Abstract
Type III protein secretion systems are unique bacterial nanomachines with the capacity to deliver bacterial effector proteins into eukaryotic cells. These systems are critical to the biology of many pathogenic or symbiotic bacteria for insects, plants, animals, and humans. Essential components of these systems are multiprotein envelope-associated organelles known as the needle complex and a group of membrane proteins that compose the so-called export apparatus. Here, we show that components of the export apparatus associate intimately with the needle complex, forming a structure that can be visualized by cryo-electron microscopy. We also show that formation of the needle complex base is initiated at the export apparatus and that, in the absence of export apparatus components, there is a significant reduction in the levels of needle complex base assembly. Our results show a substantial coordination in the assembly of the two central elements of type III secretion machines.
Keywords: bacterial pathogenesis, cryo-electron microscopy, membrane proteins, organelle assembly, protein secretion
Widespread among bacteria pathogenic for plants, animals, or humans, type III secretion systems (T3SS) are essential for the establishment of intimate bacteria/host cell interactions (1, 2). Evolutionary and structurally related to flagella (3), these systems are among the most complex protein secretion systems known. Salmonella enterica serovar Typhimurium (S. Typhimurium) encodes one of these systems within its pathogenicity island 1 (SPI-1) (4), which is essential for this pathogen's ability to invade and replicate within mammalian host cells (5). Central components of type III secretion machines are the bacterial envelope-associated needle complex (NC) and the export apparatus, which makes the NC competent for secretion. The NC is composed of a multiring base (made up in the S. Typhimurium SPI-1 T3SS by the InvG, PrgH, and PrgK proteins), a central inner rod (made up by PrgJ), and a protruding needle-like structure (made up by PrgI), which is traversed by ~28 Å-wide channel that serves as a conduit for passage of the secreted proteins through the bacterial envelope (6, 7). The recent availability of the crystal structures of soluble domains of some of the components of the NC (8–10), combined with their docking onto the NC protein density map generated by cryo-electron microscopy (7, 11), is beginning to provide a high-resolution view of this organelle (12). Previous studies have shown that assembly of the needle complex occurs in an orderly manner, in which the base substructure is assembled first, followed by the assembly of the inner rod and needle substructure (13, 14). The assembly of the last two substructures requires the base to be rendered secretion-competent by the export apparatus, thus gaining the ability to secrete the needle and inner rod subunits. After the needle complex is fully assembled, the secretion machine switches substrate specificity, becoming competent for the secretion of translocases and effector proteins. Much less is known about the arrangement of the export apparatus, which is composed of a collection of highly conserved membrane proteins (InvA, SpaP, SpaQ, SpaR, and SpaS in the S. Typhimurium SPI-1–encoded T3SS) (1–3). Although all of the components of the export apparatus are essential for type III secretion, it is unknown whether these membrane proteins exert a common function or even if they exist as a single complex within the bacterial envelope. Furthermore, their functional and topological relationship to the NC is unknown. In this paper, we have investigated in S. Typhimurium the localization of the membrane proteins that make up the export apparatus and examined their role in the assembly of the NC. We found that membrane protein components of the export apparatus are intimately associated with the base substructure of the NC, forming a defined structure that can be visualized by cryo-electron microscopy. In addition, we found that formation of the NC base is initiated at the export apparatus and that the presence of export apparatus components is required for the assembly of a functional NC. These findings revealed a substantial coordination in the assembly of the export apparatus and NC components of the T3SS.
Results
Export Apparatus Is Intimately Associated with the NC.
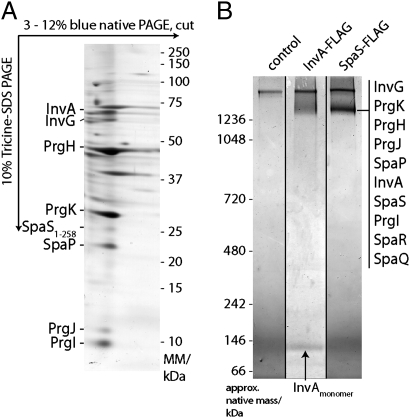
It has been previously shown that at least some components of the export apparatus are associated or cofractionate with the NC or the hook–basal body complex (a functional equivalent of the NC in flagella) (15, 16). Therefore, we investigated whether the components of the export apparatus of the S. Typhimurium SPI-1–encoded T3SS could be isolated in association with the NC. Because previously reported stringent NC isolation protocols may affect the identification of associated components (6), we developed a protocol to examine NC under milder conditions (SI Materials and Methods and Fig. S1). We first analyzed NC and associated proteins using 2D blue native PAGE (BN-PAGE). The NC and associated proteins migrated as a distinct band with a mobility that is consistent with that of a complex larger than 1,200 kDa (Fig. 1A and Fig. S2). Proteins making up this complex were identified by liquid chromatography followed by tandem MS (LC-MS/MS) and/or Western immunoblotting. In addition to the structural components of the inner and outer rings of the NC (PrgH, PrgK, and InvG) (6) and the needle and inner rod proteins (PrgI and PrgJ) (17, 18), we detected the export apparatus components InvA (19), SpaP, and SpaS (20) (Table S1). To validate the interaction of the export apparatus components with the NC base, we immunoprecipitated the complex from a strain carrying a carboxy terminal FLAG-epitope tagged allele of SpaS (SpaSN258A), which carries a mutation in its catalytic, autoprocessing site, thus preventing the cleavage of its carboxyl terminus (21, 22). Proteins affinity purified with an anti-FLAG antibody were separated by BN-PAGE, and the band corresponding to the NC was analyzed by LC-MS/MS. This band contained all needle complex and export apparatus membrane protein components, including SpaR and SpaQ (Fig. 1B, Fig. S3, and Table S1). Similar results were obtained when the complex was immunoprecipitated from a strain expressing a functional FLAG-epitope tagged allele of InvA (Fig. 1B). Unlike SpaS, however, a significant amount of free InvA was also detected, suggesting a somewhat weaker association of InvA to the NC. Taken together, these results indicate that the export apparatus is intimately associated with the NC.
Fig. 1.
The export apparatus is intimately associated with the NC. (A) Export apparatus components cofractionate with the NC. Protein complexes of purified S. Typhimurium inner membrane fractions were solubilized with n-dodecyl-β-d-maltopyranoside (DDM) and separated by 2D BN-PAGE. Indicated proteins were identified by LC-MS/MS or Western blotting. (B) The export apparatus interacts with the needle complex. DDM-solubilized inner membrane fraction proteins interacting with C-terminally FLAG-tagged InvA or SpaSN258A were immunoprecipitated, separated by BN-PAGE, and identified by LC-MS/MS.
Export Apparatus Components Associate with One Another in the Absence of the NC.
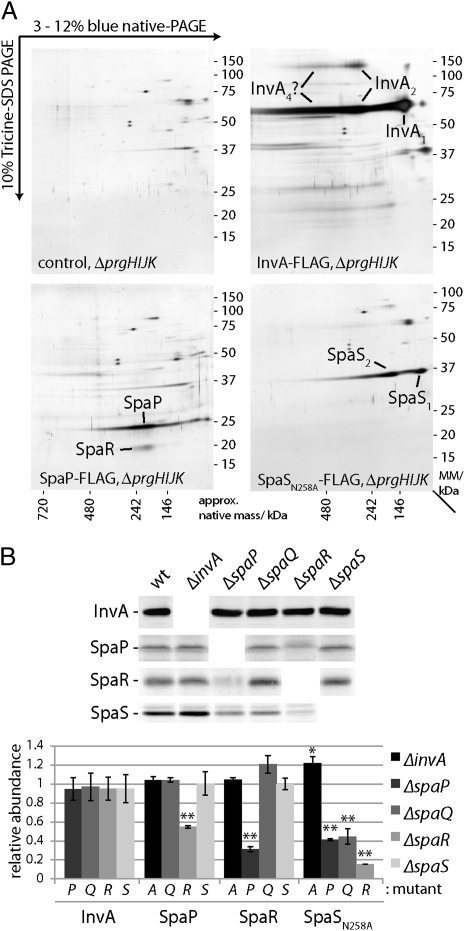
We investigated whether the components of the export apparatus themselves could form a complex in the absence of the NC. To this end, we constructed strains of S. Typhimurium expressing functional FLAG-tagged InvA, SpaP, SpaR, and SpaS (SpaQ could not be tagged in a manner that would retain function) in the context of the NC-defective ΔprgHIJK mutant. We purified inner membrane fractions from these strains and identified interacting proteins by immunoprecipitation combined with 2D BN-PAGE and LC-MS/MS analysis. We found that, in the absence of the NC, SpaP and SpaR could form a complex (Fig. 2A). In contrast, InvA and SpaS were not seen associated to any member of the export apparatus under the isolation conditions used in this assay (Fig. 2A). It is often the case that the stability of members of a multiprotein complex is compromised in the absence of other components of the complex (23). Therefore, we tested whether the stability of individual export apparatus components was compromised by the absence of other export apparatus components. Deletion of spaP resulted in reduced levels of SpaR (and vice versa) (Fig. 2B), presumably because of their reduced stability in the absence of their interacting partners. Furthermore, levels of SpaSN258A were reduced in the absence of SpaP, SpaQ, and SpaR. In contrast, the levels of InvA were not affected by the removal of any of the other export apparatus components (Fig. 2B). These results indicate that a subset of the export apparatus components can assemble into a stable complex in the absence of the NC and that the members of this complex require each other for stability.
Fig. 2.
Export apparatus components associate with one another in the absence of the needle complex. (A) The export apparatus components SpaP and SpaR form a complex in the absence of the NC. DDM-solubilized inner membrane fraction proteins of a S. Typhimurium ΔprgH ΔprgI ΔprgJ ΔprgK mutant strain expressing FLAG-epitope tagged SpaP, InvA, or SpaSN258A (as indicated) were immunoprecipitated with an anti-FLAG antibody. Precipitated proteins were separated by 2D BN-PAGE, and indicated proteins were identified by LC-MS/MS. (B) The levels of SpaP, SpaR, and SpaS (but not InvA) are significantly reduced in the absence of some export apparatus components. Purified inner membrane fractions from wild-type S. Typhimurium (wt) or the ΔspaP, ΔspaQ, ΔspaR, ΔspaS, or ΔinvA mutants expressing FLAG-epitope tagged InvA, SpaP, SpaR, or SpaSN258A (as indicated) were analyzed by Western blotting (Upper). The intensity of the bands was quantified using the Odyssey imaging system (Li-Cor), and values represent the mean ± SD of three independent experiments (Lower). Values were standardized relative to a loading control (PrgH) and across different samples relative to wild type, which was assigned an arbitrary value of 1.
Hierarchy in the Recruitment of Export Apparatus Components to the NC.
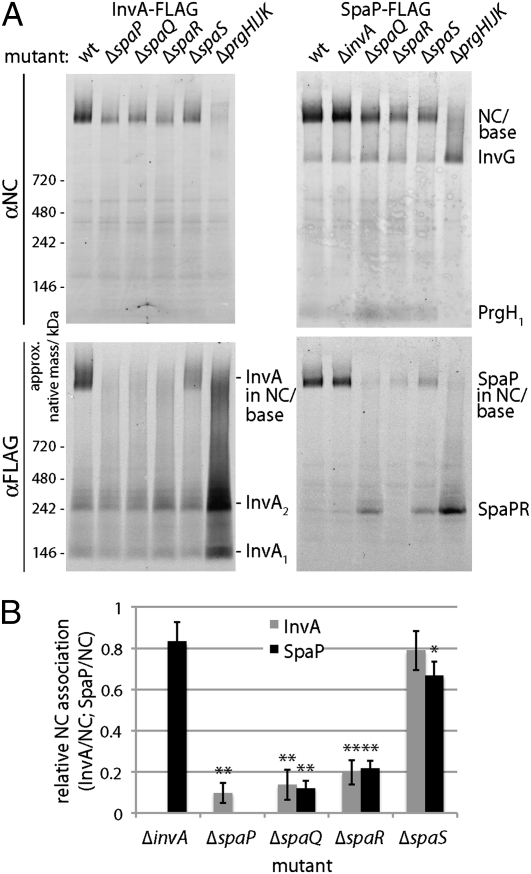
We analyzed the effect of removal of individual components of the export apparatus on the recruitment of the remaining export apparatus components to the NC using BN-PAGE combined with Western blotting and 2D BN-PAGE (Fig. 3 and Fig. S4). Deletion of invA or spaS did not significantly affect the recruitment of any of the other components of the export apparatus to the NC base. In contrast, deletion of spaP, spaQ, or spaR resulted in the abrogation of the recruitment of all components of the export apparatus to the NC base. Taken together, these results indicate that there is a hierarchy in the recruitment of export apparatus components to the NC and that SpaP, SpaQ, and SpaR act first, playing a central role in the assembly process.
Fig. 3.
Hierarchy in the recruitment to the needle complex of export apparatus components. Protein complexes of purified inner membrane fractions of wild type (wt) and the indicated mutants of S. Typhimurium expressing FLAG-epitope tagged InvA or SpaP were solubilized with DDM, separated by BN-PAGE, and analyzed by Western blotting using antibodies directed to the NC base or FLAG (to detect either SpaP or InvA; A). The intensity of the bands corresponding to SpaP or InvA associated with the NC was quantified as above (B), and values represent the mean ± SD of three independent experiments. Values were standardized relative to the levels of the NC and across different samples relative to wild type, which was assigned an arbitrary value of 1. The total levels of SpaP-FLAG and InvA-FLAG in the different mutants are shown in Fig. 2B.
Export Apparatus Is Required for Efficient NC Base Assembly.
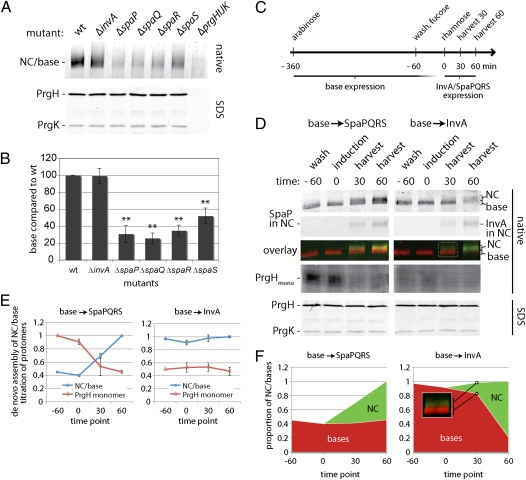
The observation that the export apparatus is assembled in a coordinated fashion with the NC prompted us to examine the effect of the absence of export apparatus components on the assembly of the NC base. We have previously shown that NC base substructures can be detected in the absence of export apparatus components (13). However, using a quantitative assay, we found that deletion of spaP, spaQ, or spaR resulted in a drastic (~80%) reduction in the levels of NC bases, although the absence of the export apparatus did not affect the total levels of the NC individual components such as PrgH, PrgK, and InvG (Fig. 4 A and B). Consequently, the absence of SpaP, SpaQ, and SpaR also resulted in an accumulation of free NC protomers (Fig. 4D). In contrast, deletion of invA had no effect on the levels of assembled NC bases (or the accumulation of free protomers), whereas deletion of spaS had an intermediate phenotype (Fig. 4 A and B). Taken together, these results indicate that components of the export apparatus play an important role in the assembly and/or stability of the NC base.
Fig. 4.
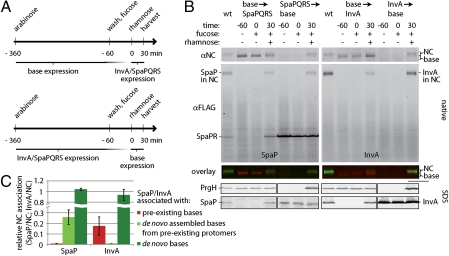
The export apparatus is required for efficient NC base assembly. (A and B) Efficient NC base assembly requires SpaP, SpaQ, SpaR, and SpaS but not InvA. DDM-solubilized protein complexes of purified inner membrane fractions of wild type (wt) and the indicated mutants of S. Typhimurium were separated by BN-PAGE and analyzed by Western blotting (A). The total levels of PrgH and PrgK in the different mutants are shown in A Lower. The quantification of the relative levels of NC in the different mutants is shown (B). The intensity of the bands was quantified as above, and values represent the mean ± SD of three independent experiments. Values were standardized across different samples relative to wild type, which was considered 100%. (C–E). The export apparatus components SpaP, SpaQ, SpaR, and SpaS promote NC base assembly. (C) The experimental setup for these experiments involved the use of engineered strains of S. Typhimurim in which expression of the base components is under the control of an arabinose-inducible promoter, whereas the expression of either spaPQRS or invA is under the control of a rhamnose-inducible promoter (details in Material and Methods). Base expression was induced by the addition of arabinose. The inducer was washed away, and expression of the base components was blocked by the addition of the repressor fucose. Sixty minutes later (indicated as time 0 in the legend), expression of export apparatus components (either SpaPFLAGQRS or InvAFLAG) was induced by the addition of rhamnose. Samples were harvested 30 min and 60 min thereafter, and DDM-solubilized proteins of whole-cell lysates were separated by BN-PAGE and analyzed by Western blotting with antibodies directed to NC base components (PrgH and PrgK) or FLAG (to detect SpaP or InvA, as indicated). SDS/PAGE of whole-cell lysates shows the absolute levels of the base components PrgH and PrgK, which remained constant throughout the experiment. D Left shows results in which expression of the base was followed by expression of SpaPQRS, whereas D Right shows results in which expression of the base was followed by expression of InvA. To minimize variation when comparing NC-associated proteins and PrgH monomers, we divided the same sample after solubilization and addition of Coomassie and loaded them onto two different gels (D). The total levels of PrgH and PrgK in different samples are shown in the bottom row (D). Quantification of these results is shown in E and F. The intensity of the bands was quantified as above, and values represent the mean ± SD of three independent experiments. The maximal values for bases/NC and PrgH protomers, respectively, were assigned an arbitrary value of 1. The proportion of bases and assembled NCs of the total base/NC signal (D) is shown in F. As an example, Inset in F shows, for a time point (D), the region corresponding to NC (green) or bases (red) in the BN-PAGE analysis.
To distinguish whether components of the export apparatus have an effect on the assembly or the stability of the NC base, we assayed to find out whether SpaP, SpaQ, SpaR, and SpaS were able to induce assembly of bases from presynthesized, free NC protomers. To this end, we constructed a strain in which expression of the NC components is controlled by an arabinose-inducible paraBAD promoter (24) through the expression of HilA, the master transcriptional regulator of the SPI-1 T3SS (25). In addition, invA and spaPQRS were deleted from this strain and expressed from a low copy plasmid under the control of the rhamnose-inducible prhaBAD promoter (26). These strains and the growth conditions used (Fig. 4C and Materials and Methods) allowed the sequential expression of NC base and export apparatus components. As shown above, in the absence of the export apparatus components SpaP, SpaQ, SpaR, and SpaS, NC base assembly occurred inefficiently and led to the accumulation of free protomers (Fig. 4 D and E). However, immediately after spaPQRS expression, free protomers were efficiently incorporated into the de novo assembled bases. This resulted in a net increase in the total levels of NC bases without a corresponding increase in the total levels of its individual subunits PrgH, PrgK, or InvG (Fig. 4 D and E). The de novo assembled bases showed a substantially higher native mass (detectable by BN-PAGE, shown in Fig. 4D) than the preassembled bases, most likely because of the incorporation of the needle and inner rod proteins, which can only occur on the recruitment of the export apparatus. The levels of the preassembled fraction of NC bases remained largely constant, and its mobility was not shifted (Fig. 4F), suggesting that SpaP, SpaQ, and SpaR may not be able to be recruited into these preassembled structures (Discussion). In contrast to what was observed with spaPQRS expression, induction of invA expression had no effect on the levels of NC bases (Fig. 4 D and E and Fig. S5), which is consistent with the observation that, in the absence of InvA, NC assembly occurs normally without accumulation of free NC base protomers (Fig. 4 D and E). Nearly all NC bases were shifted into higher mobility on expression of InvA, presumably because of the incorporation of the needle and inner rod proteins (Fig. 4F), suggesting that, in contrast to SpaP, SpaQ, and SpaR, InvA may be incorporated into preassembled NC bases. Taken together, these results indicate that the export apparatus, particularly, SpaP, SpaQ, and SpaR, can promote NC base assembly rather than increase its stability. This conclusion is also consistent with the observation that, after assembled, the stability of the NC bases isolated from the ΔspaPQRS mutant is indistinguishable from that of wild type (Fig. S6).
To further explore the role of export apparatus components on NC base assembly, we compared the efficiency of incorporation of export apparatus components into de novo synthesized or preexisting NC bases. As shown above, we found that SpaP could not be incorporated into preexisting NC bases. We also found that incorporation of InvA into de novo synthesized NC structures was more efficient than its incorporation into preexisting NC bases (Fig. 5). The recruitment of InvA to the NC required the presence of SpaP, SpaQ, SpaR, and SpaS. However, InvA was not necessary for the recruitment of SpaP (and presumably, SpaQ and SpaR) into NC bases (Fig. S5). Taken together, these results indicate that NC assembly requires prior deployment of some components of the export apparatus and that at least those components (e.g., SpaP, SpaQ, and SpaR) must be housed at a location that is inaccessible after the assembly of the NC base is complete.
Fig. 5.
NC base assembly requires the predeployment of the export apparatus. (A) Outline of the experimental setup to ensure that expression of the base components preceded expression of the export components (indicated as base > SpaPFLAGQRS or base > InvAFLAG in B) or vice versa (indicated as SpaPFLAGQRS > base or InvAFLAG > base in B). (B and C) Export apparatus components do not efficiently incorporate into preassembled bases. The incorporation of SpaP (B Left) or InvA (B Right) into preformed NC bases was examined by BN-PAGE of DDM-solubilized proteins of whole-cell lysates followed by Western blotting analysis with specific antibodies and an Odyssey (Li-Cor) infrared imaging system to simultaneously detect the indicated proteins. The merger of the two detection channels is shown in color [red, bases/NC (PrgH/PrgK); green, InvA or SpaP]. The total levels of PrgH and SpaP/InvA in the different samples are shown in Lower (B). (C) Quantification of the results shown in B. The intensity of the bands was quantified as above, and values represent the mean ± SD of three independent experiments. Incorporation of InvA or SpaP into preexisting bases, de novo assembled bases from free preexisting protomers, or de novo assembled bases from de novo synthesized protomers was determined as indicated in SI Materials and Methods. Values were standardized relative to the NC and across different samples relative to wild type, which was assigned an arbitrary value of 1.
Components of the Export Apparatus Form a Defined Structure Within the NC Base.
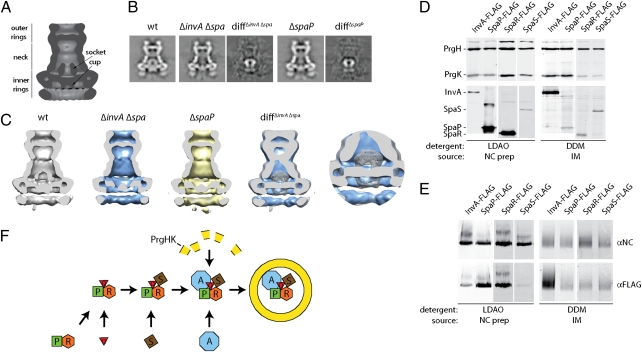
The intimate relationship between the assembly of the export apparatus and the NC base prompted us to examine the effect of removal of the export apparatus on the structure of the NC base. We have shown that NC base substructures can be obtained in the absence of the export apparatus, although in significantly lower amounts (Results). We, therefore, took advantage of this finding; we isolated NC base structures from a strain that does not express any components of the export apparatus and examined them by cryo-electron microscopy and single particle analysis (Fig. S7). Subtraction of the 2D class averages of NC bases obtained from the ΔinvA ΔspaP ΔspaQ ΔspaR ΔspaS multiple mutant strain from class averages of NC bases obtained from wild type revealed significant differences. More specifically, structures obtained from the mutant strain lacked a significant amount of density at the center of the basal side of the NC base, including the cup and socket substructures (Fig. 6 A–C). We also analyzed NC bases obtained from a ΔspaP mutant strain, which looked indistinguishable from structures obtained from the mutant lacking all of the membrane components of the export apparatus (Fig. 6 A–C). Biochemical analysis of the NC preparations used for cryo-electron microscopy isolated with a stringent isolation protocol (Material and Methods) revealed the presence of SpaP and SpaR but not InvA or SpaS (Fig. 6 D and E). Because part of the socket substructure is missing from a mutant unable to secrete the regulatory protein InvJ protein (18) (which would not be secreted in the mutant strains used in this analysis), not all of the density absent from these mutant strains can be unambiguously assigned to these membrane proteins. Rather, it is likely that just the cup substructure, which is present even in the absence of InvJ (18), is formed by SpaP, SpaQ, and SpaR. Taken together, these results indicate that a subset of the export apparatus components form a defined substructure on the basal face of the NC and may account for yet unassigned density in the NC protein density map (12). Additional studies will be required to localize InvA and SpaS, which are lost from NC preparations generated with the stringent isolation protocol that is required to obtain particles suitable for cryo-electron microscopy.
Fig. 6.
Components of the export apparatus form a defined substructure in the NC base. (A) Diagram of the S. Typhimurium NC base depicting the different substructures. (B) Total class averages of single particle analysis of cryo-electron microscopy images and density differences between the averaged images of wild-type NCs and the indicated mutant strains. (C) Longitudinal sections through the center of base complexes from wild type or the indicated mutant strains lacking SpaP, SpaQ, SpaR, SpaS, and InvA or only SpaP reveal the absence of the cup and socket substructures observed in the NC base obtained from wild type. The difference images of sections from either of the mutant strains and wild type are very similar, indicating that removal of SpaP alone is sufficient to generate this structural phenotype. The cut-away view and blow-up view of an overlay of wild-type complexes (gray/mesh) and complexes from the ΔspaPQRS ΔinvA mutant strain at a distance that allows the visualization of the entire cup and socket substructures in 3D emphasizes the difference between the complexes analyzed. (D and E) Highly purified NC bases contain SpaP and SpaR but no InvA or SpaS. NC bases either prepared in an identical manner to those used in cryo-electron microscopy studies or from purified inner membrane fractions were analyzed by SDS/PAGE (D) or BN-PAGE (E) followed by Western blotting with antibodies to detect the indicated proteins. (F) Model for the coordinated assembly of the T3SS export apparatus and NC base. SpaP forms a stable complex with SpaQ and SpaR, which is likely to be the nucleation point for PrgH/PrgK NC base ring assembly. The presence of SpaS further enhances the assembly of the PrgH/PrgK ring, although it does not seem to be essential. The presence of InvA is not required for efficient NC base formation or for the stability and/or recruitment of any of the other export apparatus components. These results suggest that InvA is the last component recruited to the export apparatus. However, InvA is not efficiently recruited into preassembled bases.
Discussion
The assembly of complex protein machines often requires the coordinated deployment of its components, both in time and space. We have shown here that two central components of T3SS machines, the NC and the export apparatus, are assembled through a coordinated process that results in the incorporation of these two subcomplexes into a single structure. It was previously assumed that the assembly of the NC is initiated by the assembly of the inner and/or outer ring substructures (13, 14). However, we found that the assembly process is initiated by the deployment of a subset of the conserved inner membrane protein components of the export apparatus, which presumably provides a platform onto which subsequent elements of the NC can assemble (Fig. 6F). The organization of this inner membrane protein complex itself must also occur in an organized and coordinated fashion, because we have shown that there is a hierarchy in its formation; in this hierarchy, SpaP, SpaQ, SpaR, and to a lesser extent, SpaS play a central role, whereas InvA seems to be less critical for this process. We have also shown that some export apparatus components are not incorporated into preassembled NC bases, indicating that they must be placed at a location that is not accessible after the NC assembly is finished. We have shown that at least a subset of these inner membrane proteins, SpaP, SpaQ, and SpaR, forms a defined substructure within the NC, where they are well-positioned to receive the T3SS substrates and presumably, facilitate their passage through the inner membrane. Our findings, therefore, indicate that the NC and at least some components of the so-called export apparatus form an indivisible holostructure, which should be considered as a single functional entity. These studies have revealed unique insight into the assembly and organization of the T3SS organelle. Given the conservation of this system among pathogenic bacteria, this information could serve as the bases for the development of broadly applicable antiinfective strategies.
Materials and Methods
General Techniques.
Detailed information about strains, plasmids, and culture conditions used can be found in SI Materials and Methods and Table S2. Western blotting was carried out using infrared fluorescent secondary antibodies (emission 680 or 800 nm). Bands were quantified using the Odyssey imaging system (Li-Cor). Cell fractionation was carried out as described before (27) with some modifications. BN-PAGE was carried out as described previously (28, 29). Identification of T3SS components by MS was carried out at the Keck Biotechnology Resource Laboratory (Yale University, New Haven, CT) according to its standardized protocol. Detailed techniques can be found in SI Materials and Methods.
Immunoprecipitation of Export Apparatus Components.
FLAG-tagged export apparatus components were immunoprecipitated from purified inner membranes using the anti-FLAG M2 affinity gel according to the recommendations of the manufacturer. Precipitated complexes were eluted and subsequently analyzed by 1D or 2D BN-PAGE and transmission electron microscopy (SI Materials and Methods).
Coordinated Expression of T3SS Components.
The coordinated expression of T3SS components was carried out by using bacterial strains in which expression of examined components was uncoupled by using two different compatible promoter/inducer systems (arabinose vs. rhamnose). Briefly, expression of HilA, which is the master regulator of the SPI-1 T3SS, was put under control of an arabinose-inducible promoter. The other components of interest were deleted from the chromosome and complemented in trans under control of a rhamnose-inducible promoter. Complex assembly was assessed by BN-PAGE and Western blotting. Details can be found in SI Materials and Methods and Fig. S8.
Stability of T3SS Bases.
The stability of needle complexes/bases was assessed by using a modified BN-PAGE/Western blotting protocol. In brief, complexes were destabilized by increasing amounts of SDS (details in SI Materials and Methods).
NC Expression, Purification, Electron Microscopy, and Image Analysis.
Electron microscopic analysis of needle complexes was carried out as described previously (7). More detailed methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Tukiet Lam for his help with the LS-MS/MS analysis and members of the Galán laboratory for critical reading of this manuscript. This work was supported by postdoctoral fellowships from the Swedish Research Council (Vetenskapsrådet), the European Molecular Biology Organization (EMBO), and the Human Frontiers Science Program (to S.W.); Zentrum für Innovation und Technology Vienna (ZIT), Center of Molecular and Cellular Nanostructure Vienna (CMCN), and Raiffeisen Landesbank Oberösterreich grants (to T.C.M.); and National Institutes of Health Grant AI30492 (to J.E.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The mass spectrometry data have been deposited on http://yped.med.yale.edu/repository (accession: Samuel Wagner).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008053107/-/DCSupplemental.
References
- 1.Galán JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 2.Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 3.Minamino T, Imada K, Namba K. Mechanisms of type III protein export for bacterial flagellar assembly. Mol Biosyst. 2008;4:1105–1115. doi: 10.1039/b808065h. [DOI] [PubMed] [Google Scholar]
- 4.Galán JE, Curtiss R., 3rd Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galán JE. Salmonella interactions with host cells: Type III secretion at work. Annu Rev Cell Dev Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 6.Kubori T, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 7.Marlovits TC, et al. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crepin VF, et al. Structural and functional studies of the enteropathogenic Escherichia coli type III needle complex protein EscJ. Mol Microbiol. 2005;55:1658–1670. doi: 10.1111/j.1365-2958.2005.04508.x. [DOI] [PubMed] [Google Scholar]
- 9.Yip CK, et al. Structural characterization of the molecular platform for type III secretion system assembly. Nature. 2005;435:702–707. doi: 10.1038/nature03554. [DOI] [PubMed] [Google Scholar]
- 10.Spreter T, et al. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nat Struct Mol Biol. 2009;16:468–476. doi: 10.1038/nsmb.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodgkinson JL, et al. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat Struct Mol Biol. 2009;16:477–485. doi: 10.1038/nsmb.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schraidt O, et al. Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 2010;6:e1000824. doi: 10.1371/journal.ppat.1000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sukhan A, Kubori T, Wilson J, Galán JE. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J Bacteriol. 2001;183:1159–1167. doi: 10.1128/JB.183.4.1159-1167.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diepold A, et al. Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 2010;29:1928–1940. doi: 10.1038/emboj.2010.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan F, Ohnishi K, Francis NR, Macnab RM. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol Microbiol. 1997;26:1035–1046. doi: 10.1046/j.1365-2958.1997.6412010.x. [DOI] [PubMed] [Google Scholar]
- 16.Zenk SF, et al. Identification of minor inner-membrane components of the Shigella type III secretion system ‘needle complex’. Microbiology. 2007;153:2405–2415. doi: 10.1099/mic.0.2007/007781-0. [DOI] [PubMed] [Google Scholar]
- 17.Kubori T, Sukhan A, Aizawa SI, Galán JE. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci USA. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marlovits TC, et al. Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature. 2006;441:637–640. doi: 10.1038/nature04822. [DOI] [PubMed] [Google Scholar]
- 19.Galán JE, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella typhimurium invasion gene invA: Homology of InvA to members of a new protein family. J Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groisman EA, Ochman H. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 1993;12:3779–3787. doi: 10.1002/j.1460-2075.1993.tb06056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferris HU, et al. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem. 2005;280:41236–41242. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- 22.Lavander M, et al. Proteolytic cleavage of the FlhB homologue YscU of Yersinia pseudotuberculosis is essential for bacterial survival but not for type III secretion. J Bacteriol. 2002;184:4500–4509. doi: 10.1128/JB.184.16.4500-4509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akiyama Y, Kihara A, Tokuda H, Ito K. FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J Biol Chem. 1996;271:31196–31201. doi: 10.1074/jbc.271.49.31196. [DOI] [PubMed] [Google Scholar]
- 24.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bajaj V, Hwang C, Lee CA. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 26.Haldimann A, Daniels LL, Wanner BL. Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J Bacteriol. 1998;180:1277–1286. doi: 10.1128/jb.180.5.1277-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner S, et al. Consequences of membrane protein overexpression in Escherichia coli. Mol Cell Proteomics. 2007;6:1527–1550. doi: 10.1074/mcp.M600431-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 29.Klepsch M, et al. Immobilization of the first dimension in 2D blue native/SDS-PAGE allows the relative quantification of membrane proteomes. Methods. 2008;46:48–53. doi: 10.1016/j.ymeth.2008.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.