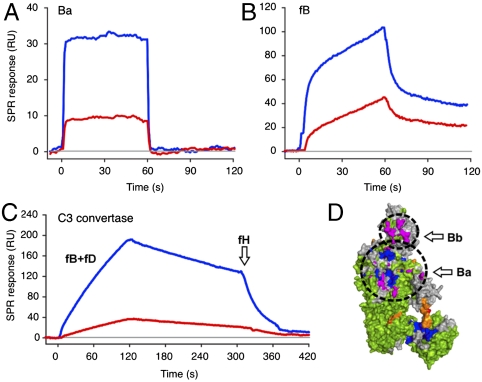
The C3 convertase is formed when fB binds to C3b and is cleaved into its fragments Ba and Bb by factor D (fD), thereby resulting in a metastable C3bBb complex that is able to cleave C3 (7, 14). In agreement with previous SPR studies (7, 36), injection of recombinant Ba onto C3b resulted in a fast kinetic profile, whereas fB bound in a more complex, biphasic manner (Fig. 4 A and B). Upon formation of the C3b/Efb-C complex, binding of Ba was significantly impaired (Fig. 4A). In the case of fB, the initial fast interaction step was also largely inhibited, whereas the secondary slow binding phase was affected to a lesser extent (Fig. 4B and Fig. S12). Together, these results indicated that Efb-C primarily disturbs the contact interface of the Ba rather than the Bb segment. In agreement with this observation, we found several contact residues for Ba on CVF that were located within or nearby peptides that experienced significant changes in HDX (corresponding to segments 110–124, 773–783, and 841–850 in C3b). Similar to the case of fH(1–4), a combination of effects may be responsible for this impairment. Efb-C not only induces conformational changes in contact areas of Ba, but also directly dislocates CUB, which forms part of the Ba site (14). On the other hand, no altered peptides were found in the Bb contact area at the C345C domain (Fig. 4D and Fig. S8F). It has to be noted that a high-resolution structure is only available for the complex of fB with CVF but not with C3b. Although individual contact residues between these homologous proteins may differ slightly (Fig. S7), the strong correlation between contact sites, HDX, and SPR analyses indicate that CVF closely resembles the situation in C3b.
Fig. 4.
Effect of Efb-C on fB binding and C3 convertase formation. (A and B) The binding of fB (A) and its fragment Ba (B) to C3b (blue) or C3b/Efb-C (red) was analyzed by SPR as in Fig. 3. (C) Formation of the C3 convertase was monitored using SPR by injecting a 1∶1 mixture of fB and fD onto either C3b (blue) or C3b/Efb-C (red). Regular convertase decay was followed for 3 min and fH was injected to accelerate decay. (D) Contact residues for fB were determined on the cocrystal structure of CVF/fB (14) and transferred on the structure of C3b (23). Peptides of differential HDX are visualized using the color code in Fig. 1 and binding areas for the Ba and Bb segments are marked with circles. A detailed binding site analysis can be found in Fig. S8F.
In order to determine whether the strong effects of Efb-C on the C3b-fB interaction have an impact on the C3 convertase, we used SPR to monitor the assembly of the convertase complex (C3bBb), its decay caused by the irreversible dissociation of Bb, and the decay acceleration by fH (25, 37) (Fig. 4C). In accordance with our hypothesis and the results obtained for fB and Ba, formation of C3b/Efb-C heavily impaired assembly of the convertase complex (Fig. 4C). The observed effect on convertase formation is also in good agreement with previous immunoblotting studies that showed a decreased level of Bb on bacterial surfaces in the presence of Efb-C (13). Furthermore, the same study excluded a direct decay acceleration effect of Efb-C on the assembled convertase (13). The extent of Efb-C-induced conformational changes in C3b, and potentially also in native C3 (9), may affect additional steps in the activation and regulation of C3 and C5 convertases. However, because rapid self-amplification on foreign surfaces is a hallmark of complement function, such a drastic impact on the assembly of the surface-bound C3 convertase likely represents the main driving force in the complement inhibitory activity of Efb.
Discussion
In this study, we identify the staphylococcal immune evasion protein Efb-C as an allosteric complement inhibitor and provide insight into its molecular mechanism. Our structural hypothesis indicates that Efb-C acts like a wedge between TED and the MG core, and thereby stabilizes an open conformer of C3b. This relocation of TED affects the conformation of several key domains in the functionally important α-chain of C3b (i.e., CUB and MG6-8), and significantly changes the ligand-binding pattern of C3b. Most importantly, the Efb-C-induced changes affect the binding of fB to surface-bound C3b and impair the formation of the major C3 convertase (C3bBb). The current model of convertase formation includes a multistep process, in which the Ba segment is essential for guiding fB toward C3b and forming initial contacts (“loading”). Mg2+-dependent binding to a secondary site on C3b and conformational rearrangements may trigger a state that allows cleavage by fD, leading to the active convertase (14). Whereas removal of Mg2+ by EDTA was shown to inhibit the kinetically slower secondary interaction phase (7), Efb-C primarily affects the fast initial binding phase of fB. Together with the impaired interaction of recombinant Ba, our data indicate that Efb-C-mediated convertase inhibition results primarily from influencing the crucial loading phase.
Among the versatile strategies of complement evasion by S. aureus, modulation of convertase-mediated C3 activation appears to be particularly effective, as it allows the bacterium to simultaneously control the effects of all initiation, amplification, and terminal pathways of complement (3). Strikingly, the Efb, SCIN, and staphylococcal immunoglobulin binding (Sbi) protein families all interact with C3 fragments and thereby target this central step in the cascade, albeit through conceptually distinct mechanisms: Whereas Sbi was reported to recruit the convertase-decaying regulator fH to the surface of S. aureus (38), SCIN traps the assembled convertase in an inactive state that does not allow cleavage of C3 (7). Based on the results of our study, Efb has evolved yet another path to achieve convertase modulation by allosterically impairing convertase formation. Interestingly, a trapping effect on the C3b/fB complex has very recently been suggested for Ehp (39), and our own results with different fH fragments indicate that Efb-C increases the overall affinity of fH to C3b and may thereby contribute to recruiting this important regulator to the bacterial surface. The contribution of each of those effects on the overall convertase evasion mechanism has to be further investigated. Intriguingly, by employing a structurally similar but functionally diverse set of inhibitors S. aureus can control the assembly (Efb), activity (SCIN), and regulation (Sbi) of the C3bBb convertase.
It has become evident that dynamic structural changes and allosteric modulation of protein function lie at the heart of a variety of biological processes (40). Yet the experimental elucidation of such phenomena still represents a major challenge, particularly for large proteins. Our approach of combining HDX-MS with other biophysical and computational analyses allows for a more detailed insight into such events. In fact, the investigated C3b/Efb-C complex ranks among the largest systems investigated by HDX-MS so far. The SAXS-derived finding that C3b coexists as a mixture of distinct conformers directly supports and extends the observations from previous EM studies (15) and further emphasizes the importance of viewing complement as a highly dynamic system. Though the implication of this intrinsic fluctuation is not yet known, it may well influence the activity and regulation of C3b. Indeed, structural studies have indicated that correct positioning of the CUB-TED interface is vital for complete degradation of C3b by factor I (25). It is likely that the dynamic fluctuation within C3b allows for Efb-C to selectively bind to the subpopulation of C3b molecules that transiently occupy a more open state. In this regard, our results support the new view of protein allostery, in which modulators stabilize preexisting intrinsic conformations rather than inducing completely new domain arrangements (40).
Compared to the increasing awareness of allosteric concepts in endogenous proteins or therapeutics (41), little is known about allosteric modulation in microbial immune escape. In the case of complement evasion, no allosteric mechanisms have been described so far and the hitherto elucidated mechanisms for Staphylococcal complement inhibitors all rely on either blockage of binding sites or trapping of complexes. Efb appears unique in this context as it uses both allosteric and competitive effects for exerting its inhibitory properties toward innate and adaptive immune responses, respectively. Though Efb also contains a fibrinogen-binding domain (Efb-N), previous studies have clearly shown that it is not essential for complement inhibition (9). In the case of the Efb-C homolog Ehp, the existence of a second C3d-binding site may lead to additional and more complex effects that might explain its stronger inhibitory potential (8, 13, 39).
Considering the current and previous findings, it is clear that binding of Efb to C3d or TED of C3 and C3b can simultaneously affect multiple complement functions via both directly competitive and allosteric phenomena. Owing to the importance of the C3 convertase in complement activation, the newly discovered mechanism of impairing formation of this complex may be considered especially important for the inhibitory activity of Efb. Our study therefore underscores the potential of Efb as a virulence factor of S. aureus, and renders Efb-C a valuable tool for studying molecular functions and therapeutic inhibition of complement.
Methods
Information about preparation and sources of all proteins and peptides, as well as more detailed descriptions of the assays used for HDX-MS, SAXS, and SPR can be found in the SI Appendix.
Hydrogen-Deuterium Exchange Mass Spectrometry.
HDX-MS experiments were performed as previously described (16, 19, 21). Briefly, C3b was incubated with either Efb-C or RANA in 50% DMSO/PBS, quenched after various time points (10–10,000 s), digested on a pepsin column, and analyzed using mass spectrometry. A difference in HDX of more then ± 10% between C3b/Efb-C and C3b/RANA was considered significant.
Structural Analysis of C3b Complexes by Small Angle X-ray Scattering.
SAXS experiments were performed based on previously described methods (26, 27). C3b alone or an equimolar C3b/Efb-C complex (0.5–1.2 mg/mL; Fig. S5) were repurified by SEC and measured by SAXS for both short (0.5 s) and long time exposure (5 s), from which the radius of gyration (RG), the pair-distance distribution functions [P(r)], and the maximum macromolecule dimension (Dmax) were calculated. In our BILBOMD strategy (31), the conformational space adopted by CUB-TED was explored by molecular dynamics, in which the MG core of C3b and TED/CUB were considered as distinct rigid bodies connected by flexible linkers. Minimal ensemble search was used to identify the minimal ensemble of two conformers required to best fit the experimental data (31).
Ligand-Binding Assays Using Surface Plasmon Resonance.
Ligand binding was assessed by injecting 1 μM fB, Ba, fH(1–4), CRIg, SCIN, and compstatin onto immobilized C3b or a saturated C3b/Efb-C complex, and comparing the relative binding intensities. The influence of Efb-C on convertase formation was monitored by injecting fB and fD on either C3b or C3b/Efb-C and observing complex formation and decay.
Correlation of HDX and SPR Data with Ligand Interaction Sites in C3b.
LIGPLOT (42) and HBPLUS (43) were used to generate lists of hydrogen-bonded and nonbonded contacts, and residues involved in ligand binding were mapped on the crystal structure of C3b along with HDX-derived peptides.
Supplementary Material
Acknowledgments.
We thank Paul N. Barlow for providing the fH(19–20) protein. This study was supported by National Institutes of Health Grants AI030040, AI068730, AI072106, GM062134, AI071028, CA099835, CA118595, AI076961, AI081982, NS070899, GM093325, RR029388, and AI2008031. Advanced Light Source beamline 12.3.1 (SIBYLS) is supported by the Department of Energy program Integrated Diffraction Analysis Technologies.
Footnotes
Conflict of interest statement: B.V.G. and J.D.L. are the inventors of a patent application about the use of Efb-C for therapeutic complement inhibition.
*This Direct Submission article had a prearranged editor.
Data deposition: Experimental and theoretical scattering profiles, P(r) functions, and SAXS atomic models have been deposited in the BIOISIS database, www.bioisis.net.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003750107/-/DCSupplemental.
References
- 1.Chavakis T, Preissner KT, Herrmann M. The anti-inflammatory activities of Staphylococcus aureus. Trends Immunol. 2007;28:408–418. doi: 10.1016/j.it.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat Rev Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ricklin D, Lambris JD. Complement-targeted therapeutics. Nat Biotechnol. 2007;25:1265–1275. doi: 10.1038/nbt1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricklin D, et al. A molecular insight into complement evasion by the staphylococcal complement inhibitor. J Immunol. 2009;183:2565–2574. doi: 10.4049/jimmunol.0901443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rooijakkers SH, et al. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat Immunol. 2005;6:920–927. doi: 10.1038/ni1235. [DOI] [PubMed] [Google Scholar]
- 7.Rooijakkers SH, et al. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nat Immunol. 2009;10:721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammel M, et al. Characterization of Ehp: a secreted complement inhibitory protein from Staphylococcus aureus. J Biol Chem. 2007;282:30051–30061. doi: 10.1074/jbc.M704247200. [DOI] [PubMed] [Google Scholar]
- 9.Hammel M, et al. A structural basis for complement inhibition by Staphylococcus aureus. Nat Immunol. 2007;8:430–437. doi: 10.1038/ni1450. [DOI] [PubMed] [Google Scholar]
- 10.Lee LYL, et al. Inhibition of complement activation by a secreted Staphylococcus aureus protein. J Infect Dis. 2004;190:571–579. doi: 10.1086/422259. [DOI] [PubMed] [Google Scholar]
- 11.Lee LYL, Liang X, Hook M, Brown EL. Identification and characterization of the C3 binding domain of the Staphylococcus aureus extracellular fibrinogen-binding protein (Efb) J Biol Chem. 2004;279:50710–50716. doi: 10.1074/jbc.M408570200. [DOI] [PubMed] [Google Scholar]
- 12.Ricklin D, Ricklin-Lichtsteiner SK, Markiewski MM, Geisbrecht BV, Lambris JD. Cutting edge: Members of the Staphylococcus aureus extracellular fibrinogen-binding protein family inhibit the interaction of C3d with complement receptor 2. J Immunol. 2008;181:7463–7467. doi: 10.4049/jimmunol.181.11.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jongerius I, et al. Staphylococcal complement evasion by various convertase-blocking molecules. J Exp Med. 2007;204:2461–2471. doi: 10.1084/jem.20070818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen BJ, et al. Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex. EMBO J. 2009;28:2469–2478. doi: 10.1038/emboj.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishida N, Walz T, Springer TA. Structural transitions of complement component C3 and its activation products. Proc Natl Acad Sci USA. 2006;103:19737–19742. doi: 10.1073/pnas.0609791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 17.Chetty PS, et al. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc Natl Acad Sci USA. 2009;106:19005–19010. doi: 10.1073/pnas.0909708106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Englander JJ, et al. Protein structure change studied by hydrogen-deuterium exchange, functional labeling, and mass spectrometry. Proc Natl Acad Sci USA. 2003;100:7057–7062. doi: 10.1073/pnas.1232301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, et al. Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen/deuterium exchange mass spectrometry. Structure. 2008;16:137–148. doi: 10.1016/j.str.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuster MC, Chen H, Lambris JD. Hydrogen/deuterium exchange mass spectrometry: Potential for investigating innate immunity proteins. Adv Exp Med Biol. 2007;598:407–417. doi: 10.1007/978-0-387-71767-8_28. [DOI] [PubMed] [Google Scholar]
- 21.Schuster MC, et al. Dynamic structural changes during complement C3 activation analyzed by hydrogen/deuterium exchange mass spectrometry. Mol Immunol. 2008;45:3142–3151. doi: 10.1016/j.molimm.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haspel N, et al. Electrostatic contributions drive the interaction between Staphylococcus aureus protein Efb-C and its complement target C3d. Protein Sci. 2008;17:1894–1906. doi: 10.1110/ps.036624.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 24.Wiesmann C, et al. Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature. 2006;444:217–220. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- 25.Wu J, et al. Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nat Immunol. 2009;10:728–733. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rambo RP, Tainer JA. Improving small-angle X-ray scattering data for structural analyses of the RNA world. RNA. 2010;16:638–646. doi: 10.1261/rna.1946310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 29.Perkins SJ, Okemefuna AI, Nan R, Li K, Bonner A. Constrained solution scattering modelling of human antibodies and complement proteins reveals novel biological insights. J R Soc Interface. 2009;6(Suppl 5):S679–696. doi: 10.1098/rsif.2009.0164.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guinier A, Fournet G. Small-Angle Scattering of X-Rays. New York: Wiley; 1955. p. 268. [Google Scholar]
- 31.Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: A web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gros P, Milder FJ, Janssen BJ. Complement driven by conformational changes. Nat Rev Immunol. 2008;8:48–58. doi: 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- 34.Janssen BJ, Halff EF, Lambris JD, Gros P. Structure of compstatin in complex with complement component C3c reveals a new mechanism of complement inhibition. J Biol Chem. 2007;282:29241–29247. doi: 10.1074/jbc.M704587200. [DOI] [PubMed] [Google Scholar]
- 35.Herbert AP, Uhrin D, Lyon M, Pangburn MK, Barlow PN. Disease-associated sequence variations congregate in a polyanion recognition patch on human factor H revealed in three-dimensional structure. J Biol Chem. 2006;281:16512–16520. doi: 10.1074/jbc.M513611200. [DOI] [PubMed] [Google Scholar]
- 36.Harris CL, Abbott RJ, Smith RA, Morgan BP, Lea SM. Molecular dissection of interactions between components of the alternative pathway of complement and decay accelerating factor (CD55) J Biol Chem. 2005;280:2569–2578. doi: 10.1074/jbc.M410179200. [DOI] [PubMed] [Google Scholar]
- 37.Ricklin D, Lambris JD. Exploring the complement interaction network using surface plasmon resonance. Adv Exp Med Biol. 2007;598:260–278. doi: 10.1007/978-0-387-71767-8_19. [DOI] [PubMed] [Google Scholar]
- 38.Haupt K, et al. The Staphylococcus aureus protein Sbi acts as a complement inhibitor and forms a tripartite complex with hostcomplement Factor H and C3b. PLoS Pathog. 2008;4:e1000250. doi: 10.1371/journal.ppat.1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jongerius I, Garcia BL, Geisbrecht BV, van Strijp JA, Rooijakkers SH. Convertase inhibitory properties of Staphylococcal extracellular complement-binding protein. J Biol Chem. 285:14973–14979. doi: 10.1074/jbc.M109.091975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai CJ, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: A classification scheme of allosteric mechanisms. Mol Biosyst. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 42.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 43.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.