Abstract
The most detrimental responses of the UV-exposed skin are triggered by cyclobutane pyrimidine dimers (CPDs). Although placental mammals rely solely on nucleotide excision repair (NER) to eliminate CPDs, none of the core NER factors are apparently able to distinguish this hazardous lesion from native DNA, raising the question of how CPDs are circumscribed to define correct excision boundaries. A key NER intermediate involves unwinding of the damaged duplex by transcription factor TFIIH, a reaction that requires xeroderma pigmentosum group D (XPD) protein. This study was prompted by the observation that the ATPase/helicase activity of XPD is necessary for an effective anchoring of this subunit to UV lesions in mammalian nuclei. The underlying mechanism by which XPD impinges on damaged DNA has been probed with a monomeric archaeal homolog, thus revealing that the collision with a single CPD inhibits the helicase but stimulates its ATPase activity. Restriction and glycosylase protection assays show that the XPD helicase remains firmly bound to a CPD situated in the translocated strand along which the enzyme moves with 5′–3′ polarity. Competition assays confirm that a stable complex is formed when the XPD helicase encounters a CPD in the translocated strand. Instead, the enzyme dissociates from the substrate after running into a CPD in the complementary 3′–5′ strand. These results disclose a damage verification and demarcation process that takes place by strand-selective immobilization of the XPD helicase and its conversion to a site-specific ATPase at DNA lesions.
Keywords: cancer, DNA damage responses, molecular recognition, ultraviolet light
The absorption of UV light by DNA results in mutagenic cross-links between adjacent bases, primarily cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6-4) photoproducts (6-4PPs) (1, 2). Of these photolesions, CPDs are responsible for a majority of the severe endpoints of UV radiation such as cutaneous erythema, hyperplasia, and cancer (3, 4). The adverse UV effects are alleviated by a plethora of DNA damage responses, but nucleotide excision repair (NER) represents the only error-free mechanism for photodimer removal in placental mammals (5–7). The relevance of DNA repair is highlighted by the inherited disorder xeroderma pigmentosum (XP) where defects in global-genome NER, operating across the entire genome, lead to a > 1,000-fold increased incidence of sunlight-induced skin cancer (8, 9). This genome-wide pathway is initiated by the XPC-Rad23B complex, which acts as a generic sensor of DNA distortions (10, 11). The XPC subunit provides a landing platform for transcription factor TFIIH, whose unwinding activity assisted by XPA and replication protein A (RPA) generates a NER intermediate in which the DNA is melted over 25–30 nucleotides. Finally, the margins of this open complex are cleaved by structure-specific endonucleases that release the offending damage by dual DNA incision (12, 13).
The problem of detecting CPDs, as opposed to 6-4PPs, resides with the minimal thermodynamic and structural changes caused by this type of lesion (14, 15). Although the core NER subunits implicated in damage recognition (XPC-Rad23B, XPA, and RPA) all display an increased affinity for 6-4PPs, they fail to discriminate between CPD sites and the native double helix (16–18). As an alternative means of detecting NER substrates, vertebrate organisms display an auxiliary factor known as UV-damaged DNA-binding (UV-DDB) protein (19, 20). This extra player provides a DNA-binding subunit (DDB2) that detects the otherwise poorly recognizable CPDs but is itself not a component of the ultimate recognition complex. In fact, upon XPC recruitment, UV-DDB leaves the CPD site (21) and DDB2 is degraded (22). Also, UV-DDB binds with high affinity to mismatched bases and abasic sites, which are not or only poorly processed by the mammalian NER system (23). Thus, the key question is how downstream factors verify damaged sites and distinguish CPDs, or other similar lesions that resemble undamaged DNA, from the native double helix.
To form open intermediates, the TFIIH complex uses two unwinding enzymes that differ in their catalytic properties. Xeroderma pigmentosum group D (XPD) represents the dominant helicase with 5′–3′ polarity (24) whose enzymatic function is required for DNA repair but not for transcription (25, 26). Instead, XPB is an ATPase with minor 3′–5′ helicase activity (26). Recent biochemical and structural studies demonstrated that archaeal homologs provide an excellent model to analyze the specialized role of the XPD subunit in the NER pathway (27). Therefore, we used the XPD protein of a mesophilic archaeon to monitor the consequences of a collision of this DNA helicase with CPD lesions. Together with the shortened residence time of an active site mutant in nuclear UV foci, our molecular analysis reveals that the XPD helicase acts as a dynamic sensor that scans DNA and thereby promotes a strand-selective lesion verification process, which culminates in site-specific lesion demarcation.
Results
Anchoring to Foci of DNA Damage.
To test the interaction of human XPD with damaged DNA in living cells, we exploited the fact that a catalytically inactive mutant carrying a K48R substitution in its ATPase motif is readily incorporated into the TFIIH complexes of CHO cells (25). The ATPase/DNA helicase activity of XPD is not essential for the actual recruitment of TFIIH to damaged sites (28). Therefore, wild-type and mutant XPD were fused with GFP to visualize their accumulation in UV-irradiated nuclear areas. Foci of UV damage were identified by immunostaining and, as observed before (28), both fusion proteins showed a faithful colocalization with CPD spots, demonstrating that TFIIH assembled with inactive XPD is engaged at damaged sites (Fig. 1A). However, by quantifying the signals in cells expressing equal levels of fusion proteins, we found that the K48R mutant yields foci with lower fluorescence intensity than the wild-type control (Fig. 1B).
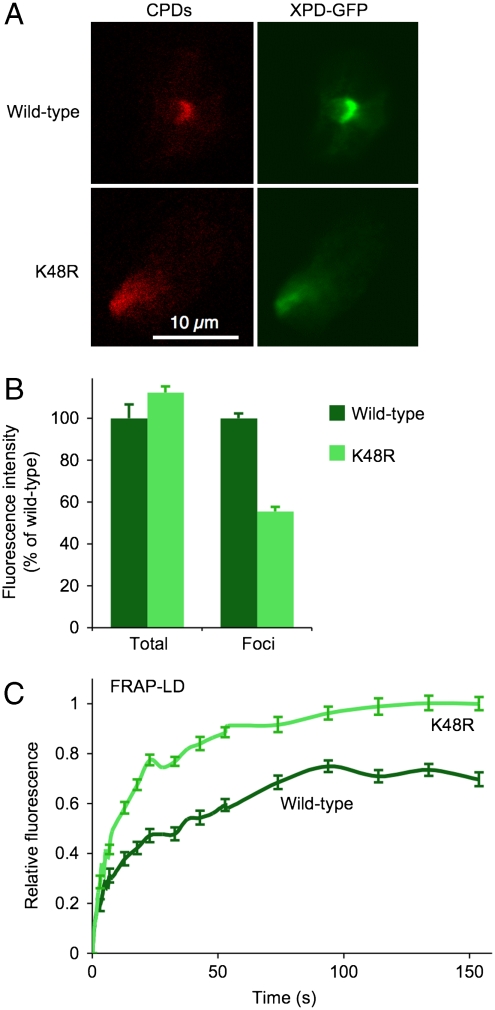
Fig. 1.
Interaction of human XPD with damaged sites in living cells. (A) Representative foci of UV damage in CHO cells detected by immunochemistry against CPDs 30 min after UV irradiation. The accumulation of XPD proteins (wild-type and K48R mutant) is visualized by measuring the fluorescence intensity in cells transfected with the respective GFP constructs. (B) Comparison between total fluorescence, reflecting the overall expression of XPD fusions, and local fluorescence intensity in UV foci (N = 30; ± SEM). See SI Text for quantification methods. (C) Dissociation of XPD-GFP proteins from UV foci determined by fluorescence recovery after photobleaching on local damage (FRAP-LD) (N = 13; ± SEM). See SI Text for a detailed description of data acquisition and analysis.
The cause of this intriguing difference between active and inactive polypeptides was examined by subjecting the XPD-GFP foci to analyses by fluorescence recovery after photobleaching on local damage. For that purpose, the fluorescence of individual foci was photobleached to reduce their intensity to the same level as the nuclear background. Subsequently, fluorescence redistributions due to the exchange of bleached molecules with nonbleached counterparts (29) were recorded over time, thus yielding distinct residence times (Fig. 1C). The diverging plateau of the fluorescence recovery profiles reveals that the wild-type enzyme persists in the UV foci, whereas the K48R mutant dissociates completely from lesion sites. Thus, the distinguishable dissociation curves indicate that the intrinsic ATPase/helicase activity of XPD protein results in the immobilization of this central TFIIH subunit onto damaged DNA.
Binding of XPD Helicase to a Site-Directed CPD.
Next, an archaeal homolog closely related in sequence to the human enzyme and active at moderate temperatures (30) was used to test how the XPD subunit interacts with native and damaged substrates. This XPD protein from Ferroplasma acidarmanus (FaXPD) was purified (Fig. 2A) and incubated with radiolabeled 51-mer oligonucleotides (Fig. 2B). When the nucleoprotein products were monitored in electrophoretic mobility shift assays, a nearly complete association with single-stranded DNA was detected after 15-min preincubations in the presence of ATP, regardless of whether the oligonucleotides were undamaged (Fig. 2C, lane 2) or modified with a site-directed CPD (lane 9).
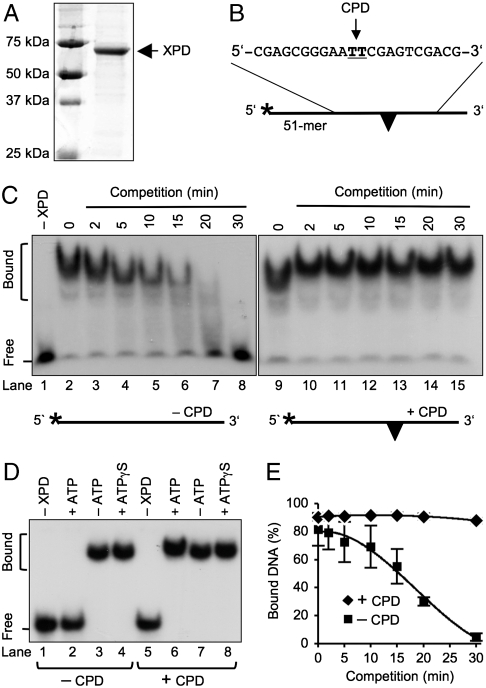
Fig. 2.
Immobilization of the XPD enzyme. (A) Electrophoretic analysis and Coomassie staining of FaXPD protein. (B) Oligonucleotide used for competition assays. The CPD is indicated by a triangle and the 32P label by an asterisk. (C) Competition in the presence of 3 mM ATP showing the dissociation of FaXPD from undamaged oligomers (lanes 3–8) and the stability of radiolabeled complexes containing a single CPD (lanes 10–15). Competitor DNA (undamaged 51-mer) was added in a 50-fold excess. Lanes: 1, incubation without protein; 2 and 9, control incubations (60 nM FaXPD and 5 nM radiolabeled oligonucleotides) without competitor DNA. F, free probes; B, protein-bound fraction. (D) ATP-dependent dissociation of FaXPD from undamaged oligonucleotides. Lanes: 4 and 8, competition assays with nonhydrolyzable ATPγS. (E) Quantification of competition assays. FaXPD (60 nM) was incubated (15 min) with radiolabeled 51-mers (5 nM). A 50-fold molar excess of unlabeled 51-mers was then added in the presence of 3 mM ATP and, after varying competition periods, the samples were analyzed in mobility shift gels. The fraction of protein-bound DNA is represented as the percentage of total radioactivity (N = 3; ± SD).
The complexes were then challenged by the addition of a 50-fold molar excess of unlabeled 51-mers. If the substrate is undamaged, ATP hydrolysis drives an unhindered XPD movement toward the 3′-terminal nucleotides, where the enzyme dissociates from the DNA ends. Subsequent reassociations take place preferentially with the surplus of unlabeled DNA, resulting in a progressive loss of radiolabeled complexes as demonstrated in Fig. 2C, lanes 3–8. After a 30-min incubation with competitor DNA, the radiolabeled probes were completely released from their interaction with the protein and migrated into the gel as free oligonucleotides (Fig. 2C, compare lanes 1 and 8). Fig. 2D shows that this dissociation from undamaged oligonucleotides does not take place in the absence of ATP (lane 3) or upon the replacement of ATP with a nonhydrolyzable analog (lane 4). On the other hand, when the labeled oligomers carry a CPD, the XPD helicase forms nucleoprotein complexes that are refractory to the challenge with contending DNA [Figs. 2C (lanes 10–15) and D (lane 6)]. The quantification of these competition assays demonstrates that the enzyme remains tightly bound to CPD-modified oligonucleotides, whereas, in the absence of damage, the complexes gradually dissociate until all radiolabeled oligomers are released (Fig. 2E). These findings indicate that the active XPD helicase discriminates CPDs by generating a stable intermediate after encountering the lesion during its 5′–3′ movement along single-stranded DNA.
Inhibition of DNA Helicase Activity.
To examine the consequences of a dynamic collision with damaged bases during the unwinding of double-stranded DNA, partial duplex substrates were constructed with a single CPD either in the 5′–3′ translocated or the 3′–5′ displaced strand (Fig. 3A and Fig. S1). In these substrates, single-stranded overhangs of 44 nucleotides are flanked by a duplex segment of 81 base pairs designed to contain the CPD lesion within a unique EcoRI sequence. Accordingly, the modification could be confirmed by EcoRI restriction, which is suppressed by a CPD in one of the two DNA strands (Fig. S2).
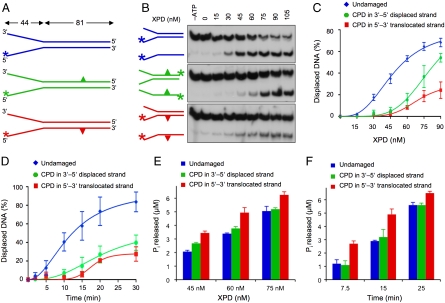
Fig. 3.
Differential impact on XPD enzyme activity. (A) Schematic view of fork substrates. The CPD is located either in the 5′–3′ translocated or the 3′–5′ displaced strand. (B) Typical autoradiographs showing the inhibition of XPD helicase by a single CPD either in the translocated (Bottom) or displaced strand (Middle) of forked substrates (5 nM). (C) Dose dependence of helicase activity. The indicated concentrations of FaXPD were incubated (15 min) with forked DNA substrate (5 nM). The CPD is located either in the translocated or the displaced strand. Oligonucleotide displacement is expressed as the percentage of total radioactivity in each reaction (N = 3; ± SD). (D) Time course experiments. FaXPD (60 nM) was incubated with forked substrates (5 nM) for the indicated time periods (N = 3; ± SD). (E) Dose-dependent stimulation of ATPase activity. The indicated concentrations of FaXPD were incubated with forked DNA (5 nM) in helicase reaction buffer containing 3 mM ATP (N = 3; ± SD). (F) Time course of Pi release upon incubation of FaXPD protein (60 nM) with forked DNA substrates (5 nM) (N = 3; ± SD).
We established by mobility shift assays that the CPD does not impede the initial association of FaXPD with forked DNA molecules. A saturating substrate binding is observed at an XPD concentration of 75 nM regardless of whether or not the partial duplexes carry a CPD lesion (Fig. S3). However, the presence of a single CPD in the translocated 5′–3′ strand led to a pronounced inhibition of DNA helicase activity and a more moderate inhibition was detected when the CPD was located in the displaced 3′–5′ strand (Fig. 3B). Dose dependence (Fig. 3C) and time course experiments (Fig. 3D) confirmed that, at all tested protein concentrations and incubation periods, damaged substrates containing a CPD result in less efficient helicase activity compared to the undamaged control. Thus, CPDs represent a barrier to the movement of the XPD helicase along DNA.
Stimulation of ATPase Activity.
The kinetics of ATP hydrolysis was tested in helicase reaction mixtures containing 45–75 nM of FaXPD and 5 nM forked DNA. The ability of the XPD enzyme to hydrolyze ATP is dependent on the presence of DNA but, surprisingly, the observed suppression of helicase activity was not paralleled by a corresponding inhibition of the ATPase reaction. On the contrary, the forked DNA substrate containing a CPD in the 5′–3′ translocated strand induces higher levels of ATP hydrolysis than the undamaged control or the counterpart with a CPD in the 3′–5′ displaced strand (Fig. 3E). Time course experiments confirmed that a CPD in the 5′–3′ strand results in increased rates of ATP hydrolysis compared to duplexes where either the 5′–3′ strand or both strands are undamaged (Fig. 3F).
It may be argued that 125-mer substrates with just one lesion still contain sufficient native DNA to stimulate ATPase activity at positions flanking the CPD. Therefore, ATPase assays were carried out with oligonucleotides of 30 or 51 residues in length. Overall, these short oligomers induce less ATP hydrolysis than forked substrates but the ones carrying a CPD were again slightly more effective than the undamaged controls (Fig. S4). Thus, even very short CPD-damaged oligonucleotides promote ATPase activity despite the fact that the helicase is blocked at the lesion sites. These findings indicate that DNA damage inhibits the XPD helicase function but not the accompanying ATPase activity.
Protection from Restriction Digestion.
After running into a CPD during DNA unwinding, the helicase may either dissociate from the duplex or, alternatively, form a tight complex as observed in Fig. 1 with single-stranded DNA. To analyze the fate of FaXPD encountering a lesion, the CPD modification in forked substrates was flanked by restriction sites for HaeIII (upstream of the CPD; Fig. 4A), SalI (in close vicinity to the CPD), and PstI (downstream of the CPD). These endonucleases were used to probe the products of 15-min helicase reactions. Efficient cleavage would show that the respective restriction sequence is protein-free whereas reduced cutting would indicate a close interaction of the XPD helicase thereby shielding the DNA substrate from digestion.
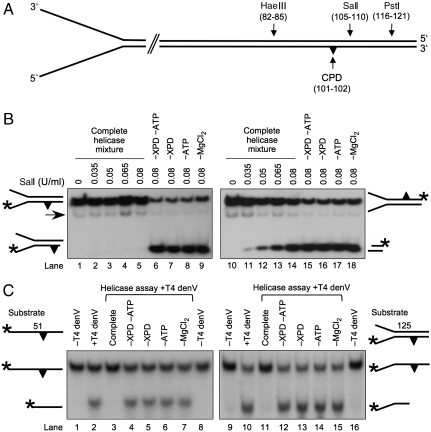
Fig. 4.
Protection assays showing that FaXPD forms a lesion demarcation complex. (A) Position of the HaeIII, SalI, and PstI recognition sequences in 125-mer forked substrates. (B) Protection from SalI cleavage. FaXPD (60 nM) was preincubated (15 min) with partial duplexes (5 nM) and ATP (3 mM), followed by treatment with SalI. The SalI site is occluded by the XPD helicase when the substrate contains a CPD in the 5′–3′ strand (Left) but not if the CPD is situated in the 3′–5′ strand (Right). Lanes: 6–9 and 15–18, control reactions with incomplete helicase mixtures. The arrows indicate the position of the displaced strand. (C) Glycosylase protection assay with single-stranded DNA (Left) and forked substrates (Right). Helicase reaction products were probed by incubation with T4 denV and resolved on denaturing polyacrylamide gels. Lanes: 4–7 and 12–15, control incubations with incomplete helicase mixtures.
When the preceding helicase reaction was performed with a CPD in the 5′–3′ translocated strand, the SalI cleavage was completely inhibited (Fig. 4B, lanes 2–5). This SalI site is occluded only when the preincubation mixture is supplied with all helicase reagents. No protection was detected when either the XPD enzyme itself or one of the cofactors (ATP or MgCl2) was omitted during the preincubation (Fig. 4B, lanes 6–9). These results confirm that the 5′–3′ movement of the XPD helicase is arrested by a CPD lesion, resulting in a stable nucleoprotein complex at damaged sites. Instead, the XPD helicase fails to protect from SalI cleavage if the CPD is located in the 3′–5′ displaced strand (Fig. 4B, lanes 11–14), indicating that the collision with a lesion in the opposing strand triggers dissociation of the enzyme from DNA.
Contrary to the findings with SalI, the helicase was unable to prevent the digestion by HaeIII (Fig. S5A), indicating that the respective site located 16 nucleotides upstream of the CPD remains free of protein even though the adjacent SalI region is obstructed by a stalled XPD. Similarly, preincubation with XPD did not protect from the cleavage by PstI, whose restriction site is located 15 nucleotides downstream of the lesion (Fig. S5B). In summary, these endonuclease protection assays indicate that, by forming stable nucleoprotein interactions precisely at the damaged position, the XPD helicase demarcates CPD lesions in a strand-selective and site-specific manner.
Protection from Glycosylase Digestion.
Single-stranded or forked substrates were preincubated for 15 min with XPD and the reaction products were challenged by T4 denV, a DNA glycosylase that catalyzes the incision of CPD sites in both single- and double-stranded DNA (31). With the CPD-modified 51-mer oligonucleotide, the activity of T4 denV generates a radiolabeled fragment of 27 residues (Fig. 4C, lane 2). However, this CPD-specific cleavage was prevented when damaged substrates were preincubated with FaXPD in the presence of ATP and MgCl2 (lane 3). In control reactions, cleavage by T4 denV, producing the 27-mer fragment, was restored when either the helicase, ATP, or MgCl2 was omitted from the preincubation mixture (Fig. 4C, lanes 4–7).
When the same protection assay is applied to forked substrates with a CPD in the translocated 5′–3′ strand, cleavage by T4 denV yields a radiolabeled fragment of 101 residues (Fig. 4C, lane 10). This CPD-specific cleavage was suppressed after a 15-min preincubation with the complete helicase mixture (lane 11). Instead, no inhibition of T4 denV cleavage occurred when either the XPD helicase, ATP, or MgCl2 was missing during the preincubation period (Fig. 4C, lanes 12–15). Also, no inhibition of denV cleavage was detected when XPD was incubated with forked substrates containing a CPD in the opposing displaced strand (Fig. S6). These glycosylase protection assays show that the XPD helicase makes very close contacts with DNA lesions that obstruct its 5′–3′ movement.
Discussion
This report bears on the discovery that global-genome NER is initiated by a versatile sensor that detects the single-stranded character of unpaired bases rather than the target lesions themselves (10, 11). The XPC-Rad23B initiator, like its UV-DDB partner, even binds to mismatched bases that, in the absence of chemical modifications, fail to induce NER activity (18, 23). Such a lesion-independent action implies that the NER pathway must include a follow-up step to verify the presence of base modifications (18, 32). Although the nature of this verification process remained poorly defined, a previous reconstitution assay suggested that the loading of XPC onto DNA results in a NER intermediate that searches for base damage in the 5′–3′ direction (33). One key finding of the present study is that the enzymatic activity of XPD promotes its own anchoring to damaged DNA in living cells, thus supporting the conclusion that XPD is directly responsible for the predicted lesion verification step.
Earlier studies on the Rad3 protein of Saccharomyces cerevisiae indicated that the helicase activity of this yeast XPD homolog is suppressed after substrate treatment with various DNA-damaging agents (34). Although archaeal XPD homologs provide an excellent model to study the function of this evolutionary conserved protein (34–37), recently, Rudolf et al. (38) reported that damaged bases do not inhibit the DNA unwinding by such an archaeal enzyme. Our present study was focused on CPDs as an example of abundant and highly mutagenic lesions for which an effective verification process appears critical because they evade recognition by the core damage sensors including XPC-Rad23B (6, 18). It is important to note that UV-DDB, which is required for CPD recognition in the global-genome pathway, is displaced from the repair target after recruitment of XPC-Rad23B (21). Similarly, in transcribed sequences, DNA damage is detected by RNA polymerase II, which is released from repair sites before excision can occur (39, 40). Thus, in both subpathways, at least one additional player must take over a lesion verification function in order to demarcate CPDs and define the correct positions as well as orientation of DNA cleavage. Being a component of the TFIIH complex (24–28), XPD is strategically placed at the crossroad between global-genome and transcription-coupled repair (12, 13).
Here, the interplay between the XPD helicase and DNA lesions has first been examined in competition assays that challenge the stability of nucleoprotein complexes formed when this enzyme collides with a single CPD. Second, the effect of CPDs on DNA helicase activity was tested across a wide range of enzyme concentrations using substrates with a long 81-mer duplex region. Third, we determined how CPDs influence the rate of DNA-dependent ATP hydrolysis. Finally, the unwinding junctions were probed by digestion with endonucleases and a CPD-specific glycosylase. In combination, our findings demonstrate that XPD forms a tight complex with the DNA substrate after encountering a lesion during its directional 5′ → 3′ movement, thus providing a dynamic mechanism for strand-selective and site-specific lesion demarcation in the NER pathway. Our current data do not contradict the previously mentioned experiments by Rudolf et al. (38), who concluded that there is no inhibition of DNA unwinding by a CPD located in a 19-base-pair segment. Indeed, if the XPD enzyme senses the lesion and remains in place at the damaged site, as demonstrated in our study, it would still destabilize the short DNA duplex of that earlier report to a sufficient degree to cause strand separation.
As observed for UvrB, the ultimate recognition subunit in the prokaryotic NER system (41), the ATPase activity of XPD is stimulated by CPDs. Thus, the enzyme is not “paralyzed,” but retains the ability to hydrolyze ATP when encountering damaged sites. We propose a model whereby the XPD helicase is arrested by lesions situated in the translocated DNA strand and, thereafter, changes its catalytic properties to cooperate with the XPB partner (26, 28) as a site-specific ATPase. The combined action of these two unwinding enzymes generates the local bubble transition necessary for dual incision. An attractive feature of this model is that a stable nucleoprotein intermediate that allows for DNA incision can only be formed by damage-induced immobilization of the XPD subunit, such that its activity is focused on the lesion site without further translocation of the TFIIH complex. Instead, native DNA regions that fail to inhibit the XPD helicase are bypassed and will not be presented as a substrate to the NER system. This inherent verification mechanism serves as a decision point in the NER pathway to ensure that DNA incision only takes place at sites of true base damage.
Materials and Methods
Analysis of Nuclear XPD Foci.
Local areas of DNA damage were generated in CHO nuclei by UV irradiation (150 J/m2) through polycarbonate filters. Fluorescent protein accumulation and protein dynamics at lesion sites were monitored as described in SI Text.
Enzymes.
The F. acidarmanus XPD was expressed with an N-terminal His6 tag in Escherichia coli (BL21-Codon Plus, Stratagene) using a pET28a vector kindly provided by M. Spies (University of Illinois, Urbana, IL). The helicase was purified by affinity (HisTrap HP and HiTrap Heparin columns, GE Healthcare) and anion exchange chromatography (HiTrap Q XL) as described (30). Restriction enzymes were from New England Biolabs and T4 denV was from Epicentre.
Oligonucleotides.
The CPD building block was purchased from GlenResearch. The oligomers 5′-GCCTGCAGTCAGCGTCGACTCGAATTCCCG-3′ and 5′-CATGATTACGGCCATATCGAGCGGGAATTCGAGTCGACGCTGACTGCAGGC-3′, with a CPD at the position of the underlined thymines, were provided by Trilink Biotechnologies.
DNA-Binding Assays.
The indicated concentrations of FaXPD were incubated (25 °C, 15 min) with radiolabeled substrates (5 nM), either forked duplexes (without ATP) or 51-mer oligonucleotides (in the presence of 3 mM ATP). The buffer (15 μL) consisted of 20 mM Tris·HCl (pH 7.5), 5 mM MgCl2, and 2 mM dithiothreitol (DTT). Competition was started by adding 250 nM unlabeled 51-mers. After different incubation periods, the samples were transferred on ice, supplemented with 5 μL loading buffer [100 mM Tris·HCl (pH 8.3), 10% (vol/vol) glycerol, and 0.05% (wt/vol) orange G], and resolved on 5% (wt/vol) polyacrylamide gels in 45 mM Tris·HCl (pH 8.3), 45 mM boric acid, and 1 mM EDTA. The radioactive bands were visualized by autoradiography and quantified in a GS-800 Densitometer using the Quantity One software (Bio-Rad).
Helicase and ATPase Assay.
The construction of partial duplex substrates is outlined in Fig. S1. Helicase activity was tested by incubating (25 °C) the indicated amounts of FaXPD with forked 125-mer substrates (5 nM) in a volume of 15 μL containing 20 mM Tris·HCl (pH 7.5), 5 mM MgCl2, 2 mM DTT, and 3 mM ATP (30). The reactions were stopped by the addition of 5 μL loading buffer containing 20% (vol/vol) glycerol, 0.2 M EDTA, 2% (wt/vol) sodium dodecyl sulfate, 0.25% (wt/vol) xylene cyanol, 0.25% (wt/vol) bromphenol blue, 2 mg/mL proteinase K, and 250 nM of unlabeled 125-mers. The products were separated on native 5% (wt/vol) polyacrylamide gels and quantified as described before. The Pi release was measured using a colorimetric kit (Innova Bioscience) following the manufacturer’s instructions.
Protection Assays.
To subject helicase reaction products to restriction digestion, the buffer (15 μL) was adjusted by the addition of appropriate stock solutions provided for each restriction enzyme by the manufacturer. The incubations with SalI, PstI, or HaeIII were carried out at 30° for 40 min in a final volume of 20 μL. Digested samples were supplemented with 5 μL of loading buffer and resolved on native 5–12% (wt/vol) polyacrylamide gels. Alternatively, the reactions were adjusted to 50 mM Tris·HCl (pH 7.5), and 5 mM EDTA in a final volume of 20 μL. This mixture was incubated for 30 min at 30 °C with 0.015 unit of T4 denV and the reaction was stopped by the addition of 5 μL of 20 mM Tris·HCl (pH 8.0), 0.8 M NaCl and 80% (vol/vol) formamide. After heating to 95 °C for 10 min, the samples were chilled in an ice-cold water bath and resolved on denaturing 6% (wt/vol) polyacrylamide gels.
Supplementary Material
Acknowledgments.
We thank M. Spies and M. Stefanini for XPD expression vectors. We also thank C. Kisker for critical reading of the manuscript. This work is supported by the Swiss National Science Foundation Grant 31003A-127499 and the Internationale Bodensee-Hochschule.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.R.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004339107/-/DCSupplemental.
References
- 1.Mitchell DL. The relative cytotoxicity of (6-4) photoproducts and cyclobutane dimers in mammalian cells. Photochem Photobiol. 1988;48:51–57. doi: 10.1111/j.1751-1097.1988.tb02785.x. [DOI] [PubMed] [Google Scholar]
- 2.Mouret S, et al. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc Natl Acad Sci USA. 2006;103:13765–13770. doi: 10.1073/pnas.0604213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jans J, et al. Powerful skin cancer protection by a CPD-photolyase transgene. Curr Biol. 2005;15:105–115. doi: 10.1016/j.cub.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Garinis GA, et al. Transcriptome analysis reveals cyclobutane pyrimidine dimers as a major source of UV-induced DNA breaks. EMBO J. 2005;24:3952–3962. doi: 10.1038/sj.emboj.7600849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedberg EC, et al. DNA Repair and Mutagenesis. Washington, DC: American Society for Microbiology; 2006. pp. 865–894. [Google Scholar]
- 6.Reardon JT, Sancar A. Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Genes Dev. 2003;17:2539–2551. doi: 10.1101/gad.1131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoeijmakers JH. DNA damage, aging, and cancer. New Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 8.Friedberg EC. How nucleotide excision repair protects against cancer. Nat Rev Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 9.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer. 2005;5:564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 10.Trego KS, Turchi JJ. Pre-steady-state binding of damaged DNA by XPC-hHR23B reveals a kinetic mechanism for damage discrimination. Biochemistry. 2006;45:1961–1969. doi: 10.1021/bi05196t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min J-H, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 12.Evans E, Moggs JG, Hwang JR, Egly J-M, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mu D, Wakasugi M, Hsu DS, Sancar A. Characterization of reaction intermediates of human excision repair nuclease. J Biol Chem. 1997;272:28971–28979. doi: 10.1074/jbc.272.46.28971. [DOI] [PubMed] [Google Scholar]
- 14.Kim J-K, Patel D, Choi B-S. Contrasting structural impacts induced by cys-syn cyclobutane dimer and (6-4) adduct in DNA duplex decamers: Implication in mutagenesis and repair activity. Photochem Photobiol. 1995;62:44–50. doi: 10.1111/j.1751-1097.1995.tb05236.x. [DOI] [PubMed] [Google Scholar]
- 15.McAteer K, Jing Y, Kao J, Taylor J-S, Kennedy MA. Solution-state structure of a DNA dodecamer duplex containing a cis-syn thymine cyclobutane dimer, the major UV photoproduct of DNA. J Mol Biol. 1998;282:1013–1032. doi: 10.1006/jmbi.1998.2062. [DOI] [PubMed] [Google Scholar]
- 16.Jones CJ, Wood RD. Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- 17.Burns JL, Guzder SN, Sung P, Prakash S, Prakash L. An affinity of human replication protein A for ultraviolet-damaged DNA. J Biol Chem. 1996;271:11607–11610. doi: 10.1074/jbc.271.20.11607. [DOI] [PubMed] [Google Scholar]
- 18.Sugasawa K, et al. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nichols AF, et al. Human damage-specific DNA-binding protein p48. J Biol Chem. 2000;275:21422–21428. doi: 10.1074/jbc.M000960200. [DOI] [PubMed] [Google Scholar]
- 20.Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell. 2000;5:737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugasawa K, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 22.Rapic’-Otrin V, McLenigan MP, Bisi DC, Gonzales M, Levine AS. Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation. Nucleic Acids Res. 2002;30:2588–2598. doi: 10.1093/nar/30.11.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.C Wittschieben BO, Iwai S, Wood RD. DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J Biol Chem. 2005;280:39982–39989. doi: 10.1074/jbc.M507854200. [DOI] [PubMed] [Google Scholar]
- 24.Coin F, et al. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interactions between XPD and the p44 subunit of TFIIH. Nat Genet. 1998;20:184–188. doi: 10.1038/2491. [DOI] [PubMed] [Google Scholar]
- 25.Winkler GS, et al. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J Biol Chem. 2000;275:4258–4266. doi: 10.1074/jbc.275.6.4258. [DOI] [PubMed] [Google Scholar]
- 26.Coin F, Oksenych V, Egly J-M. Distinct roles for the XPB/p52 and XPD7p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell. 2007;26:245–256. doi: 10.1016/j.molcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Lehman AR. XPD structure reveals its secrets. DNA Repair. 2008;7:1912–1915. doi: 10.1016/j.dnarep.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 28.Oksenych V, de Jesus BB, Zhovmer A, Egly J-M, Coin F. Molecular insights into the recruitment of TFIIH to sites of DNA damage. EMBO J. 2009;28:2971–2980. doi: 10.1038/emboj.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alekseev S, et al. Cellular concentrations of DDB2 regulate dynamic binding of DDB1 at UV-induced DNA damage. Mol Cell Biol. 2008;28:7402–7413. doi: 10.1128/MCB.01108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pugh RA, et al. The iron-containing domain is essential in Rad3 helicases for coupling of ATP hydrolysis to DNA translocation and for targeting the helicase to the ssDNA-dsDNA junction. J Biol Chem. 2007;283:1732–1743. doi: 10.1074/jbc.M707064200. [DOI] [PubMed] [Google Scholar]
- 31.Gordon LK, Haseltine WA. Comparison of the cleavage of pyrimidine dimers by the bacteriophage T4 and Micrococcus luteus UV-specific endonucleases. J Biol Chem. 1980;255:12047–12050. [PubMed] [Google Scholar]
- 32.Buschta-Hedayat N, Buterin T, Hess MT, Missura M, Naegeli H. Recognition of nonhybridizing base pairs during nucleotide excision repair of DNA. Proc Natl Acad Sci USA. 1999;96:6090–6095. doi: 10.1073/pnas.96.11.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugasawa K, Akagi J, Nishi R, Iwai S, Hanaoka F. Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol Cell. 2009;36:642–653. doi: 10.1016/j.molcel.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 34.Naegeli H, Bardwell L, Friedberg EC. Inhibition of Rad3 DNA helicase activity by DNA adducts and abasic sites: Implications for the role of a DNA helicase in damage-specific incision of DNA. Biochemistry. 1993;32:613–621. doi: 10.1021/bi00053a029. [DOI] [PubMed] [Google Scholar]
- 35.Liu H, et al. Structure of the DNA repair helicase XPD. Cell. 2008;133:801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolski SC, et al. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 2008;6:e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan L, et al. XPD helicase structures and activities: Insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudolf J, Rouillon C, Schwarz-Linek U, White MF. The helicase XPD unwinds bubble structures and is not stalled by DNA lesions removed by the nucleotide excision repair pathway. Nucleic Acids Res. 2009;38:931–941. doi: 10.1093/nar/gkp1058. 10.1093/nar/gkp1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lainé J-P, Egly J-M. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. 2006;25:387–397. doi: 10.1038/sj.emboj.7600933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 41.Skorvaga M, Theis K, Mandavilli BS, Kisker C, Van Houten B. The β-hairpin motif of UvrB is essential for DNA binding, damage processing, and UvrC-mediated incisions. J Biol Chem. 2002;277:1553–1559. doi: 10.1074/jbc.M108847200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.