Abstract
The mammalian oocyte possesses powerful reprogramming factors, which can reprogram terminally differentiated germ cells (sperm) or somatic cells within a few cell cycles. Although it has been suggested that use of oocyte-derived transcripts may enhance the generation of induced pluripotent stem cells, the reprogramming factors in oocytes are undetermined, and even the identified proteins composition of oocytes is very limited. In the present study, 7,000 mouse oocytes at different developmental stages, including the germinal vesicle stage, the metaphase II (MII) stage, and the fertilized oocytes (zygotes), were collected. We successfully identified 2,781 proteins present in germinal vesicle oocytes, 2,973 proteins in MII oocytes, and 2,082 proteins in zygotes through semiquantitative MS analysis. Furthermore, the results of the bioinformatics analysis indicated that different protein compositions are correlated with oocyte characteristics at different developmental stages. For example, specific transcription factors and chromatin remodeling factors are more abundant in MII oocytes, which may be crucial for the epigenetic reprogramming of sperm or somatic nuclei. These results provided important knowledge to better understand the molecular mechanisms in early development and may improve the generation of induced pluripotent stem cells.
Keywords: germinal vesicle, metaphase II, zygote, protein, reprogramming
Reprogramming of patient-specific somatic cells into pluripotent stem cells has attracted wide scientific and public interest because of the great potential value in both research and therapy. Recent advances in induced pluripotent stem cell (iPSC) research have clearly indicated that a small number of transcription factors can reverse the cell fate of differentiated somatic cells; however, the reprogramming process remains slow, and the efficiency is low. Typically, 1% of cells are reprogrammed, but this process requires at least 7 d to 2 wk (1–8). In contrast, reprogramming during somatic cell nuclear transfer (SCNT) occurs within one or two cell cycles and often in a majority of embryos (9–14). The oocyte-derived transcripts that promote this more efficient reprogramming remain unidentified; however, it has been suggested that their inclusion with the four transcription factors (Oct4, Sox2, Klf4, and c-Myc) may increase the speed and efficiency of the reprogramming process (15). As a step to identification of these factors, this project sought to define the proteome of mouse oocytes at three stages of development, which will also provide us important information on the factors regulating developmental competence of oocytes.
During mammalian oogenesis, the oocyte undergoes two cell cycle arrests at the dictyate or germinal vesicle (GV) stage and the metaphase II (MII) stage (16, 17). MII oocytes have been widely used to reprogram somatic cell nuclei, because during normal reproduction, sperm and oocyte nuclei are reprogrammed by the MII oocyte to produce totipotent zygotes. By contrast, results from our previous nuclear transfer studies have shown that the cytoplasm of GV stage oocytes have no reprogramming activity (18). Zygotes have lost the ability to reprogram during most of their developmental stages; however, M-phase zygote cytoplasm has a very low reprogramming efficiency (19). A comparison of the proteomes of oocytes at stages with different reprogramming abilities may aid in the identification of the factors responsible for reprogramming.
In the present study, semiquantitative MS was applied to identify the protein composition of each type of oocyte. We successfully identified numerous proteins in oocytes that had not been previously identified, and we discovered a larger number of differentially expressed proteins among the oocytes at different developmental stages.
Results
Identification of Total Proteins Expressed in GV and MII Oocytes and Zygotes.
Overall, 7,000 oocytes at each developmental stage were collected from the mouse strain most commonly used as a recipient for somatic cell nuclear transfer (Fig. 1A). From these oocytes, we successfully identified 132,127 peptides present in the GV oocyte, 185,643 peptides in the MII oocyte, and 85,369 peptides in the zygote using an LTQ Orbitrap mass spectrometer. According to this criterion, the total protein numbers identified in GV, MII, and fertilized oocytes (zygotes) were 2,781, 2,973, and 2,082, respectively. The detailed information of all identified proteins as well as the peptides in all three types of oocytes is shown in Datasets S1 and S2. We categorized the proteins identified in the present study according to their molecular functions. Overall, the distribution of proteins between these different functions seemed to be similar in each type of oocyte (Fig. 1B). The details of the gene ontology categories of the identified proteins are shown in Dataset S3. Approximately 1,000 proteins with unknown functions were identified in each group of oocytes.
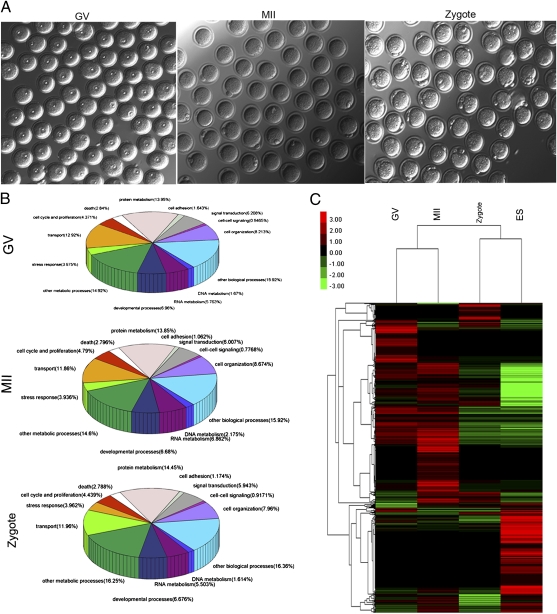
Fig. 1.
Total proteins identified in mouse oocytes and zygotes. (A) Representative images of mouse oocytes at different developmental stages including GV, MII, and zygote. As shown, fully grown GV oocytes with visible germinal vesicles, MII oocytes with the first polar bodies, and zygotes with two pronuclei were carefully selected. (B) Functional categorization of the identified proteins in GV and MII oocytes and zygotes. The identified proteins were grouped into 14 categories according to their molecular functions, which include involvement in the cell cycle and proliferation, transport, stress response, developmental processes, RNA metabolism, DNA metabolism, cell organization and biogenesis, cell–cell signaling, signal transduction, cell adhesion, protein metabolism, death, other metabolic processes, and other biological processes. We then calculated the percentage of every group of proteins. (C) Clustering of proteins in GV and MII oocytes, zygotes, and ES cells with a color gradient for gene abundance ranks. The cluster of more abundantly expressed proteins is highlighted in red. The total number of observed spectra assigned to each protein in each cell type was used as the basis for clustering. Clustering showed that the protein expression profile is similar for MII oocytes and GV oocytes, whereas the profile of zygotes is more similar to ES cells.
To gain insight into the change between each stage, we compared four groups of proteins: GV and MII oocytes, zygotes, and ES cells. We used the number of identified peptides as the measure of protein abundance; for the comparative number of proteins in GV and MII oocytes, zygotes, and ES cell proteins, see Dataset S4. The results showed that GV and MII oocytes are more similar than zygotes and ES cells (Fig. 1C). Compared with oocytes, ES cells express some specific or highly up-regulated proteins, mostly involved in metabolism, such as isoform M1 of pyruvate kinase isozymes M1/M2 (PKM2), GAPDH, α-enolase (ENO1), β-enolase (ENO3), fructose-bisphosphate aldolase A (ALDOA), l-lactate dehydrogenase A chain (LDHA), phosphoglycerate kinase 1 (PGK1), adenosylhomocysteinase (AHCY), and GST ω-1 (GSTO1). More importantly, oocytes and zygotes possess specific protein families, which are more than in ES cells. The majority of these unique protein families are involved in self-renewal and cell cycle regulation; they include the ADP ribosylation factor (ARF) family, the maternal antigen that embryos require (MATER) family, the Tudor family, and the F-box family.
Six ARF family proteins were detected in mouse oocytes (Fig. S1). ADP ribosylation is a protein posttranslational modification that involves the addition of one or more ADP and ribose moieties. Except for the ribosylation of proteins, ARF1 has been shown to play an important role in regulating asymmetric division in meiosis during oocyte maturation (20). The ARF family gene is overrepresented in mouse oocytes and zygotes compared with the ES cells, which may partly explain the asymmetric division of oocytes.
The NACHT, leucine rich repeat and PYD containing (NLRP) family proteins are known as oocyte-specific proteins and are necessary for embryo development. Nlrp5 (also known as Nalp5 or Mater) is a maternal effect gene essential for embryonic development past the two-cell stage (21). Several Nlrp genes are preferentially expressed in germ cells, including Nlrp14, Nlrp4f, and Nlrp4c. We identified 12 NLRP proteins in mouse oocytes (Fig. S1).
TUDOR family proteins are detected in oocytes but not in ES cells. Our present proteomics data confirm that tudor domain containing protein (TDPD), TDRD1, TDRD3, TDRD7, and TDKB are expressed in mouse oocytes. Tdrd1−/− mice showed male sterility with postnatal spermatogenic defects. During spermatogenesis, TDRD1 interacts with the Piwi protein, Mili, and they are colocalized in the cytoplasm of spermatogenic cells, where they associate with the nuage or its derivative, the chromatoid body (22). The full function of the TUDOR family proteins in oocytes remains to be resolved.
A large group of F-box proteins was identified in mouse oocytes and zygotes. The Skp1-Cullin-F-box (SCF) complex functions as an E3 ligase in the ubiquitin protein degradation pathway. Transcripts and proteins must change in composition and abundance during the transition from oocyte to embryo. The SCF protein–ubiquitin ligase complex member F-box proteins are highly abundant in oocytes and two-cell embryos (23). For example, Fbxw15/Fbxo12J is an F-box protein-encoding gene that is selectively expressed in oocytes of the mouse ovary (24). We identified that 19F-box family proteins are overpresented in the oocyte (Fig. S1). These F-box proteins may play important roles in protein degradation after fertilization, and different F-box complexes can selectively degrade specific target proteins.
Overrepresented Pathways in GV Oocytes.
Compared with MII and fertilized oocytes, GV oocytes contain a greater number of metabolism-related proteins responsible for supporting oocyte maturation. SLC family proteins are amino acid transporters, and they play important roles in transporting various molecules across the membranes of the oocyte and the cumulus cells. The SLC family proteins are much more abundant in GV oocytes compared with MII oocytes (Fig. 2A). Primary active transporters and cation channel family members are also more abundant in GV oocytes than in MII oocytes (Fig. 2A). Among the primary active transporters expressed, only the calcium-transporting ATPases are more abundant in MII oocytes.
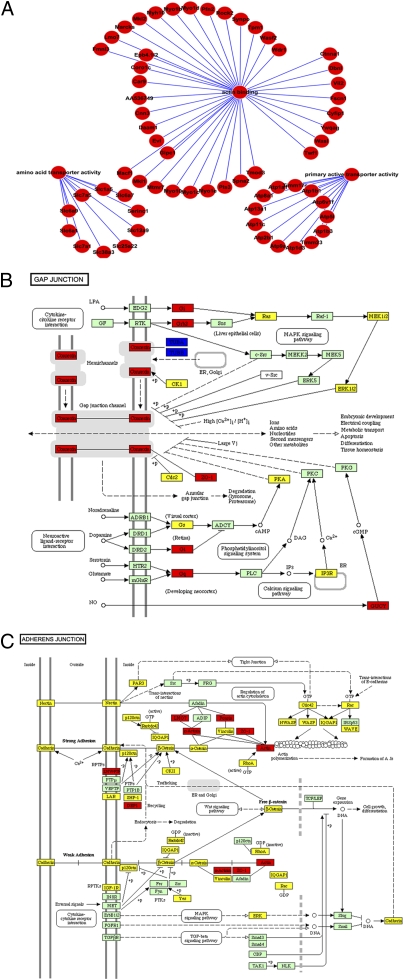
Fig. 2.
Highly expressed proteins in GV oocytes. (A) Actin binding proteins (shown at the top), primary transporters (shown at the left), and amino acid transporters (shown at the right) are more abundantly expressed in GV oocytes compared with MII oocytes and the zygotes. (B) The gap junction pathway is overrepresented in the GV oocyte. Red rectangles represent the gap junction proteins expressed more abundantly in GV oocytes compared with MII oocytes. Yellow rectangles represent the comparable expression level of gap junction proteins between GV and MII oocytes. Green is the color of Kyoto Encyclopedia of Genes and Genomes (KEGG) database. (C) The adhesion junction pathway is overrepresented in the GV oocyte. Red rectangles represent the adhesion junction proteins expressed more abundantly in GV oocytes compared with MII oocytes. Yellow rectangles represent the comparable expression level of adhesion junction proteins between GV and MII oocytes. Green is the color of KEGG database.
In addition to the proteins involved in metabolism, more microfilament motors were identified in the GV oocyte (Fig. 2A). We identified 5 gap junction proteins and 10 adhesion junction pathway proteins that were more abundant in GV oocytes than in MII oocytes (Fig. 2 B and C). The GV oocyte is tightly connected with cumulus cells, and these proteins may play a critical role in gap and adhesion junctions during GV oocyte maturation (25).
Overrepresented Pathways in MII Oocytes.
Transcription factors.
The MII oocyte contains a large number of proteins participating in the regulation of cell cycle events (26). We identified more than 70 transcription factors expressed in the mouse oocyte and zygote. 43 are more prevalent in MII oocytes than in GV oocytes (Fig. 3A), among which Rela, an important factor in stem cells, was identified. Transcriptional activators and repressors are also more abundant in the MII oocyte than the GV oocyte. Some of these proteins, such as Sin3a and Trim28, play important roles in preventing stem cell differentiation.
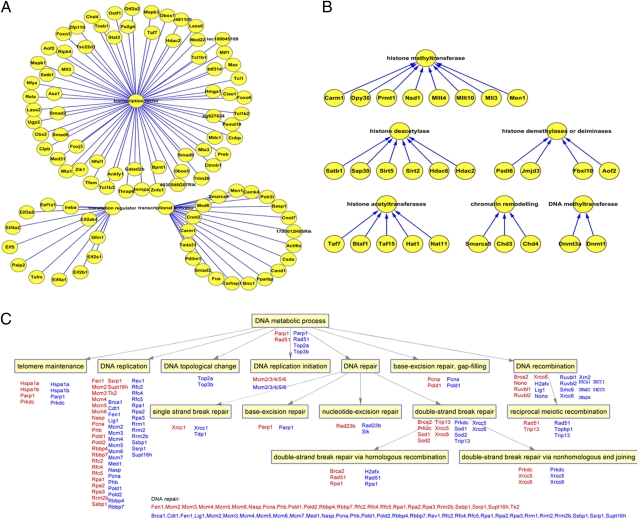
Fig. 3.
Highly expressed proteins in MII oocytes. (A) Transcription factors, translation factors, and transcriptional activators expressed in the MII oocyte. Forty-three transcription factors are more prevalent in MII oocytes than in GV oocytes, and they include TCL family proteins TCL1, TCL1b1, TCL1b2, and TCL1b3; 12 translation factors and 20 transcriptional activators are expressed in MII oocytes. (B) Epigenetic modification-related proteins are expressed more abundantly in MII oocytes. The MII oocytes express four histone demethylases, including Padi6, Jmjd3, Fbxl10, and Aof2, five histone acetyltransferases, including Nat11, Hat1, Taf15, Btaf1, and Taf7, six histone deacetylases, including Hdac2, Hdac6, Sirt2, Sirt5, Sap30, and Satb1, and eight histone methyltranferases, including Men1, Mll3, Mllt10, Mllt4, Nsd1, Prmt1, Dpy30, and Carm1. (C) DNA metabolic pathway in MII oocytes. More DNA repair proteins are expressed in MII oocytes than in GV oocytes; they include the single-strand break repair and nucleotide-excision repair proteins. DNA recombination pathway and DNA replication protein families are also more abundant in MII oocytes. Proteins involved in DNA metabolic pathways are highlighted in blue for MII oocytes and red for GV oocytes.
The T cell leukemia/lymphoma 1 (TCL) family is specifically expressed in oocytes and stem cells. Importantly, Tcl1 functions as a downstream target for Oct4 (27), which is one of the four factors that reprogram differentiated somatic cells to become pluripotent cells (1). A previous study showed that TCL family members occur in a cluster on the chromosome (28). We identified four TCL family factors, TCL1, TCL1b1, TCL1b2, and TCL1b3, in MII oocytes; however, the role of TCL family members in early development remains to be investigated.
Epigenetic modification.
Epigenetic modification enzymes changed dynamically during oocyte maturation, and proteins involved in epigenetic modification were more enriched in the MII oocyte (Fig. 3B). This accumulation of proteins may allow embryonic transcription to initiate correctly, because the GV oocyte is transcriptionally inactive. We also identified three chromatin remodeling enzymes in MII oocytes, including Smarca5, Chd3, and Chd4. Histone demethylases, methylases, acetylases, and histone deacetylases are all more abundant in MII oocytes compared with GV oocytes (Fig. 3B). MII oocytes express eight histone methyltransferases: Men1, Mll3, Mllt10, Mllt4, Nsd1, Prmt1, Dpy30, and Carm1. The trithorax complex can activate transcription by inducing the trimethylation of lysine 4 of histone H3 (H3K4me3) at specific regulatory sites at the target chromatin. TrxG family proteins are highly enriched in MII oocytes, including Smarcc1, Smarca5, Zswim3, Mll3, Mllt10, and Mllt4. Moreover, Hsp90 is also highly expressed in MII oocytes, and Hsp90 cooperates with TrxG at chromatin to maintain the active expression state of targets (29). In addition to histone methyltransferase, MII oocytes also express four histone demethylases (Padi6, Jmjd3, Fbxl10, and Aof2), five histone acetyltransferases (Nat11, Hat1, Taf15, Btaf1, and Taf7), and six histone deacetylases (Hdac2, Hdac6, Sirt2, Sirt5, Sap30, and Satb1). Among the histone modification enzymes, the histone deiminase/demethylase PADI6 is highly expressed in both GV and MII oocytes. PADI6 was identified by 25,365 spectrophotometry counts, comprising 13.6% of the total peptides identified in the MII oocyte. Although PADI6 is highly expressed in the oocyte and zygote, it is totally absent from ES cells. PADI6 is believed to function in the ribosome storage, but its relationship with histone modification is still unclear. A previous study showed that embryonic genome activation (EGA) is defective in Padi6−/− two-cell embryos (30). Another histone modification enzyme attracting our attention was the histone methyltransferase CARM1. CARM1 can catalyze H3 arginine methylation, and the overexpression of CARM1 can influence the formation of the inner cell mass (ICM) (31).
DNA metabolism.
In total, 53 proteins involved in the DNA repair process were identified in both MII oocytes and zygotes. Furthermore, 35 of these proteins are up-regulated in the MII oocyte, including both the single-strand break repair proteins and the nucleotide-excision repair proteins. The MII oocyte contains more proteins in the DNA recombination and DNA replication pathway than either the GV oocyte or the zygote. For example, the base-excision repair, gap-filling protein proliferating cell nuclear antigen is overrepresented in the MII oocyte compared with the GV oocyte. Topological enzymes are all up-regulated in the MII oocyte too (Fig. 3C). Heterochromatin formation proteins are down-regulated in the zygote. SIRT2, SIRT5, and DNA methyltransferases 3a were not detected in the zygote but were identified in the MII oocyte.
Signaling pathways in MII oocytes.
We found that MII oocytes express many genes involved in pluripotency regulation (Fig. S2A), and 11 genes in the MAPK pathway are more highly expressed in MII oocytes (Fig. S2B). The MAPK pathway is known to play an important role in stem cell differentiation, and inhibiting the MAPK pathway may elevate the efficiency of deriving iPSCs (32). NOT2, NOT3, and NOT5 are highly expressed in MII oocytes. The CCR4–NOT complex is a nuclear complex that can negatively regulate the basal and activated transcription of many genes (33). Trim28 and Cnot3 have been shown to be important regulators of the pluripotency of ES cells (34).
Members of the TGF superfamily are potent regulators of cell proliferation and differentiation in a number of organisms (35–38). Consistent with a previous study showing that TGF-β pathway proteins are highly expressed in human oocytes (39), TGF-β pathway proteins are more abundant in the MII oocyte than in the GV oocyte. In this pathway, Smad2 and Smad3 are important factors in maintaining the pluripotency of stem cells (40) (Fig. S3A). The IL-6 signaling pathway is also up-regulated in GV and MII oocytes. Leukemia inhibitory factor (LIF) is an archetypal self-renewal factor for mouse ES cells and belongs to the family of IL-type cytokines (41). We identified the expression of the common IL-6 receptor IL6st, also called gp130, in MII oocytes. Additionally, the LIF pathway member STAT3 is more abundant in the MII oocyte than in the zygote (Fig. S3B). Stat3 activation has been shown to be necessary and sufficient to maintain self-renewal of mouse ES cells in the presence of serum (42).
MII oocytes contain more proteins involved in RNA processing and Golgi transportation than zygotes (Fig. S4 A and B). Undoubtedly, RNA processing is essential for the oocyte–embryo transition, but surprisingly, the Golgi transportation-related proteins are more abundant in MII oocytes than in either GV oocytes or zygotes, although the Golgi is disassembled at metaphase.
Significantly Altered Pathways in Zygotes.
Ubiquitination pathway.
After fertilization, maternal proteins are quickly degraded. As a result, 185,643 peptides were identified in MII oocytes, whereas only 85,369 peptides were identified in zygotes; however, the same number of oocytes and zygotes were analyzed. Proteins involved in the ubiquitination pathway are highly enriched in the zygote, where they may play essential roles in degrading the maternal proteins inherited from the MII oocytes (33). Zygotes express more key factors involved in protein degradation, including ubiquitin B (UBB) and ubiquitin C (UBC). UBC is up-regulated about 1,000-fold in zygotes compared with MII oocytes, which suggests that protein degradation is highly active in zygotes (Fig. 4A). The expression of proteasome members is also up-regulated in zygotes. The ubiquitination pathway not only is responsible for protein degradation but also plays an important role in epigenetic regulation. We identified an E3 ligase, HECT, UBA and WWE domain containing 1 (Huwe1), expressed in the oocyte and zygote, but isoform 2 of HUWE1 was only identified in the zygote. HUWE1 has an important role in ubiquitinating histones (43). In addition, we identified another E3 ubiquitin–protein ligase Uhrf1 (also named NP95), which is highly expressed in the oocyte/zygote compared with ES cells. UHRF1 plays an important role in the ubiquitination pathway and in DNA methylation maintenance. Therefore, in addition to degrading maternal proteins, the ubiquitination pathway may play an important role in reprogramming.
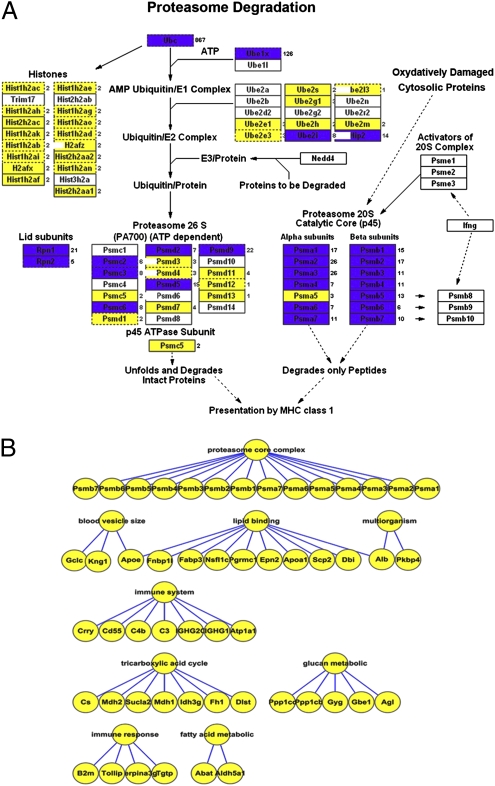
Fig. 4.
Highly expressed proteins in zygotes. (A) Ubiquitination pathway proteins expressed in the zygote. The blue rectangles represent proteins that were identified by more than five spectrophotometry counts. The yellow rectangles represent proteins that were identified by fewer than five spectrophotometry counts. UBC is expressed much more abundantly in zygotes than in MII oocytes. (B) Proteasome complexes and several pathways involved in embryo development were expressed in the zygote. New metabolic pathways begin to be expressed in the zygote, which include the TCA (citrate cycle) pathway and fatty acid and glucan metabolic proteins.
Other pathways.
Mitochondrial components of the MII oocyte differ significantly from the components of the zygote (Fig. S4C). MII oocytes contain a group of mitochondrial proteins, which are totally absent in the zygote; however, the zygote contains more up-regulated mitochondrial proteins including Cytochrome C Oxidase, the last enzyme in the respiratory electron transport chain of mitochondria.
Our data showed that zygotes start to express genes involved in immune system development and certain genes involved in organogenesis. The expression of certain metabolism pathway genes is initiated at the zygote stage, including the citrate cycle pathway, glucan metabolism, lipid binding proteins, and fatty acid metabolism-related genes (Fig. 4B).
Discussion
1D or 2D electrophoresis combined with MS was performed in several studies to identify the total proteins and differentially expressed proteins in oocytes at different developmental stages; however, only a few proteins were identified because of the limited quantity of oocytes used and the limitations of the technology (26, 44, 45). In a more recent report, only 625 proteins were found to be differentially presented, although the quantity of the oocytes was increased to several thousand (46). Compared with the methods previously used, the semiquantitative MS analysis in the present study is more accurate and has been widely applied to investigate the proteome of different organisms (47–49). We successfully identified 2,781 proteins in the GV stage oocyte, 2,973 proteins in the MII stage oocyte, and 2,082 proteins in the fertilized oocyte (zygote).
In general, the proteome characteristics of mouse oocytes discovered in the present study are consistent with previous findings on gene expression profiles of oocytes and early embryos analyzed by either microarray or sequencing of the ESTs from mouse cDNA libraries (50–52). For example, the gene expression analysis of GV oocytes has revealed that abundant expression of genes involved in cell communication and metabolism is the major characteristic of GV oocytes, which is in agreement with the proteome characteristic of GV oocytes detected in the present study (52). Similarly, the proteome of MII oocytes has shown that DNA damage and repair-related proteins are overrepresented, which is consistent with the gene expression data obtained previously (51). Meanwhile, we noticed that close to 3,000 proteins could be detected in the mouse oocytes, whereas the total transcripts detected in mouse oocyte are over 9,000 (51). Such difference might be caused by either low abundant expression of some proteins or the nontranslation of some mRNAs in oocytes.
In summary, our present study provides an invaluable resource for scientists to further investigate the functions of proteins expressed specifically in oocytes at different developmental stages. Such efforts will enable us to better understand the molecular mechanism of early embryo development and possibly improve the efficiency of iPSC generation.
Materials and Methods
We collected 7,000 GV, MII, and zygote oocytes separately from a total of 1,000 B6D2 F1 female mice. The oocytes were then lysed and prepared for the LC-MS/MS analysis. Tandem mass spectra were searched against the European Bioinformatics Institute International Protein Index mouse protein database. Expression Analysis Systematic Explorer (EASE) was used to analyze gene ontology and KEGG pathways. Additional details are in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the laboratory members for helpful comments on the manuscript. The work was supported by Ministry of Science and Technology Grants 2008AA022311, 2010CB944900, and 2011CB964800.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013185107/-/DCSupplemental.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 3.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 7.Kang L, Wang J, Zhang Y, Kou Z, Gao S. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell. 2009;5:135–138. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Zhao XY, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 9.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 10.Inoue K, et al. Effects of donor cell type and genotype on the efficiency of mouse somatic cell cloning. Biol Reprod. 2003;69:1394–1400. doi: 10.1095/biolreprod.103.017731. [DOI] [PubMed] [Google Scholar]
- 11.Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 12.Eggan K, et al. Mice cloned from olfactory sensory neurons. Nature. 2004;428:44–49. doi: 10.1038/nature02375. [DOI] [PubMed] [Google Scholar]
- 13.Hochedlinger K, Jaenisch R. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature. 2002;415:1035–1038. doi: 10.1038/nature718. [DOI] [PubMed] [Google Scholar]
- 14.Sung LY, et al. Differentiated cells are more efficient than adult stem cells for cloning by somatic cell nuclear transfer. Nat Genet. 2006;38:1323–1328. doi: 10.1038/ng1895. [DOI] [PubMed] [Google Scholar]
- 15.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. Intercellular communication in the mammalian ovary: Oocytes carry the conversation. Science. 2002;296:2178–2180. doi: 10.1126/science.1071965. [DOI] [PubMed] [Google Scholar]
- 17.Sugiura K, Pendola FL, Eppig JJ. Oocyte control of metabolic cooperativity between oocytes and companion granulosa cells: Energy metabolism. Dev Biol. 2005;279:20–30. doi: 10.1016/j.ydbio.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 18.Gao S, et al. Germinal vesicle material is essential for nucleus remodeling after nuclear transfer. Biol Reprod. 2002;67:928–934. doi: 10.1095/biolreprod.102.004606. [DOI] [PubMed] [Google Scholar]
- 19.Egli D, Rosains J, Birkhoff G, Eggan K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447:679–685. doi: 10.1038/nature05879. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Hu J, Guo X, Liu JX, Gao S. ADP-ribosylation factor 1 regulates asymmetric cell division in female meiosis in the mouse. Biol Reprod. 2009;80:555–562. doi: 10.1095/biolreprod.108.073197. [DOI] [PubMed] [Google Scholar]
- 21.Tong ZB, et al. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet. 2000;26:267–268. doi: 10.1038/81547. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Saxe JP, Tanaka T, Chuma S, Lin H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol. 2009;19:640–644. doi: 10.1016/j.cub.2009.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knowles BB, Evsikov AV, de Vries WN, Peaston AE, Solter D. Molecular control of the oocyte to embryo transition. Philos Trans R Soc Lond B Biol Sci. 2003;358:1381–1387. doi: 10.1098/rstb.2003.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De La Chesnaye E, et al. Fbxw15/Fbxo12J is an F-box protein-encoding gene selectively expressed in oocytes of the mouse ovary. Biol Reprod. 2008;78:714–725. doi: 10.1095/biolreprod.107.063826. [DOI] [PubMed] [Google Scholar]
- 25.Memili E, et al. Bovine germinal vesicle oocyte and cumulus cell proteomics. Reproduction. 2007;133:1107–1120. doi: 10.1530/REP-06-0149. [DOI] [PubMed] [Google Scholar]
- 26.Ma M, et al. Protein expression profile of the mouse metaphase-II oocyte. J Proteome Res. 2008;7:4821–4830. doi: 10.1021/pr800392s. [DOI] [PubMed] [Google Scholar]
- 27.Hu T, et al. Octamer 4 small interfering RNA results in cancer stem cell-like cell apoptosis. Cancer Res. 2008;68:6533–6540. doi: 10.1158/0008-5472.CAN-07-6642. [DOI] [PubMed] [Google Scholar]
- 28.Paillisson A, et al. Identification, characterization and metagenome analysis of oocyte-specific genes organized in clusters in the mouse genome. BMC Genomics. 2005;6:76. doi: 10.1186/1471-2164-6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tariq M, Nussbaumer U, Chen Y, Beisel C, Paro R. Trithorax requires Hsp90 for maintenance of active chromatin at sites of gene expression. Proc Natl Acad Sci USA. 2009;106:1157–1162. doi: 10.1073/pnas.0809669106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yurttas P, et al. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135:2627–2636. doi: 10.1242/dev.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemberger M, Dean W. Epigenetic arbitration of cell fate decisions: Tipping the bias. Dev Cell. 2007;12:176–178. doi: 10.1016/j.devcel.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Xu J, et al. MAPK/ERK signalling mediates VEGF-induced bone marrow stem cell differentiation into endothelial cell. J Cell Mol Med. 2008;12:2395–2406. doi: 10.1111/j.1582-4934.2008.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki A, Igarashi K, Aisaki K, Kanno J, Saga Y. NANOS2 interacts with the CCR4-NOT deadenylation complex and leads to suppression of specific RNAs. Proc Natl Acad Sci USA. 2010;107:3594–3599. doi: 10.1073/pnas.0908664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu G, et al. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z, Sui L, Toh WS, Lee EH, Cao T. Stage-dependent effect of TGF-beta1 on chondrogenic differentiation of human embryonic stem cells. Stem Cells Dev. 2009;18:929–940. doi: 10.1089/scd.2008.0219. [DOI] [PubMed] [Google Scholar]
- 36.Xu RH, et al. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang J, Ng HH. TGFbeta and SMADs talk to NANOG in human embryonic stem cells. Cell Stem Cell. 2008;3:127–128. doi: 10.1016/j.stem.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 38.Yang W, et al. Extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase pathway is involved in myostatin-regulated differentiation repression. Cancer Res. 2006;66:1320–1326. doi: 10.1158/0008-5472.CAN-05-3060. [DOI] [PubMed] [Google Scholar]
- 39.Kocabas AM, et al. The transcriptome of human oocytes. Proc Natl Acad Sci USA. 2006;103:14027–14032. doi: 10.1073/pnas.0603227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 41.Dani C, et al. Paracrine induction of stem cell renewal by LIF-deficient cells: A new ES cell regulatory pathway. Dev Biol. 1998;203:149–162. doi: 10.1006/dbio.1998.9026. [DOI] [PubMed] [Google Scholar]
- 42.Raz R, Lee CK, Cannizzaro LA, d'Eustachio P, Levy DE. Essential role of STAT3 for embryonic stem cell pluripotency. Proc Natl Acad Sci USA. 1999;96:2846–2851. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z, Oughtred R, Wing SS. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol Cell Biol. 2005;25:2819–2831. doi: 10.1128/MCB.25.7.2819-2831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vitale AM, et al. Proteomic profiling of murine oocyte maturation. Mol Reprod Dev. 2007;74:608–616. doi: 10.1002/mrd.20648. [DOI] [PubMed] [Google Scholar]
- 45.Meng Y, et al. The protein profile of mouse mature cumulus-oocyte complex. Biochim Biophys Acta. 2007;1774:1477–1490. doi: 10.1016/j.bbapap.2007.08.026. [DOI] [PubMed] [Google Scholar]
- 46.Zhang P, et al. Proteomic-based identification of maternal proteins in mature mouse oocytes. BMC Genomics. 2009;10:348. doi: 10.1186/1471-2164-10-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malmström J, et al. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature. 2009;460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petyuk VA, et al. Characterization of the mouse pancreatic islet proteome and comparative analysis with other mouse tissues. J Proteome Res. 2008;7:3114–3126. doi: 10.1021/pr800205b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- 51.Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 52.Evsikov AV, et al. Cracking the egg: Molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev. 2006;20:2713–2727. doi: 10.1101/gad.1471006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.