Abstract
X chromosome dosage compensation in female eutherian mammals is regulated by the noncoding Xist RNA and is associated with the differential acquisition of active and repressive histone modifications, resulting in repression of most genes on one of the two X chromosome homologs. Marsupial mammals exhibit dosage compensation; however, they lack Xist, and the mechanisms conferring epigenetic control of X chromosome dosage compensation remain elusive. Oviparous mammals, the monotremes, have multiple X chromosomes, and it is not clear whether they undergo dosage compensation and whether there is epigenetic dimorphism between homologous pairs in female monotremes. Here, using antibodies against DNA methylation, eight different histone modifications, and HP1, we conduct immunofluorescence on somatic cells of the female Australian marsupial possum Trichosurus vulpecula, the female platypus Ornithorhynchus anatinus, and control mouse cells. The two marsupial X's were different for all epigenetic features tested. In particular, unlike in the mouse, both repressive modifications, H3K9me3 and H4K20Me3, are enriched on one of the X chromosomes, and this is associated with the presence of HP1 and hypomethylation of DNA. Using sequential labeling, we determine that this DNA hypomethylated X correlates with histone marks of inactivity. These results suggest that female marsupials use a repressive histone-mediated inactivation mechanism and that this may represent an ancestral dosage compensation process that differs from eutherians that require Xist transcription and DNA methylation. In comparison to the marsupial, the monotreme exhibited no epigenetic differences between homologous X chromosomes, suggesting the absence of a dosage compensation process comparable to that in therians.
Keywords: DNA methylation, histone modifications, X chromosome inactivation
In mammals, X inactivation has evolved to solve the difference in X chromosome gene dosage between homogametic female mammals and heterogametic male mammals. Inactivation of one of the two female X chromosomes provides an equal dose in eutherian mammals and marsupials. In the mouse, X-chromosome inactivation (XCI) is imprinted in early development and is then reprogrammed at the blastocyst stage to become random, where, in response to expression of Xist RNA from the future inactive X (Xi), repressive histone modifications, histone variants, and DNA methylation are acquired (1, 2). This results in transcriptional repression of most X-linked genes. In particular, the eutherian Xi is late in replicating and is distinguished from the active X by a specific set of covalent histone modifications. The Xi is hypoacetylated on the NH2-terminal lysines of nearly all histones (3, 4) and lacks H3K4me2 and H3K4me3 but carries H3K9me2, H3K9me3, and H3K27me3 (1). In addition, Xi chromatin is enriched in the histone variants macroH2A1 and macroH2A2 (5). In human and bovine models, H3K27me3 and H3K9me3 are spatially distributed in nonoverlapping regions (6, 7). These stable histone modifications appear shortly after the accumulation of Xist RNA along the X with H3K9 methylation as the first modification occurring at the onset of XCI (8, 9). The Xist gene, together with the noncoding transcripts that contribute to its regulation, Tsix and Xite, is located within an X-inactivation center (Xic) that also regulates X chromosome counting and choice (10).
In marsupials, less is known about XCI; however, aspects are comparable but not identical to those of eutherian species. In female marsupials, dosage compensation also involves chromosome-wide X inactivation, but marsupials do not have an ortholog of the eutherian Xist gene (11, 12) and XCI is not random but imprinted, with the paternally inherited X being inactivated. The eutherian Xist inactivation system therefore evolved within a time interval of 28 million years: after marsupial divergence about 148 Mya and before the major radiation of placental mammals 120 Mya (13). The marsupial therefore provides a useful model for studying the evolution of dosage compensation and epigenetic silencing mechanisms in mammals. Interestingly, it has recently been suggested that preimplantation eutherian imprinted X inactivation is less dependent on Xist than the later random inactivation process (14). In the tammar wallaby, Macropus eugenii, analyses indicating H4 hypoacetylation of one of the two X chromosomes suggested mechanistic similarities with eutherians; however, a more recent study by the same group suggested absence of repressive histone modifications and proposes that the Xi chromosome in marsupials fails to recruit repressive modifications as seen in eutherians (15, 16).
Monotremes have a peculiar sex chromosome constitution. Monotremes have an X-Y system that appears unrelated to that of the marsupial and eutherian (17, 18). They diverged 166 Mya at the base of the mammalian branch (19). Strikingly, the male platypus has five different X chromosomes and five different Y chromosomes (20), and the male echidna has five different X chromosomes and four different Y chromosomes (17). Hence, the female platypus and echidna have the five X chromosomes in pairs and no Y chromosomes. In monotremes, the sex chromosomes form a chain at male meiosis linked by their pairing regions (17, 20, 21). These pairing regions do not cover the sex chromosomes entirely but are separated by a differential region. In particular, platypus X1 and X5 contain large differential regions. Monotreme sex chromosomes share considerable homology with chicken chromosomes Z, 2, 12, 13, and 17 and with certain human autosomes (17). Genes orthologous to the human Xq are found on platypus chromosome 6 (22) and echidna chromosome 16 (17); hence, the platypus does not require dosage compensation. Nonetheless, the five X-differential regions that account for about 12% of the platypus genome contain genes whose products are predicted to be more abundant in the female platypus than in the male platypus unless some form of dosage compensation takes place. Such genes remain to be characterized, and, to our knowledge, no assessment of epigenetic state has been conducted for the more than 400 protein-coding genes thus far recognized as X-linked in monotremes (Ensembl release 55, Wellcome Trust Sanger Institute-EMBL-EBI).
The discovery that the platypus has multiple X's that are unrelated to human X, together with the finding that the mammalian X-inactivation gene Xist evolved after monotremes and marsupials diverged from the eutherian lineage, makes a comparison of monotreme, marsupial, and eutherian mammals of value with respect to the evolution of sex chromosome-related dosage compensation in mammals. Here, we investigate the pattern of multiple histone modifications and DNA methylation on metaphase chromosomes from the female marsupial Trichosurus vulpecula, the female platypus Ornithorhynchus anatinus, and control mouse cells. DNA methylation studies were also performed on female Potorous tridactylus and male Monodelphis domestica metaphase chromosomes. Our results provide important insights into the similarities and differences in the epigenetic mechanisms of dosage compensation between euarchontoglires, metatherians, and monotremata.
Results
Active Modifications.
H4K8ac, H4K16ac, H3K9ac, and H2AK5ac.
In mouse control cells (Fig. S1 A, D, G, and J), one underacetylated X chromosome was present in all metaphases scored, presumably the Xi, as previously reported for H4K8, H4K16, and H3K9 (8, 23). Similarly, for H2AK5, we show hypoacetylation of one X, which, to our knowledge, has only been reported previously for human cells (3).
Marsupials similarly showed one hypoacetylated X in the majority of metaphases analyzed, with values of >90% for H4K9ac/H2AK5ac and 50–55% for H4K8ac/H4K16ac (Fig. S1 B, E, H, and K). These results are consistent with data from the mouse. On autosomes, all histone-acetylated isoforms tested resulted in evenly distributed staining patterns in marsupials and eutherians, with the exception of centromeres, as previously described. In contrast to marsupials, mouse H3K9ac showed a chromosome-specific banding pattern, as previously described (8).
In monotremes, no differences in acetylation were observed on either the larger X1 and X5 or the smaller X-chromosome pairs (Fig. S1 C, F, I, and L). However, unlike the situation in the therian mammals, region-specific staining of all chromosomes was observed for acetylated histones. This remarkable banding pattern was consistent for the acetylated isoforms and corresponded to the R-banded regions of euchromatin. In particular, we noted a clear absence of staining on centromeres and pericentric regions and on the satellite region of chromosome 6, which represents the nucleolar organizing region (NOR) in the platypus. However, a region near the centromere of platypus 6 is hyperacetylated for H4K8 (Fig. S1C, pale blue arrows). The distal portion of X5p in the platypus is hyperacetylated on both homologs. In the male platypus, this is the region that specifically pairs with the Y4 (where it is also hyperacetylated; Fig. S2) during formation of the meiotic chain. Compared with autosomes, the male platypus sex chromosomes do not differ in their levels of staining compared with the autosomes (Fig. S2). This suggests that sex chromosome dosage compensation in the platypus is unlikely to be attributable to hyperactivity of the single X chromosome, as has been described in Drosophila. Interestingly, in eutherians, both male and female animals can up-regulate X-linked genes from their single active X to balance dosage compared with autosomes (24, 25). This process may be unrelated to canonical dosage compensation and may not be associated with a change in acetylation state, suggesting that other mechanisms can act on mammalian X chromosomes to control levels of gene expression.
In contrast to the staining in mouse and T. vulpecula, H2AK5ac on the platypus showed weaker staining on both sex chromosomes and autosomes, suggesting that this modification is underrepresented in platypus metaphase chromosomes. However, the telomeric regions of several platypus autosomes exhibited hyperacetylation of H2AK5 (Fig. S1L).
H3K4me2.
H3K4me2 is a clear mark that distinguishes the active X in all (>90%) observed mouse female metaphases from the Xi. The Xi exhibits hypomethylation of this modification compared with the other X and with autosomes (8). Consistent with this, in mouse primary cells, constitutive heterochromatin at centromeres and facultative heterochromatin on the Xi showed a diminished presence of H3K4me2 (Fig. S1M). Similarly, in the possum (Fig. S1N), the inactivated X could be recognized in all observed metaphases by a diminished presence of H3K4me2, consistent with previously published data for the Xi in the tammar wallaby (15). Thus, this is a modified histone mark for the possum Xi. Some possum autosomal regions contain an enrichment of H3K4me2, suggesting regions of hyperactivity (Fig. S1N).
For H3K4me2 (Fig. S1O), no differential pattern was observed between female platypus X1 and X5. The distal portion of X5 (p-arm) in the platypus is hypermethylated on both homologs (Fig. 1).
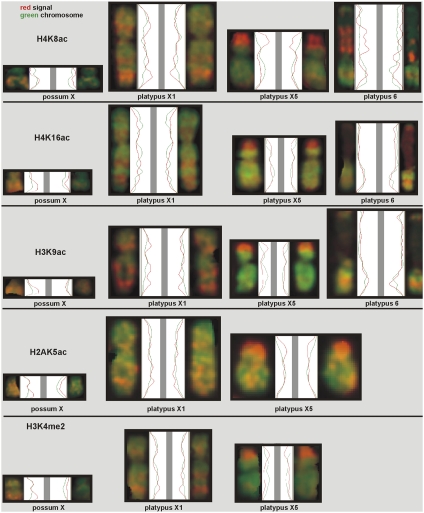
Fig. 1.
Line scans of distributions of canonical active histone modifications. The green curves correspond to the DAPI staining along the length of the chromosomes. The red curves show the distribution of each active epigenetic mark. As indicated, each row corresponds to a specific modified histone, the first column represents the possum X's, and the other three columns show platypus X1 and X5 and chromosome 6.
Fig. 1 shows line scans of the proximal-distal distribution of active histone modifications along the presented chromosomes. For the possum, the X in the right column is always dimmer than its homolog on the left: The red curve representing the epigenetic mark has lower amplitude. For the platypus, the red curves for both X-pairs indicate identical distributions of activating histone modifications (i.e., the enrichment and depletion regions for H4K8ac on both homologs of platypus X1 and X5 are the same). Note the strong enrichment of H4K8ac on the top region of the p-arm of platypus X5. Platypus chromosome 6 shows a differential pattern in all cells that is likely attributable to the variable expression of the rRNA from the two parental chromosomes in the NOR. This feature appears specific for H4K8ac.
Repressive Modifications.
H3K27me3, H3K9me3, H4K20me3, and HP1α.
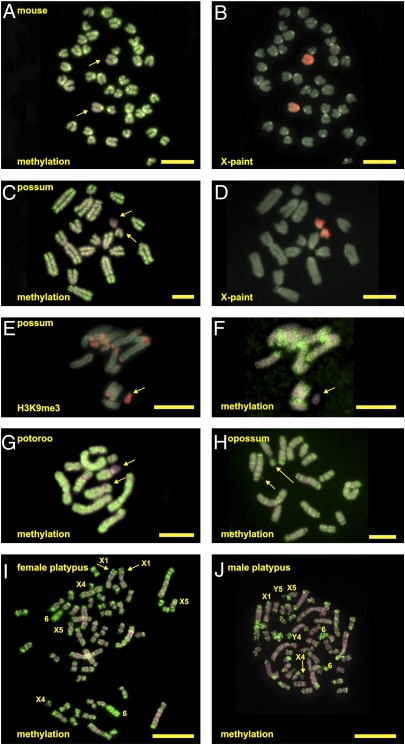
Only H3K27me3 was a clear mark for the inactivated X in mouse control cells (26), Fig. 2A). H3K9me3, H4K20me3, and HP1α were not present differentially on the mouse X chromosomes and, in our study, were mainly found at mouse centromeres (Fig. 2 D, G, and J), which is in agreement with the observation that they are present at constitutive pericentric heterochromatin in eutherians (27). Chadwick and Willard (6) describe enrichments of these modified histones on human Xi in addition to pericentric chromatin. In mouse metaphases and, in contrast to all interphase nuclei where it was present, HP1α could only be detected in less than 30% of the spreads (Fig. 2J). Fig. 2L shows that platypus chromosome 6 X1 and X5 can be recognized by its size, centromere position, and banding pattern.
Fig. 2.
(A–K) Repressive modifications in the female mouse, possum, and platypus. The columns represent each species, and the rows present each epigenetic modification. In all images, chromosomes are shown in light green and antibody staining is shown in red. The arrow in the mouse column points to the inactivated X. The arrows in the possum column point to both X homologs. In the platypus column, relevant chromosomes are indicated. HP1α did not stain platypus metaphases. (L) DAPI-stained platypus chromosome 6 and X1 and X5 chromosomes. Each chromosome can be recognized by its specific size, centromere position, and banding pattern. (Scale bar, 10 μm.)
In marsupials, the inactivated X was recognized by all four repressive marks. In contrast to the mouse control cells, H3K9me3, H4K20me3, and HP1α were clearly enriched on one of the X chromosomes in all observed metaphases (Fig. 2 E, H, and K). H3K27me3 was found in 50% of observed metaphases on the possum Xi (Fig. 2B). This is consistent with recent work showing H3K27me3 on the M. domestica Xi (28). All four repressive marks were enriched at pericentric and telomeric regions of the possum chromosomes. Because H3K9me3 was epigenetically dimorphic on the possum X chromosomes, it was not surprising to detect an enrichment of HP1α on the possum Xi, because H3K9me3 binds HP1α (29). As in humans (30), HP1α may recognize and facilitate heterochromatin maintenance and gene silencing in the possum Xi.
In the platypus, repressive histone modifications were evident on both chromosome homologs for all sex chromosomes and autosomes. Unfortunately, for the experiment with platypus H4K20me3, we could not unambiguously distinguish either X1 or X5, although all chromosomes were equally stained. However, both H3K9me3 and H4K20me3 were strongly enriched at blocks of constitutive heterochromatin, especially at the satellite region of the platypus NOR-bearing chromosome 6 (Fig. 2 F and I), and differential distributions were not observed. The strong enrichment at constitutive heterochromatic blocks is in contrast to H3K27me3, whose pattern corresponded more to R-banding and which was depleted on the satellite region of platypus chromosome 6 (Fig. 2C). No metaphases with HP1α protein were detected. However, interphase nuclei showed clusters of this protein, suggesting that HP1α does colocalize to heterochromatic DNA in a cell cycle-dependent manner.
Fig. S3 shows line scans of the distribution of repressive histone modifications. The inactivated possum X homolog is brighter for all repressive marks, as shown by the higher amplitude of the red curve. Similar to the active histone modifications (see above) the platypus X1 homologs do not show a differential pattern for the repressive epigenetic marks.
DNA Methylation.
Immunostaining with a 5-methylcytosine (meC) antibody was used to visualize the overall DNA methylation state of mouse, possum, potoroo, opossum, and platypus metaphase chromosomes. The two X chromosomes in female mouse metaphases showed a generally equivalent reduction in methylated DNA compared with the autosomes (Fig. 3A). The two X chromosomes were identified by chromosome painting (Fig. 3B).
Fig. 3.
DNA methylation in the female mouse, female possum, male potoroo, male opossum, and female and male platypuses. Chromosomes are shown in light gray, and antibody staining is shown in red or green. X-chromosome paints (B and D) to identify the X chromosomes (A and C) are shown. (E) Enrichment of H3K9me3 on the possum Xi. (F) This X homolog has a strong reduction in DNA methylation. (G) Potoroo metaphase shows hypomethylation of one of the X regions of the P. tridactylous-specific X-autosome translocation. (H) DNA methylation on opossum male chromosomes. The arrows in A, C, E, F, G, and H point to the X chromosome (long arrow) and Y chromosome (short arrow). (I and J) DNA methylation on female and male platypus metaphases with relevant chromosomes indicated. (Scale bar, 10 μm.)
In all observed metaphases, one of the two X's in the female possum showed a clear reduced level of DNA methylation (Fig. 3C); both homologs were identified by chromosome painting (Fig. 3D). The autosomes showed even distributions of meC. To determine the identity of the dim X chromosome, we performed sequential immunofluorescence. The procedure was followed to detect the distribution of H3K9me3, and the slides were then washed in 2× SSC, after which the procedure was followed for detection of meC (Materials and Methods) but with a denaturing temperature of 58 °C. Although the methylation detection result is less optimal (because of the sequential immunofluorescence procedure), Fig. 3E (H3K9me3) and Fig. 3F (DNA methylation) both show that it is the Xi marked by an enrichment of H3K9me3 that shows global DNA hypomethylation.
This hypomethylation was confirmed in a different female marsupial, the potoroo (P. tridactylus). In the potoroo, the X chromosome(s) is fused to an autosome. A reduced level of DNA methylation was seen on the X region of one of the translocation homologs; the autosome part had an equal level when compared with its homolog (Fig. 3G). Thus, the hypomethylation section was restricted to one of the X-chromosome regions of the X-autosome translocations specific for this species, consistent with marsupial hypomethylation of one X.
In contrast to the paler staining female X in the marsupial, the male X chromosome of the opossum M. domestica (long arrow in Fig. 3H, fourth lane, small arrow points to the Y) has a methylation distribution equal to that of the autosomes. This supports our finding by sequential immunofluorescence that it is the inactive female X that is hypomethylated in marsupials. The male opossum was chosen because a male potoroo cell culture was unavailable and our cell culture of male possum T. vulpecula contained an extra copy of the X chromosome (XXY).
The X chromosomes in female platypus metaphases did not show a differential level of DNA methylation (Fig. 3I). However, the satellite regions of platypus chromosome 6 and other heterochromatic regions showed enhanced levels of DNA methylation. This latter pattern was also seen on male platypus chromosomes; the Y chromosomes had methylation levels comparable to other chromosomes (Fig. 3J).
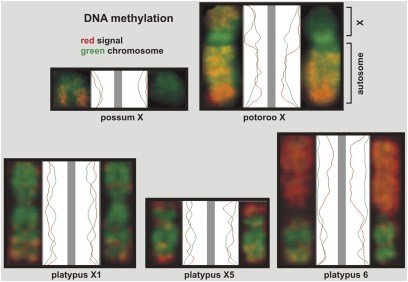
Fig. 4 shows line scans of the distribution of DNA methylation. Hypomethylation of the inactivated possum X is clearly indicated by the lower amplitude of the red signal curve. A diminished presence of methylated DNA can also be seen on the p-arm of one of the potoroo X-autosome chromosomes. The q-arm represents the autosome of the translocated chromosome (see above), has an identical distribution of methylated DNA on both homologs, and functions as an internal control. The platypus homologs have identical distributions, with strong enrichment of DNA methylation along the satellite region of platypus chromosome 6.
Fig. 4.
Line scans of the distribution of DNA methylation on X chromosomes of the female possum, potoroo, and platypus. The green curves correspond to the DAPI staining along the length of the chromosomes. The red curves show the distribution of methylated DNA. The potoroo contains an X-autosome translocation (described in the text).
Discussion
In eutherians, the evolution of XCI is logically considered with respect to the evolution and differentiation of the mammalian X and Y chromosomes, and X inactivation is thought to be a natural consequence of this differentiation. The slow attrition of the Y chromosome led to the loss of functional genes, with only the X-linked copies remaining for the majority (31). The development of the X-inactivation system was completed before the major radiation of therian mammals, because no therian species (so far studied) exists without this dosage compensation system. Therefore, this mechanism was likely to have been established at the start of the Y-attrition process, suggesting that species using this process could cope better with the loss of subsequent genes, and thus were outcompeting those mammalian species that did not have this system. X inactivation may even have accelerated the process of Y attrition (31).
The epigenetic modifications studied here in general have a conserved role in vertebrates for chromatin structure and heterochromatin formation. We show specificity for X inactivation in that modified histones are enriched or depleted on a chromosome-wide scale. XCI is initiated in eutherians by the production of the noncoding Xist RNA that interacts with and spreads in cis across the chromosome, resulting in monoallelic expression of X-linked genes (except escapees). This feature is specific for eutherian mammals and is not seen in Drosophila melanogaster, Caenorhabditis elegans, or marsupials.
The common brushtail possum (T. vulpecula: Phalangeridae) is one of the Australian marsupials studied for this project. Despite the fact that marsupial X inactivation is not initiated by a Xist non-coding RNA (ncRNA), all 10 studied epigenetic modifications are shown to be differential marks for marsupial X inactivation. Seven of those were clear indicators, although three (H3K27me3, H4K8ac, and H4K16ac) showed depletion/enrichment in only a fraction of the observed metaphases. Nonetheless, these results show that in contrast to the mouse, marsupial X inactivation is accompanied by a full battery of active and repressive histone modifications. Our marsupial findings differ from a previous study in the tammar wallaby in which only differential active marks (H3K9Ac, H3K4me2, and H4Kac) were observed (15). This inconsistency might be explained by a technical difference in the sensitivity of protocols used, a difference in antibody batch, or a species-specific difference. The cell types (fibroblasts) used are the same in both studies.
H3K9ac, H3K27me3, H4K8ac, H4K16ac, H3K4me2, and H2AK5ac were epigenetically dimorphic marks on mouse and possum female X's in our experiments. Unlike marsupials, the trimethylated forms of H3K9 and H4K20 did not show Xi specificity in mouse. However, Chadwick and Willard (6) described an enrichment of these modified histones on human Xi and noted that they are spatially distributed in regions that are nonoverlapping with regions of elevated H3K27me3. Also, in bovine models, H3K27me3 and H3K9me3 are spatially distributed in nonoverlapping regions (7). This observed pattern of the mutually exclusive presence of H3K27me3 and H3K9me3 supports our findings in the mouse and possum that although H3K27 has an enhanced presence on the mouse Xi, it is not such a clear XCI mark in the possum. In contrast, H3K9me3 and H4K20me3 are strongly enriched on the possum Xi but are less so on the mouse Xi.
Although not exhibiting epigenetic dimorphism on sex chromosomes, monotremes also show these mutually exclusive features elsewhere. The satellite region of platypus chromosome 6 is strongly enriched with H3K9me3 and H4K20me3, but the same region is depleted for H3K27me3. This one-or-the-other pattern is thus not necessary correlated to X inactivation but, instead, may reflect different repertoires of histone modifications regulating repressive chromatin.
DNA methylation of CpG island promoters of Xi-linked genes has been reported as a late event in the XCI process (32). Cell cycle- and cell type-dependent dynamic differences in methylation have been observed in mice, suggesting a role for this modification in gene repression on the Xi, with chromosome-wide hypomethylation of Xi observed in some instances (33). In our study, mouse metaphases did not exhibit methylation dimorphism between the two X chromosomes consistent with the discrepancies in the literature. However, a reduction in global chromosome DNA methylation was clearly observed in our study on the possum and potoroo Xi compared with the active X and autosomes. This may be associated with the strong epigenetic dimorphism seen for the repressive marks H3K9Me3, HP1, and H4K20Me3, which might preclude the need for a DNA methylation-mediated mechanism of maintenance of the inactive state. However, in the human and mouse (note that the latter has no dimorphism for these three repressive marks in this study), it has also been reported that the Xi is globally hypomethylated (33, 34).
The reduction in global chromosome DNA methylation of the marsupial Xi cannot be readily explained by a differential distribution of CpGs, because marsupial and eutherian X chromosomes have similar CpG contents [around 1.4% (35)]. However, an enrichment of Line/L1 elements seen on the eutherian X chromosome is not observed on the marsupial X chromosome (35). LI elements have been suggested to play a role in Xist-mediated X inactivation (36). We show here that marsupial X inactivation is accompanied by a full set of active and repressive histone modifications despite the lack of Xist. Hence, Line/L1 elements are not directly correlated with these epigenetic modifications but, instead, may be related to Xist-mediated random XCI or DNA methylation-mediated maintenance of silencing; indeed, the accumulation of L1 elements on eutherian X chromosomes may require alternative explanations (37).
Although we report specific differences in the epigenetic state between the mouse and marsupial XCI, in general, the presence of active and repressive histone modifications suggests a related underlying mechanism of dosage compensation. Although marsupial XCI is not regulated by the Xist ncRNA, a role for a currently unidentified alternative ncRNA cannot be ruled out. This RNA might mediate a related repertoire of repressive histone modifications as used in eutherian XCI but confined to the paternal X. The variable dependence of mouse X-linked genes on Xist for the establishment of paternal-specific XCI during preimplantation development (14) may indicate that this ancestral RNA might also contribute to eutherian imprinted X inactivation.
That repressive chromatin modifications are used in both eutherians and marsupial XCI suggests that these epigenetic tools are very suitable for gene silencing over a whole chromosome and were used in the marsupial-eutherian ancestor to establish X-chromosome dosage compensation. It will be of interest to compare underlying mechanisms regulating the early events in XCI, which are mediated by Xist in eutherians. For example, H3K9 methylation and H3K27me3 are the first epigenetic modifications to emerge after the appearance of the Xist ncRNA in eutherians (8, 9, 38, 39). What initiates this process in marsupials? Recent work has suggested that, like eutherians, meiotic sex chromosome inactivation in marsupials is not a persistent state that survives fertilization (28); hence, the XCI initiating mechanisms may be similar. In normal ES cells, the Xist gene is regulated by Oct4 and Sox 2. Xist only becomes active on the future Xi after levels of Oct 4 and Sox2 decrease on differentiation (40–42). To what extent might these pluripotency factors regulate paternal X inactivation in marsupials or, indeed, the early imprinted X inactivation in eutherians when their levels are high?
The epigenetic modifications we studied did not show a differential presence on either of the sex chromosomes in female or male platypus sex chromosomes, which strongly indicates that there is no chromosome-wide epigenetic dosage compensation mechanism in the platypus. The evolution of a chromosome-wide dosage compensation mechanism in monotremes involving a ncRNA similar to that in eutherians would be surprising because it would be expected to involve either multiple ncRNAs acting in cis on all five different X chromosomes or a transacting mechanism involving a single ncRNA. More likely, monotreme X-linked genes may be dosage-compensated on a gene-by-gene basis that may or may not involve RNA-mediated processes. Recently, Deakin et al. (43) investigated platypus dosage compensation at six genes by quantitative RT-PCR, RNA FISH, and SNP analysis. Findings were consistent with some form of compensation at the transcriptional level for some but not all genes assessed. These results support our conclusion that there is no chromosome-wide X inactivation in monotremes.
Materials and Methods
Cell Culture.
The platypus [O. anatinus, diploid chromosome number 52 (2n = 52)] and brush-tailed possum (T. vulpecula, 2n = 20) fibroblast cultures [established for our previous studies (44)] were grown at 32 °C in standard medium; for platypus cell cultures, the medium was supplemented with an equal volume of Amniomax (GIBCO, Invitrogen) to promote cell growth. Female fibroblast metaphase preparations from the potoroo (P. tridactylous, 2n = 12 female and 2n = 13 male), whose normal sex chromosome constitution involves an X:autosome translocation, and from a male opossum (M. domestica, 2n = 18) were generated as described previously (45, 46). Mouse (2n = 40) primary adult (CBAxC57BL6) lung and embryo (129Sv) fibroblasts were generated and grown at 37 °C in standard medium. All cells were harvested at around 60–70% confluency. After hypotonic treatment (SI Materials and Methods), mouse cells were added to the harvested marsupial/monotreme cells, pelleted, and spun onto slides so that cells from the control and test species underwent identical processing. Marsupial and monotreme metaphase chromosomes were identified by cytogenetic analysis, as described previously (20, 45). Mouse sex chromosomes were identified by X-chromosome paints when appropriate (47).
Antibodies and Immunofluorescence.
Antibodies used to detect epigenetic modifications, HP1, and DNA methylation are listed in Table S1, along with methodology for their use on chromosome preparations using protocols adapted from published procedures (3, 48) (SI Materials and Methods). Chromosome preparations were conducted as described previously (45) and are further outlined in SI Materials and Methods.
Fluorescence Microscopy.
For each experiment, 20–40 metaphase spreads were scored for each species, including the mouse. Images were captured using Leica QFISH software (Leica Microsystems) and a cooled CCD camera (Photometrics Sensys; Photometrics) (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank T. Jenuwein (Max Planck Institute of Immunobiology, Freiburg, Germany) for providing an antibody against H3K27me3 and Leica Microsystems for provision of CW4000 CytoFISH software. Work was performed at the Cambridge Resource Centre for Comparative Genomics and supported by grants from the Wellcome Trust (to M.A.F.-S. and W.R.) and the Medical Research Council and Wellcome Trust (to A.C.F.-S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0910322107/-/DCSupplemental.
References
- 1.Heard E. Recent advances in X-chromosome inactivation. Curr Opin Cell Biol. 2004;16:247–255. doi: 10.1016/j.ceb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Heard E, Disteche CM. Dosage compensation in mammals: Fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 3.Belyaev N, Keohane AM, Turner BM. Differential underacetylation of histones H2A, H3 and H4 on the inactive X chromosome in human female cells. Hum Genet. 1996;97:573–578. doi: 10.1007/BF02281863. [DOI] [PubMed] [Google Scholar]
- 4.Jeppesen P, Turner BM. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 5.Chadwick BP, Willard HF. Histone H2A variants and the inactive X chromosome: Identification of a second macroH2A variant. Hum Mol Genet. 2001;10:1101–1113. doi: 10.1093/hmg/10.10.1101. [DOI] [PubMed] [Google Scholar]
- 6.Chadwick BP, Willard HF. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci USA. 2004;101:17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coppola G, Pinton A, Joudrey EM, Basrur PK, King WA. Spatial distribution of histone isoforms on the bovine active and inactive X chromosomes. Sex Dev. 2008;2:12–23. doi: 10.1159/000117715. [DOI] [PubMed] [Google Scholar]
- 8.Heard E, et al. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 9.Mermoud JE, Popova B, Peters AH, Jenuwein T, Brockdorff N. Histone H3 lysine 9 methylation occurs rapidly at the onset of random X chromosome inactivation. Curr Biol. 2002;12:247–251. doi: 10.1016/s0960-9822(02)00660-7. [DOI] [PubMed] [Google Scholar]
- 10.Augui S, et al. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- 11.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 12.Elisaphenko EA, et al. A dual origin of the Xist gene from a protein-coding gene and a set of transposable elements. PLoS ONE. 2008;3:e2521. doi: 10.1371/journal.pone.0002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishihara H, Maruyama S, Okada N. Retroposon analysis and recent geological data suggest near-simultaneous divergence of the three superorders of mammals. Proc Natl Acad Sci USA. 2009;106:5235–5240. doi: 10.1073/pnas.0809297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalantry S, Purushothaman S, Bowen RB, Starmer J, Magnuson T. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature. 2009;460:647–651. doi: 10.1038/nature08161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koina E, Chaumeil J, Greaves IK, Tremethick DJ, Graves JA. Specific patterns of histone marks accompany X chromosome inactivation in a marsupial. Chromosome Res. 2009;17:115–126. doi: 10.1007/s10577-009-9020-7. [DOI] [PubMed] [Google Scholar]
- 16.Wakefield MJ, Keohane AM, Turner BM, Graves JA. Histone underacetylation is an ancient component of mammalian X chromosome inactivation. Proc Natl Acad Sci USA. 1997;94:9665–9668. doi: 10.1073/pnas.94.18.9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rens W, et al. The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol. 2007;8:R243. doi: 10.1186/gb-2007-8-11-r243. 1–R243.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veyrunes F, et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008;18:965–973. doi: 10.1101/gr.7101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bininda-Emonds OR, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 20.Rens W, et al. Resolution and evolution of the duck-billed platypus karyotype with an X1Y1X2Y2X3Y3X4Y4X5Y5 male sex chromosome constitution. Proc Natl Acad Sci USA. 2004;101:16257–16261. doi: 10.1073/pnas.0405702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grützner F, et al. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature. 2004;432:913–917. doi: 10.1038/nature03021. [DOI] [PubMed] [Google Scholar]
- 22.Waters PD, et al. Autosomal location of genes from the conserved mammalian X in the platypus (Ornithorhynchus anatinus): Implications for mammalian sex chromosome evolution. Chromosome Res. 2005;13:401–410. doi: 10.1007/s10577-005-0978-5. [DOI] [PubMed] [Google Scholar]
- 23.Keohane AM, O'Neill LP, Belyaev ND, Lavender JS, Turner BM. X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- 24.Lin H, et al. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 2007;5:e326. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 26.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 27.Schotta G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahadevaiah SK, et al. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr Biol. 2009;19:1478–1484. doi: 10.1016/j.cub.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 29.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 30.Chadwick BP, Willard HF. Chromatin of the Barr body: Histone and non-histone proteins associated with or excluded from the inactive X chromosome. Hum Mol Genet. 2003;12:2167–2178. doi: 10.1093/hmg/ddg229. [DOI] [PubMed] [Google Scholar]
- 31.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norris DP, Brockdorff N, Rastan S. Methylation status of CpG-rich islands on active and inactive mouse X chromosomes. Mamm Genome. 1991;1:78–83. doi: 10.1007/BF02443782. [DOI] [PubMed] [Google Scholar]
- 33.Bernardino J, Lombard M, Niveleau A, Dutrillaux B. Common methylation characteristics of sex chromosomes in somatic and germ cells from mouse, lemur and human. Chromosome Res. 2000;8:513–525. doi: 10.1023/a:1009271706488. [DOI] [PubMed] [Google Scholar]
- 34.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 35.Mikkelsen TS, et al. Broad Institute Genome Sequencing Platform Broad Institute Whole Genome Assembly Team Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 36.Lyon MF. X-chromosome inactivation: A repeat hypothesis. Cytogenet Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- 37.Cantrell MA, Carstens BC, Wichman HA. X chromosome inactivation and Xist evolution in a rodent lacking LINE-1 activity. PLoS ONE. 2009;4:e6252. doi: 10.1371/journal.pone.0006252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zakharova IS, Shevchenko AI, Zakian SM. Monoallelic gene expression in mammals. Chromosoma. 2009;118:279–290. doi: 10.1007/s00412-009-0206-8. [DOI] [PubMed] [Google Scholar]
- 39.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–132. doi: 10.1038/nature08098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Navarro P, Avner P. When X-inactivation meets pluripotency: An intimate rendezvous. FEBS Lett. 2009;583:1721–1727. doi: 10.1016/j.febslet.2009.03.043. [DOI] [PubMed] [Google Scholar]
- 42.Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 43.Deakin JE, Hore TA, Koina E, Marshall Graves JA. The status of dosage compensation in the multiple X chromosomes of the platypus. PLoS Genet. 2008;4:e1000140. doi: 10.1371/journal.pgen.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rens W, et al. Reversal and convergence in marsupial chromosome evolution. Cytogenet Genome Res. 2003;102:282–290. doi: 10.1159/000075764. [DOI] [PubMed] [Google Scholar]
- 45.Rens W, O'Brien PC, Yang F, Graves JA, Ferguson-Smith MA. Karyotype relationships between four distantly related marsupials revealed by reciprocal chromosome painting. Chromosome Res. 1999;7:461–474. doi: 10.1023/a:1009249813617. [DOI] [PubMed] [Google Scholar]
- 46.Rens W, et al. Karyotype relationships between distantly related marsupials from South America and Australia. Chromosome Res. 2001;9:301–308. doi: 10.1023/a:1016646629889. [DOI] [PubMed] [Google Scholar]
- 47.Rabbitts P, et al. Chromosome specific paints from a high resolution flow karyotype of the mouse. Nat Genet. 1995;9:369–375. doi: 10.1038/ng0495-369. [DOI] [PubMed] [Google Scholar]
- 48.Bisoni L, Batlle-Morera L, Bird AP, Suzuki M, McQueen HA. Female-specific hyperacetylation of histone H4 in the chicken Z chromosome. Chromosome Res. 2005;13:205–214. doi: 10.1007/s10577-005-1505-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.