Abstract
No-go decay and nonstop decay are mRNA surveillance pathways that detect translational stalling and degrade the underlying mRNA, allowing the correct translation of the genetic code. In eukaryotes, the protein complex of Pelota (yeast Dom34) and Hbs1 translational GTPase recognizes the stalled ribosome containing the defective mRNA. Recently, we found that archaeal Pelota (aPelota) associates with archaeal elongation factor 1α (aEF1α) to act in the mRNA surveillance pathway, which accounts for the lack of an Hbs1 ortholog in archaea. Here we present the complex structure of aPelota and GTP-bound aEF1α determined at 2.3-Å resolution. The structure reveals how GTP-bound aEF1α recognizes aPelota and how aPelota in turn stabilizes the GTP form of aEF1α. Combined with the functional analysis in yeast, the present results provide structural insights into the molecular interaction between eukaryotic Pelota and Hbs1. Strikingly, the aPelota·aEF1α complex structurally resembles the tRNA·EF-Tu complex bound to the ribosome. Our findings suggest that the molecular mimicry of tRNA in the distorted “A/T state” conformation by Pelota enables the complex to efficiently detect and enter the empty A site of the stalled ribosome.
Keywords: X-ray crystallography, small G protein, dual specificity
The fidelity of gene expression is ensured by many checkpoints in the replication, transcription, and translation of genetic information. The quality control of mRNA is one of the important steps to ensure the fidelity of protein biosynthesis (1, 2). Recent studies revealed three mRNA quality-control mechanisms in the eukaryotic cell, which detect and degrade defective mRNA. Nonsense-mediated decay (3) prevents translation of mRNA with premature termination (nonsense) codon, which would produce a truncated protein if translated. Nonstop decay (NSD) (4, 5) detects and degrades mRNAs lacking an in-frame termination codon (nonstop mRNA). No-go decay (NGD) (6) detects a ribosome blocked in translational elongation, and the mRNA is then endonucleolytically cleaved near the stalled site. The resulting mRNA, with free 3′ and 5′ termini, is further degraded by the mRNA clearance machinery, including the exosome and Xrn1 nuclease.
In NSD and NGD processes, the detection of a stalled ribosome requires at least two protein factors, Pelota (Dom34 in yeast) and Hbs1 (6, 7). An amino acid sequence alignment revealed that Pelota is homologous to eRF1, which binds to the termination codon at the ribosomal A site by presumably mimicking an aminoacyl-tRNA (aa-tRNA). Therefore, Pelota is proposed to bind to the empty A site of the stalled ribosome (7). Hbs1 is a member of the translational small GTPase family (8), including bacterial elongation factor Tu (EF-Tu), eukaryotic and archaeal elongation factor 1α (e/aEF1α), and eukaryotic translation-termination factor 3 (eRF3). e/aEF1α and its bacterial homologue, EF-Tu, deliver aa-tRNA to the ribosomal A site in the translation elongation cycle (9). Similarly, eRF3 delivers eRF1 to the ribosomal A site in the translation-termination process (10). Hbs1 was shown to interact with Pelota in vivo (11), and thus Hbs1 is considered to bind Pelota and deliver it to the empty A site of the stalled ribosome (6, 7). These tRNA or tRNA-mimicry proteins stabilize the GTP-bound state of their cognate GTPase carriers.
The crystal structures of the archaeal homologue of Pelota (aPelota) (12) and yeast Dom34 (13) revealed that Pelota has three domains, with the middle and C-terminal domains sharing quite similar structures to those of eRF1 (14, 15), whereas the N-terminal domain is completely different, with weak similarity to Sm-fold proteins (16). Sm-fold proteins have RNA-binding ability and the complex structures with RNA were reported (17, 18). However, because the key residues for RNA binding in Sm-fold proteins are not conserved in Dom34, the function of the N-terminal domain as well as the molecular mechanism of the mRNA surveillance by Pelota remains unknown. Moreover, it remains unknown how Pelota interacts and stabilizes the GTP-bound state of Hbs1.
Pelota homologues are widely conserved in archaea (19), suggesting the conservation of a similar surveillance mechanism in archaea. In contrast, the archaeal counterpart of Hbs1 has not yet been identified in the archaeal genome, which has precluded the elucidation of aPelota’s function. Recently, we discovered that archaeal EF1α (aEF1α) functionally interacts not only with aa-tRNA but also with archaeal RF1 (aRF1) and aPelota (20). Our in vitro pull-down analysis showed that aEF1α binds to both aRF1 and aPelota in a GTP-dependent manner. Therefore, in archaea, it is quite likely that aEF1α performs three different functions: translational elongation and termination as well as quality control of mRNA.
Here we report the complex structure of aPelota and GTP-bound aEF1α determined at 2.3-Å resolution. The structure revealed the recognition mechanism of aPelota by GTP-bound aEF1α in atomic detail and explains the dual specificity of aEF1α for both RNA and protein (i.e., aa-tRNA and aPelota). Our findings provide an insight into how the complex detects and enters the empty A site of the stalled ribosome.
Results and Discussion
Overall Structure.
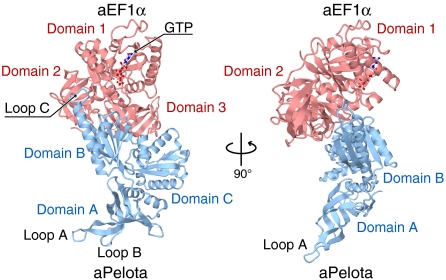
To understand the structural mechanism of NGD and NSD, we solved the complex structure of aPelota and GTP-bound aEF1α from Aeropyrum pernix at 2.3-Å resolution, by the single-wavelength anomalous diffraction method (Fig. 1; see also SI Appendix, Tables SI and SII). The asymmetric unit of the crystal contains four ternary complexes with essentially the same conformation, with rms deviations from 0.28 to 0.85 Å for the Cα atoms.
Fig. 1.
Overall structure of the aPelota·aEF1α·GTP complex, viewed from two perpendicular directions. aEF1α is colored red and aPelota is colored light blue. The bound GTP is shown in a stick model.
The overall structure of aEF1α in the complex consists of three domains, domains 1, 2, and 3 (Fig. 1). The arrangement of the three domains of aEF1α is quite similar to that of EF-Tu in complex with a nonhydrolyzable GTP analog, GMPPNP (guanylyl imidodiphosphate) (21, 22), and their structures superimpose well on each other (rms deviation of 1.6 Å over 363 Cα atoms). Domain 1 of aEF1α, which shares structural similarity with the small GTPases, recognizes a GTP molecule through one Mg2+ ion and several water molecules (SI Appendix, Fig. S1). The GTP recognition manner of aEF1α is also essentially the same as that of GDPNP by EF-Tu. A water molecule is present in-line with the γ-phosphate of GTP (SI Appendix, Fig. S2A). However, the side chain of the conserved His94, which may act as a general base in the catalysis, is flipped away from this in-line water molecule (SI Appendix, Fig. S1), and thus the bound GTP molecule was not hydrolyzed in the present structure.
The overall structure of aPelota in the complex also consists of three domains, domains A, B, and C (Fig. 1). The domain arrangement is similar to that in the isolated form of aPelota from Thermoplasma acidophilum (TaPelota) (12), with an rms deviation of 3.1 Å for 298 Cα atoms. However, as compared to TaPelota, the domain orientations are different; domains A and B are respectively rotated by 22.4° and 17.2° relative to domain C. Furthermore, the three loops that are disordered in the TaPelota structure (two in domain A and one in domain B) are clearly visible in the present complex structure (SI Appendix, Figs. S2B and C).
Interactions Between aPelota and aEF1α.
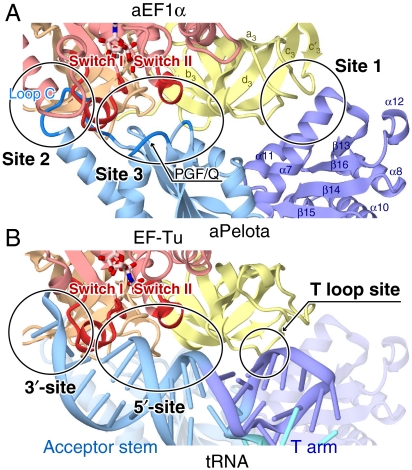
In the complex structure, all three domains of aEF1α are involved in intermolecular interactions, although the N-terminal domain A of aPelota does not interact with aEF1α (Fig. 1). The interaction interface between aPelota and aEF1α can be divided into three sites (sites 1, 2, and 3) (Fig. 2A; a comprehensive list of the interactions in sites 1, 2, and 3 is in SI Appendix, Table SIII).
Fig. 2.
Comparison of the interaction interfaces between (A) aPelota and aEF1α and (B) aa-tRNA and EF-Tu. Proteins are shown by ribbon models with the domains color-coded (turquoise for domain A, light blue for domain B, and purple for domain C of aPelota; and red for domain 1, brown for domain 2, and yellow for domain 3 of aEF1α). In B, the ribbon model of aPelota superimposed on the aa-tRNA and EF-Tu complex is shown with semitransparent coloring. The superposition of the two complexes was calculated based on the similarity between aEF1α and EF-Tu, using the SSM algorithm (30).
In site 1, domain C of aPelota interacts with domain 3 of aEF1α by shape complementarity and several hydrogen bonds (SI Appendix, Fig. S3A). In particular, the side chain of Tyr273 of aPelota, which is widely conserved among archaea and eukaryotes (SI Appendix, Fig. S4), forms a hydrogen bond with the side chain of Gln378 of aEF1α (SI Appendix, Fig. S3A). These interactions involve only aPelota domain C and aEF1α domain 3 (Fig. 2A) and seem to be possible even in the GDP-bound aEF1α (23). Therefore, these residues in site 1 may contribute to the initial association for complex formation.
In site 2, the binding site for domain B of aPelota is formed by a narrow cleft between domains 1 and 2 of aEF1α (Fig. 2A and SI Appendix, Fig. S3B). Loop C of aPelota domain B is sandwiched between the switch I region of domain 1 and the β-strand a2 of domain 2 of aEF1α and forms an extensive hydrogen-bonding network (SI Appendix, Figs. S3B and C). Consequently, the disordered region of Loop C, in the isolated forms of both eukaryotic and archaeal Pelota (12, 13), becomes clearly visible in the electron density map of the present structure (SI Appendix, Fig. S2B). The side chain of the conserved Lys162 in loop C resides in a small cavity formed by the side chains of Glu65 and Thr75, and the main chain oxygen of Asp73, in the switch I region of aEF1α (SI Appendix, Fig. S3B).
In site 3, a shallow pocket, formed at the junction of the three aEF1α domains, accommodates a small bulge on the surface of aPelota domain B (Fig. 2A and SI Appendix, Fig. S3D). This bulge in aPelota is formed by the turn between strands β8 and β9 (residues 135–138), and the two loops between strand β11 and helix α4 (residues 199–201), and between strand β12 and helix α5 (residues 224–227). Notably, the loop between strand β11 and helix α4 harbors the conserved PGF/Q motif (SI Appendix, Fig. S3D). The main chain nitrogen of Gln201 forms a hydrogen bond with the carboxyl group of the conserved Glu64 in the aEF1α switch I region, and the side chains of these residues stack on each other. This position is replaced by Phe in the Pelota proteins from other species (SI Appendix, Fig. S4), which enables a similar interaction. The main chain nitrogen of Gly200 forms a hydrogen bond with a well-ordered water molecule, which is further fixed by the side chains of the conserved Arg68 and Asp96 residues of the aEF1α switch I region (SI Appendix, Fig. S3D). In turn, this water molecule contacts the side chain of His94, the putative general base for GTP hydrolysis. In concert with the hydrophobic gate (21, 24) formed by Val15 and Ile70 of aEF1α, these interactions may sequester the side chain of His94 from the GTPase catalytic site, thereby preventing the activation of the in-line water molecule and stabilizing the GTP form of aEF1α. In addition, Pro199 of the PGF/Q motif contributes to form part of the small bulge of aPelota, and its shape is complementary to that of the switch I and II regions of aEF1α (SI Appendix, Fig. S3D). Likewise, the conserved Ser225 residue in the loop between strand β12 and helix α5 also forms part of the small bulge of aPelota, and its shape is complementary to the cleft between domains 1 and 3 of aEF1α (SI Appendix, Fig. S3D). The turn between strands β8 and β9 provides conserved acidic residues, among which the side chains of Asp135 and Asp137 form salt bridges with Lys99 in the aEF1α switch II region (SI Appendix, Fig. S3E). Thus, these interactions recognize and stabilize the GTP-bound form of aEF1α, by fixing the spatial arrangement of the switch I and II regions.
Altogether, domain B of aPelota is specifically docked on the aEF1α surface, which involves the three domains of aEF1α in the GTP binding state. Especially, the interactions with the switch I and II regions of domain 1 of aEF1α may stabilize its GTP-bound conformation. The resultant complex of aPelota and the GTP form of aEF1α would be favorably recruited to the ribosomal A site, thereby initiating the processes of NGD and NSD.
tRNA-Mimicry Mechanism by aPelota.
The overall structure of the aPelota·aEF1α complex resembles that of the aa-tRNA·EF-Tu·GDPNP ternary complex (25). All three domains of aEF1α superimposed quite well on those of the complexed EF-Tu (rms deviation of 1.6 Å over 363 Cα atoms). Thus it is likely that aEF1α also binds the aa-tRNA in a similar manner to the aa-tRNA·EF-Tu·GDPNP complex. In fact, the basic residues of EF-Tu involved in tRNA recognition are well conserved in aEF1α (SI Appendix, Fig. S5). Furthermore, the intermolecular interfaces of aPelota·aEF1α and tRNA·EF-Tu complexes overlap each other well (Fig. 2 A and B), even though their interacting partners (i.e., aPelota and tRNA) are different. In particular, sites 2 and 3 of aPelota correspond to the 3′ and 5′ termini of tRNA, respectively (Fig. 2 A and B). This significant conservation of the molecular interfaces of the elongation factors with RNA and protein is in good agreement with our recent findings that aEF1α is a multifunctional factor, acting in the processes of translational elongation and termination as well as mRNA surveillance (20). The present crystal structure provides the structural mechanism by which aEF1α exerts its dual specificity for protein and tRNA.
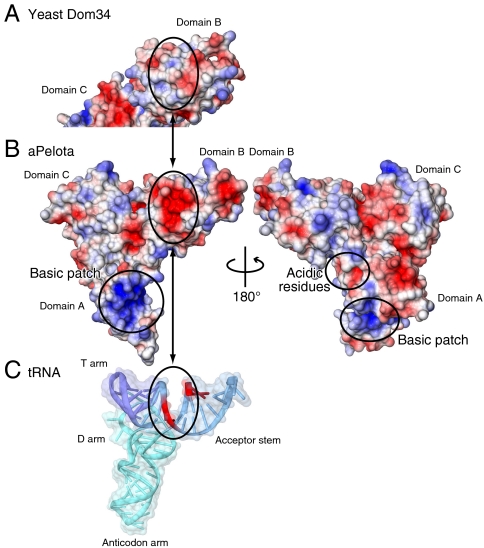
Interestingly, some of the basic residues of aEF1α in the putative tRNA binding site (K99 and K394) interact with the conserved acidic residues of aPelota domain B (Asp135, Asp137, Glu138, and Asp154) (SI Appendix, Fig. S3E). These aPelota residues form a negatively charged strip on its surface, which coincides well with the phosphate groups of the nucleotide residues at positions 1, 2, 66, and 67 in the tRNA acceptor stem (Fig. 3 B and C). In addition, the shape of the surface of aPelota domain B resembles that of the acceptor stem of tRNA (Fig. 3 B and C). Therefore, in site 3, domain B of aPelota mimics both the shape and charge distribution of the surface of the tRNA molecule, thereby enabling the dual specificity of aEF1α for both protein and RNA.
Fig. 3.
Comparison of the molecular surfaces of (A) yeast Dom34, (B) aPelota, and (C) tRNA. In A and B, the molecular surfaces of the positively charged regions are colored blue and those of the negatively charged regions are colored red, with the intensity of the color being proportional to the local potential (range -10 kT/e to +10 kT/e). The electrostatic potential was calculated by the program APBS (31). In C, the tRNA is depicted by a ribbon model, and the solvent accessible surface is shown with semitransparent coloring. The phosphate backbones of positions 1, 2, 66, and 67 are highlighted in red.
On the other hand, there may be differences in the interaction surface of aEF1α for aPelota and aa-tRNA. First, in site 2, the aminoacylated A76 (aa-A76) of tRNA is accommodated in a deep pocket formed between domains 1 and 2 of EF-Tu, which is also conserved in aEF1α (Fig. 2B). This putative pocket for aa-A76 is empty in the aPelota·aEF1α complex and is not used for aPelota recognition (SI Appendix, Fig. S3B). On the other hand, Lys162 of aPelota contacts switch I of aEF1α, whereas the corresponding interaction is not observed in the aa-tRNA·EF-Tu complex. Second, domain C of aPelota has a bulkier and more protruded structure as compared to the corresponding T loop of tRNA (Fig. 3 B and C). Consequently, site 1 interactions in the aPelota·aEF1α complex do not precisely overlap with those between EF-Tu domain 3 and the tRNA T loop (Fig. 2 A and B). Taken together, in sites 1 and 2, the dual specificity is achieved by distinctive interactions with the different binding partners.
Structural Insight into the Interaction Between Dom34 and Hbs1.
The present structure of the aPelota·aEF1α complex provides insight into the interaction between their eukaryotic counterparts, Pelota/Dom34 and Hbs1. The arrangements of domains B and C in the crystal structure of yeast Dom34 (13) are considerably different from those of the present structure (SI Appendix, Fig. S6). The linker helices connecting domains B and C (α5, 6, and 7) adopt a more stretched conformation in the structure of yeast Dom34 (SI Appendix, Fig. S6), which causes this domain arrangement difference. However, the amino acid sequences of these linker helices are well conserved between the eukaryotic and archaeal Pelota proteins (SI Appendix, Fig. S4), suggesting that the observed structural differences in these helices are not species-specific. Therefore, it is likely that domains B and C of yeast Dom34 also adopt a similar arrangement to that of aPelota, upon binding to Hbs1, and that the interaction sites between Pelota/Dom34 and Hbs1 are similar to those in the aPelota·aEF1α complex.
In contrast, the negatively charged strip in aPelota, which mimics the phosphate moieties of the tRNA acceptor stem, is absent from the surface of yeast Dom34 (Fig. 3 A and B). The cluster of acidic residues at the β-turn between strands β8 and β9 in aPelota is replaced by neutral residues in Pelota/Dom34 (SI Appendix, Fig. S4). This difference in the surface charge distribution is reasonable because Hbs1 interacts only with Pelota/Dom34, but not with tRNA, so that Pelota/Dom34 no longer needs to mimic the surface property of a tRNA. Accordingly, the basic residues (Lys99 and Arg309) of aEF1α located near this β-turn of aPelota are also replaced by neutral/hydrophobic residues in Hbs1 (SI Appendix, Fig. S5). Therefore, the interface residues of the Pelota/Dom34·Hbs1 complex are specialized for protein–protein interaction and maintain a similar interaction manner to that in the aPelota·aEF1α complex.
To test the Pelota/Dom34·Hbs1 interaction, we designed mutants of yeast Dom34 and Hbs1 and evaluated their functions by two different in vivo analyses (Fig. 4 and Fig. S8). The first analysis detected the cleaved mRNA produced by the putative endonuclease in the NGD process, which is triggered by the mRNA stem-loop structure (6) (NGD assay). The second detected the truncated polypeptides released from the ribosome stalled with the nonstop mRNA, which may directly represent the ribosome disassembly activity that is prerequisite for the degradation of nonstop mRNA (NSD assay; see SI Appendix).
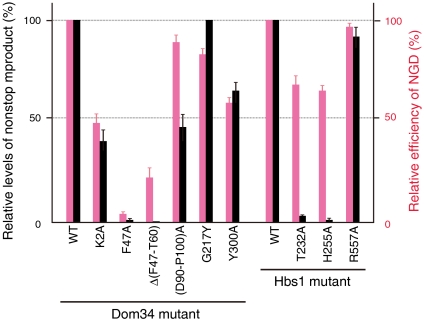
Fig. 4.
Translation of nonstop mRNA and NGD efficiencies of yeast Dom34 and Hbs1 mutants. The activities of Dom34 and Hbs1 mutants in nonstop mRNA translation and NGD were measured using yeast mutant strains, W303ski2Δdom34Δ or W303ski2Δhbs1Δ, harboring a plasmid expressing the indicated mutants and the p416GPDp-GFP-Rz-FLAG-HIS3 or p416GPDp-GFP-SL-FLAG-HIS3 plasmids. Detailed procedures for the activity measurements are described in Materials and Methods. Error bars, SD of three independent experiments.
For GTPase catalytic site, yeast Hbs1 mutants deficient in the GTPase (T232A and H255A corresponding to Thr71 and His94 of aEF1α, respectively) significantly impaired the NSD activity (Fig. 4). These results suggest that the Hbs1 departure from Pelota/Dom34, following the GTP hydrolysis, is essential for ribosome disassembly that is required for ribosome clearance (see SI Appendix).
The interactions in site 1 may form the initial binding site for the complex formation, but their importance has never been examined. We tested the Ala mutants of Tyr300 of yeast Dom34 and Arg557 of yeast Hbs1, which corresponds to Tyr273 of aPelota and Gln378 of aEF1α (SI Appendix, Fig. S3A), respectively. The result showed that the Tyr300 mutant of yeast Dom34 caused a moderate reduction in both the NGD and NSD activities (Fig. 4), whereas the Arg557 mutant of yeast Hbs1 has no effects. Therefore, this conserved Tyr300 residue is likely involved in the formation of the initial binding site between the eukaryotic Pelota/Dom34 and Hbs1; however, its interaction manner is possibly different from that of the present aPelota·aEF1α complex.
Moreover, our Pelota/Dom34·Hbs1 complex model is in good agreement with the results of the previous functional analyses. The mutational analysis of yeast Dom34 suggested the importance of the Arg/Lys cluster of Loop C for the NGD activity (7). The corresponding loop of eRF1 habors the GGQ motif, which is proposed to interact with the peptidyl transferase center (PTC) in the ribosome and catalyzes the release of the nascent peptide in translation termination (26). In contrast, loop C of Pelota/Dom34 has a divergent sequence lacking the GGQ motif (SI Appendix, Fig. S4). The present crystal structure revealed that the structure formed by loop C and the following helix α3 of aPelota is significantly shorter than the corresponding structure of eRF1 (SI Appendix, Fig. S7), supporting the previous hypothesis that loop C of Pelota cannot interact with PTC in the ribosome and thus does not hydrolyze the peptidyl-tRNA at the ribosomal P site (13). The present crystal structure showed that Lys162 in loop C of aPelota contacts the switch I region of the aEF1α domain 1 (SI Appendix, Fig. S3B). This Lys162 residue may correspond to one of the basic residue in the Arg/Lys cluster of eukaryotic Dom34/Pelota (SI Appendix, Fig. S4). Therefore, one role of this Arg/Lys cluster may be to interact with the switch I region of Hbs1.
In addition, the (Ala)3 replacement of the conserved PGF/Q motif in yeast Dom34 moderately reduced the NGD efficiency (7). The Tyr mutation of Gly217, which introduces a bulky side chain in the PGF/Q motif, also showed a similar tendency (Fig. 4). Based on the present crystal structure, it is likely that the PGF/Q motif of Pelota/Dom34 contacts the switch I region of Hbs1 (SI Appendix, Fig. S3D). Therefore, the mutations in this motif altered the conformation of the interaction interface on domain B of Dom34, which may have disrupted the interaction with the switch I region of Hbs1.
Docking of the aPelota·aEF1α Complex to the Ribosome.
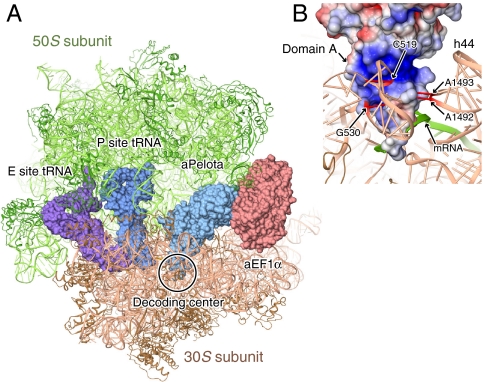
Although the atomic resolution structures of eukaryotic 80S and archaeal 70S ribosomes are unavailable, recent progress in cryoelectron microscopy of eukaryotic ribosome suggested that the core structure as well as the fundamental protein synthesis mechanism is essentially conserved among all phylogenetic domains (27, 28). Therefore, we constructed a docking model of the aPelota·aEF1α complex bound to the ribosomal A site, based on the complex structure of the bacterial ribosome, EF-Tu and tRNA (24) (Fig. 5A).
Fig. 5.
Docking model of the aPelota·aEF1α complex to the ribosome. (A) Overall view of the model. The 50S and 30S subunits of the ribosome (24) (PDB ID codes 2WRR and 2RWQ) are shown in green and brown ribbon representations, respectively. The tRNA in the E and P sites, aPelota, and aEF1α are shown in surface representations colored purple, dark blue, light blue, and pink, respectively. (B) Possible interactions between rRNA and aPelota domain A. aPelota is shown in a surface representation colored as in Fig. 4B. The 16S rRNA and mRNA are shown in brown and green ribbon representations, respectively. The rRNA residues discussed in the main text are highlighted in red.
This docking model showed the striking similarity between the structures of aPelota and tRNA in the distorted “A/T state” conformation (see SI Appendix). Moreover, this docking model revealed that the aPelota·aEF1α complex can bind to the ribosomal A site without any structural changes or steric hindrance, except for loop A of aPelota domain A (Fig. 5). In the present docking model, loop A, which is located at the tip of domain A, clashes with the mRNA in the decoding center. Loop A is disordered in the previous crystal structures of isolated Pelota/Dom34 and has higher B factors than the other parts in the present complex structure as well, indicating its intrinsic flexibility. Therefore, this loop may fit to the surrounding structure of the ribosome when bound to the A site. Furthermore, although loop A can contact the mRNA codon at the A site, its divergence in both sequence and length rules out the possibility of sequence-specific mRNA recognition by Pelota/Dom34.
Recognition of the A Site of the Stalled Ribosome.
This docking model further suggests the function of the N-terminal domain (domain A) of Pelota. The surface of domain A contains two regions with conserved amino acid residues. One region is formed by the acidic residues (Glu18, Asp22, and Asp34; SI Appendix, Fig. S4), which are located near the interface between domains A and C (Fig. 3B). A previous study suggested that Pelota domain A by itself has nuclease activity arising from these acidic residues (12). However, the proposed nucleolytic site is quite far from any rRNA and mRNA residues around the A site in the docking model (Fig. 5).
The other region is a large positively charged patch around the two loops (loops A and B) at the tip of domain A (Fig. 3B), which is formed by conserved basic residues (Arg2, Arg42, Arg53, and Arg75). Interestingly, in the docking model, this positively charged patch is located near the nucleotide residues 519, 530 (530 loop), 1492 and 1493 (helix h44) of 16S rRNA, which constitute the decoding center of the 30S ribosomal subunit (Fig. 5B). Actually, the Ala mutation of Phe47 (Arg42 in aPelota) and the deletion of loop A almost completely abolished the NGD and NSD activities, and the Ala mutation of Lys2 (Arg2 in aPelota) and the poly(Ala) replacement of loop B also caused a reduction in both activities (Fig. 4). These results are consistent with the previous functional analysis of yeast Dom34, in which the mutations in both loops A and B reduced the NGD efficiency (7). Therefore, it is likely that this positively charged patch of Pelota domain A (Figs. 3B and 5B) interacts with the decoding center, to specifically recognize the empty A site of the stalled ribosome. The events that might occur after the binding of the Pelota/Dom34·Hbs1 complex to the stalled ribosome, leading to the ribosome disassembly, will be discussed in the SI Appendix. The complete understanding of the recognition mechanism of the stalled ribosome awaits the determination of the Pelota/Dom34·Hbs1 complex or its archaeal counterpart bound to the ribosome.
Materials and Methods
Sample Preparation and Structure Determination.
The genes encoding full-length Aeropyrum pernix aEF1α and aPelota were cloned into pET15b vectors (Novagen). The proteins were overexpressed in the Escherichia coli BL21(DE3)CodonPlus strain and were purified by heat treatment, followed by chromatography on Ni-chelating, ion-exchange, and size-exclusion chromatography columns. SeMet-labeled aEF1α was expressed in the B834(DE3)CodonPlus strain and was purified by the same procedure as for the native protein. Crystals of the aPelota·aEF1α·GTP complex were grown from the reservoir solution containing 100 mM imidazole (pH 8.0) and 2.2 M NaCl. The crystal structure was determined by the single-wavelength anomalous diffraction method. Detailed procedures for sample preparation and structure determination are described in SI Appendix.
Measurement of the Activities of Dom34 and Hbs1 Mutants in Nonstop mRNA Translation and NGD.
The activities of Dom34 and Hbs1 mutants in nonstop mRNA translation and NGD were measured using yeast mutant strains, W303ski2Δdom34Δ or W303ski2Δhbs1Δ, harboring a plasmid expressing the indicated mutants and the p416GPDp-GFP-Rz-FLAG-HIS3 or p416GPDp-GFP-SL-FLAG-HIS3 plasmids (29). Detailed procedures for the activity measurements are described in SI Appendix.
Supplementary Material
Acknowledgments.
We are grateful to the beam-line staffs at NW12A and NE3A of KEK PF-AR and BL41XU of SPring-8 for assistance in data collection, RIKEN BioResource Center (Ibaraki, Japan) for providing genomic DNA of Aeropyrum pernix, and to Drs. Hiroshi Nishimasu and Luc Bonnefond (University of Tokyo) for helpful comments on the manuscript. This work was supported by a Strategic International Cooperative Program grant from Japan Science and Technology to O.N., by a grant for the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) to O.N., by grants from MEXT to R.I. and O.N., and by grants from The Uehara Memorial Foundation to O.N.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3AGJ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009598107/-/DCSupplemental.
References
- 1.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 2.Isken O, Maquat LE. Quality control of eukaryotic mRNA: Safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 3.Maquat LE. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat Rev Mol Cell Bio. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 4.Frischmeyer PA, et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 5.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 6.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Passos DO, et al. Analysis of Dom34 and its function in no-go decay. Mol Biol Cell. 2009;20:3025–3032. doi: 10.1091/mbc.E09-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inagaki Y, Blouin C, Susko E, Roger AJ. Assessing functional divergence in EF-1 alpha and its paralogs in eukaryotes and archaebacteria. Nucleic Acids Res. 2003;31:4227–4237. doi: 10.1093/nar/gkg440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riis B, Rattan SIS, Clark BFC, Merrick WC. Eukaryotic protein elongation-factors. Trends Biochem Sci. 1990;15:420–424. doi: 10.1016/0968-0004(90)90279-k. [DOI] [PubMed] [Google Scholar]
- 10.Frolova L, et al. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- 11.Carr-Schmid A, Pfund C, Craig EA, Kinzy TG. Novel G-protein complex whose requirement is linked to the translational status of the cell. Mol Cell Biol. 2002;22:2564–2574. doi: 10.1128/MCB.22.8.2564-2574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee HH, et al. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Mol Cell. 2007;27:938–950. doi: 10.1016/j.molcel.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 13.Graille M, Chaillet M, van Tilbeurgh H. Structure of yeast Dom34—A protein related to translation termination factor eRF1 and involved in no-go decay. J Biol Chem. 2008;283:7145–7154. doi: 10.1074/jbc.M708224200. [DOI] [PubMed] [Google Scholar]
- 14.Song HW, et al. The crystal structure of human eukaryotic release factor eRF1—Mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 15.Cheng Z, et al. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009;23:1106–1118. doi: 10.1101/gad.1770109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khusial P, Plaag R, Zieve GW. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem Sci. 2005;30:522–528. doi: 10.1016/j.tibs.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc Natl Acad Sci USA. 2009;106:19286–19291. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: A bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ragan MA, Logsdon JM, Sensen CW, Charlebois RL, Doolittle WF. An archaebacterial homolog of pelota, a meiotic cell division protein in eukaryotes. FEMS Microbiol Lett. 1996;144:151–155. doi: 10.1111/j.1574-6968.1996.tb08522.x. [DOI] [PubMed] [Google Scholar]
- 20.Saito K, et al. An omnipotent role of archaeal EF1α in translation elongation, termination and quality control of protein synthesis. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1009599107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berchtold H, et al. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993;365:126–132. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- 22.Kjeldgaard M, Nissen P, Thirup S, Nyborg J. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993;1:35–50. doi: 10.1016/0969-2126(93)90007-4. [DOI] [PubMed] [Google Scholar]
- 23.Vitagliano L, Masullo M, Sica F, Zagari A, Bocchini V. The crystal structure of Sulfolobus solfataricus elongation factor 1 alpha in complex with GDP reveals novel features in nucleotide binding and exchange. EMBO J. 2001;20:5305–5311. doi: 10.1093/emboj/20.19.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nissen P, et al. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 26.Frolova LY, et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spahn CMT, et al. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 28.Taylor DJ, et al. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 2009;17:1591–1604. doi: 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 31.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.