Abstract
Nasopharyngeal carcinoma (NPC) is a rare malignancy with unique genetic, viral and environmental characteristic that distinguishes it from other head and neck carcinomas. The clinical management of NPC remains challenging largely due to the lack of early detection strategies for this tumor. In the present study we have sought to identify novel genes involved in the pathogenesis of NPC that might provide insight into this tumor's biology and could potentially be used as biomarkers. To identify these genes, we studied the epigenetics of NPC by characterizing a panel of methylation markers. Eighteen genes were evaluated by quantitative methylation-specific PCR in cell lines as well as in tissue samples including 50 NPC tumors and 28 benign nasopharyngeal biopsies. Significance was evaluated using Fisher's exact test and quantitative values were optimized using cut off values derived from receiver-operator characteristic curves. The methylation status of AIM1, APC, CALCA, DCC, DLEC, DLC1, ESR, FHIT, KIF1A, and PGP9.5 was significantly associated with NPC compared to controls. The sensitivity of the individual genes ranged from 26 to 66% and the specificity was above 92% for all genes except FHIT. The combination of PGP9.5, KIF1A, and DLEC had a sensitivity of 84% and a specificity of 92%. Ectopic expression of DCC and DLC1 lead to decrease in colony formation and invasion properties. Our results indicate that methylation of novel biomarkers in NPC could be used to enhance early detection approaches. Additionally, our functional studies reveal previously unknown tumor suppressor roles in NPC.
INTRODUCTION
Nasopharyngeal carcinoma (NPC) is a relatively rare head and neck malignancy in the United States. The global age adjusted incidence is 1 per 100,000 per year representing 0.7% of all cancers1. In contrast, certain southern regions of China have an incidence as high as 50 cases per 100,000 per year2. In fact in Hong Kong, NPC is the leading cancer in men age 20 to 44 years old representing 25% of all cancers3. NPC is particularly interesting because of its unique genetic, viral, and environmental factors. Epstein-Barr virus (EBV) is a critical factor for malignant transformation in NPC4. However only a small fraction of widespread EBV-infected patients will develop a NPC and not all NPC are EBV positive5. EBV is present in over 95% of the endemic type I non keratinizing lesions, while 30 to 50% of the type II keratinizing lesions in the non endemic regions are EBV negative6. Aside from EBV, environmental exposure to food preservatives in food, salted fish, and occupational exposures have been related to increased NPC incidence3.
The initial presentation of NPC varies widely, but rarely is the disease localized to the nasopharynx. Commonly, patients present with advanced disease that has metastasized to the neck lymph nodes rather than with signs and symptoms of the initial nasopharyngeal involvement7. Surveillance of the nasopharyngeal cavity in high risk patients remains a challenge due to anatomy of the region where the least invasive approach for tissue biopsies or inspection involves endoscopy procedures in the operating room. Additionally, current tumor-node-stage classification fails to predict which NPC patients will later progress to develop recurrence or metastasis. Due to the extensive lymphatic drainage of the nasopharynx, from 70 to 90% of the NPC patients develop metastatic neck nodes8. There is a need for improving early diagnosis and clinical stratification methods for the management of NPC patients.
A better understanding of the molecular alterations in NPC is likely to contribute to improve clinical management. Molecular events like loss of heterozygosity9, EBV viral loads10, and EGFR amplifications and over-expression11 have shown promising results in NPC. Abnormal expression of genes and in particular tumor suppressor gene silencing is a common event in cancer12. The field of epigenetics offers alternative oncogenic mechanism for gene silencing than gene mutations or deletions. Epigenetic alterations are events that alter gene expression without altering genes' DNA sequence. The most studied epigenetic phenomenon in cancer is DNA methylation, by which binding of methyl group to CpG dinucleotides to promoter regions of genes lead to silencing of expression13. Gene-specific methylation of promoter regions of tumor suppressor genes has been described in a variety of human cancers14. Gene methylation has the potential of being used as a detection marker and as a prognostic marker. For example, in head and neck cancer gene methylation has been studied in body fluids like saliva and oropharyngeal swabs in an effort to improve cancer diagnosis15. Furthermore, MGMT methylation in glioblastoma is an example of a prognostic marker currently being implemented in clinical practice for the management of patients16.
In NPC, several studies have reported promoter hypermethylation of individual genes by bisulfite sequencing and conventional methylation-specific PCR (MSP)3, 17–20. Several studies have started to analyze the presence of methylation markers in serum of NPC patient21, 22. Conventional MSP has the limitation of not being able to distinguish between the different levels of positivity hence increasing the number of false positive methylation markers in benign or premalignant lesions. In the head and neck region, false positive results as a result of field cancerization upon exposure to alcohol, tobacco, and other carcinogens, are of significant concern23. Histologically normal epithelial cells exposed to carcinogenic insults undergo molecular alterations including p53 mutations and promoter methylation24. Overcoming the challenge of false positives might be possible through quantitative methylation-specific PCR (QMSP).
In the present study, we sought to analyze by QMSP the promoter methylation status of a panel of 18 genes in NPC. To our knowledge this is the first study using QMSP in NPC. The initial panel of genes (AIM, APC, CALCA, DCC, DLC1, DLEC, ESR, FHIT, GSTP, HIC, KIF1α, NISCH, PAK3, PGP9.5, S100, TGFβ, TIG1, and TIMP3) was evaluated in NPC cell lines and normal nasopharynx tissues. Genes showing differential methylation patterns and were subsequently evaluated in sample set of NPC tumors and controls. Relationship between methylation levels and clinicopathologic parameters were assessed. To further elucidate the function and role of methylation, 5 genes were tested for expression levels after demethylation treatment with 5-aza-20-deoxycytidine (Aza) treatment. Ectopic expression of two of these genes, DCC and DLC1, was then carried out to evaluate tumor suppressor properties in the NPC cell lines.
MATERIALS AND METHODS
Tissue samples
We collected a total of 50 primary tumor NPC samples and 28 nasopharyngeal biopsies samples with benign diagnoses for which paraffin embedded samples were available. The tumor sample set had 30 samples from the Chinese University of Hong Kong and 19 samples from the Johns Hopkins Hospital. The tumors were classified by histopathological grading according to the WHO classification of 1991: Type I, keratinizing squamous cell carcinoma; Type II, nonkeratinizing carcinoma. Of the 28 nasopharyngeal biopsies, 10 were obtain from head and neck cancer patients with primaries outside of the nasopharynx for which biopsies were available as part of the screening protocol for unknown primaries. This particular subset of samples is enriched for the field cancerization effect. Demographic and clinical information was obtained from the computer records at the Chinese University of Hong Kong and the Johns Hopkins Healthcare system. A summary of the patient population is available in Table 1. Collection of tissue and demographic data was performed in accordance to the Johns Hopkins University Institutional Review Board under protocol 03-11-12-06e.
Table 1.
Demographic and clinical characteristics of nasopharyngeal carcinoma patients (n=50)
| Characteristic | NPC patients No. (%) | Control patients No. (%) |
|---|---|---|
| Gender | ||
| Female | 10 (20%) | 20 (71%) |
| Male | 26 (52%) | 8 (29%) |
| Unknown | 14 (28%) | |
| Age (y) | ||
| Median (range) | 48 (17–78) | 41 (21–71) |
| Race | ||
| Asian | 30 (60%) | 6 (21%) |
| Caucasian | 12 (24%) | 12 (42%) |
| African American | 5 (10%) | 6 (23%) |
| Other | 3 (6%) | 0 |
| Stage | ||
| I | 4 (8%) | |
| II | 12 (24%) | |
| III | 8 (16%) | |
| IV | 11 (22%) | |
| Unknown | 15 (30%) | |
| Grade | ||
| II: non keratinizing | 50 (100%) | |
| EBV status | ||
| Positive | 50 (100%) | |
| Local recurrence | ||
| Yes | 3 (6%) | |
| No | 31 (62%) | |
| Unknown | 16 (32%) | |
| Metastasis developed | ||
| Yes | 4 (8%) | |
| No | 29 (58%) | |
| Unknown | 16 (32%) | |
| Median follow-up (m) (range) | 42.5 (6–154) | |
Nasopharyngeal carcinoma cell lines
The cell lines C-666 and CNE1 were kindly provided by Dr. Qian Tao from the Chinese University of Hong Kong, and HONE1 and HNE by Dr. Maria Lung from the Hong Kong University of Science and Technology. C-666 is the only EBV positive cell line, the rest of the cell lines are EBV negative. Cells were cultured in RPMI 1640 or DMEM (Life Technologies, Inc., Carlsbad, CA) supplemented with 10% calf serum, 1 mm sodium pyruvate, 0.1 mm nonessential amino acids, and penicillin & streptomycin in 5% CO2.
DNA extraction
DNA was extracted from paraffin embedded tissue after xylene deparaffinization and culture cells were extracted by digestion with 50 μg/mL proteinase K (Boehringer) in the presence of 1% SDS at 48°C followed by phenol/chloroform extraction and ethanol precipitation. Extracted DNA was dissolved in either LoTE (2.5 mM EDTA, 10 mM Tris–HCl [pH 8]) or molecular grade water and stored at −20°C.
EBV Status Evaluation
Conventional PCR was used to evaluate the presence of two EBV regions with the previously published primers25. PCR amplification was performed using 200ng of DNA as template. Cell line C-666 was used as a positive control and HNE as a negative control. The following primers were used: BamHI region F 5'-CCCAACACTCCACCACACC-3' and R 5'-TCTTAGGAGCTGTCCGAGGG-3' and EBNA-1 region F 5'-TCATCATCATCCGGGTCTCC-3' and R 5'-CCTACAGGGTGGAAAAATGGC-3'.
Gene selection
A total of 18 tumor suppressor genes or candidate tumor suppressors were selected for QMSP-based examination of methylation abnormalities. They include 3 tumor suppressor genes (DLC1, DLEC, and TIG1) reported as hypermethylated in more than 70% of the NPC and not in normal nasopharyngeal samples and/or immortalized nasopharyngeal cell lines17, 18, 20. The remaining fifteen genes had never been evaluated in NPC (AIM, APC, CALCA, DCC, ESR, FHIT, GSTP, HIC, KIF1α, NISCH, PAK3, PGP9.5, S100, TGFβ, and TIMP3). They were selected because these genes are known to be involved in other tumor types. Each gene was selected because of its cancer-specific methylation pattern or its potential biologic relevance. For example, PGP9.5 shows a cancer specific methylation pattern in bladder and esophageal cancer26, 27. Furthermore, for esophageal cancer PGP9.5 is associated with poor 5 year survival and with lymph node metastasis27.
Sodium Bisulfite Treatment
All tissues were subjected to bisulfite treatment, which converts unmethylated cytosine residues to uracil residues and preserves methylated cytosines as such, as described previously28. The EpiTect Bisulfite kit (Qiagen, Valencia, CA) was used according to the manufacturer's instructions. Converted DNA was stored at −80°C.
Methylation analysis
Bisulfite treated DNA was used for gene-specific QMSP reactions. The details of the design of methylation-specific primers for QMSP have been published15. PCR reactions were performed in a 384 well plate TaqMan 7900HT (Applied Biosystems) and analyzed by a sequence detector system (SDS 2.3; Applied Biosystems). Briefly, fluorogenic PCRs were carried out in duplicate in with 3μL of bisulfite-modified DNA. The anhealing temperature was 60°C for all genes. Supplementary Table 1 shows the primer and probe sequences used. Leukocytes from a healthy individual were methylated in vitro with excess SssI methyltransferase (New England Biolabs Inc., Beverly, MA) to generate positive controls for calibration of the reactions. Molecular grade water was used as a non-template control. The β-actin gene was used to normalize and as an internal loading control. The methylation ratio is the ratio of values for the gene-specific PCR products to those of the β-actin and then multiplied by 1,000 for easier tabulation.
5-Aza-2'-deoxycytidine (Aza) treatment of NPC cell lines
NPC cells lines, HNE and CNE, freshly seeded at 1×105 cells/ml were allowed to grow overnight. The culture medium was then replaced with fresh medium containing Aza at a final concentration of 5μM (Sigma-Aldrich Corporation, St Louis, MO, USA). Cells were allowed to grow for 120 hrs, with changing of Aza containing medium every 24 h, and then harvested for DNA and RNA extraction. We handled control cells the same way, but adding phosphate buffer saline (PBS, pH 7.5) instead of Aza as Aza was dissolved in this solution.
RNA extraction, cDNA synthesis, and reverse transcription- PCR
Total cellular RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA) according to the instructions of the manufacturer. Total RNA was adjusted to the same amount, and cDNA synthesis was done using SuperScript First-Strand Synthesis kit (Invitrogen, Frederick, MD). The final cDNA products were used as the templates for subsequent reverse transcription-PCR (RT-PCR). Specific primers were designed across the intron-exon boundaries to prevent genomic amplification. RT-PCR primers are shown in supplementary Table 2. The GAPDH gene was used as a control.
Plasmids and transfection protocol
For transfection, 1 × 106 HNE cells were seeded per well using six-well plates at a confluence of 50% to 70%. Cells were transfected with 1 μg of the expression for DCC (cytomegalovirus (CMV) + DCC)29, DLC1 (pcDNA 3.1+ DLC1)18 or of the mock, and 3 μL of Fugene 6 (Roche Diagnostics) diluted in 100 μL serum-free medium following the instructions of the manufacturer. Incubation of cells with the transfection complex was allowed for 4 hours followed by change for fresh medium. The expression of the gene of interest was measured by RT-PCR following transfection to ensure successful transfection.
Colony formation assay
At 48-h post-transfection, G-418 (500 μg/ml) was added as a selection antibiotic. After 2 weeks, cells were stained with 0.4% crystal violet solution (MeOH/acetic acid, 3:1). Colonies were photographed under the microscope and counted. Numbers of colonies (with >50 cells/colony) were counted and analyzed. The experiment was repeated twice independently and each experiment was done in triplicate. Statistical analysis was performed with Student's t-test, P<0.05 was considered as statistically significant difference.
Cell invasion/Matrigel assay
Cells (1 × 104) in 0.5 mL of serum-free media were added to each well of 24-well/8-μm pore invasion membrane chambers coated with Matrigel (BD Discovery Labware, Bedford, MA). The lower chambers contained 10% fetal bovine serum (FBS) in media to serve as a chemo attractant. Cells were allowed to migrate or invade over the course of 48 hours. Cells that failed to penetrate the filters were removed by scrubbing with cotton swabs. Chambers were fixed and stained with 100% methanol and 0.5% crystal violet. Cells per membrane were counted under the microscope (20×objective) and represented the average of three independent experiments. Statistical analysis was performed with Student's t-test, P<0.05 was considered as statistically significant difference.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) bromide assay
The assay was based on a previously described colorimetric assay for cellular growth and survival30. At 24, 48, 72, 98, and 123 hours after transfection, medium was changed, and MTT was added. The plates were read on a microplate reader (Molecular Device, CA) at 550 nm with a reference wavelength of 650 nm. The absorbance was reported, where a higher absorbance means a higher concentration of live cells.
Statistical analysis
Non parametric receiver-operator characteristic (ROC) curves to distinguish malignant from control tissues were constructed based on the methylation levels. Cutoff values that maximized sensitivity and specificity were selected from the ROC curves and applied to determine methylation frequencies. Cross-tabulation and Fisher's exact test were used to evaluate differences in gene methylation.
The regression analysis began with univariate logistic regression analysis between the tumor/normal occurrence and each of the predictor variables, i.e., the log methylation value of a gene in the model where p is the probability of tumor/normal and X is the predictor variable.
For the multivariate model, variables which have a p-value <0.25 in the univariate tests were selected as candidates. A stepwise procedure in which a forward selection with a test for a backward elimination was then used to select the variables with the entry and stay p-values set at 0.15 and 0.20. The final model selection was determined by the likelihood ratio test based on the model of each intermediate step vs the previous model with a significance level set at α=0.1. The importance of each variable included in the fitted model was verified by examining the Wald statistic for each variable. The model fitting was checked with Hosmer 1-df, Goodness of fit test, Pearson Chi-square, Deviance and other diagnostics such as Pearson residuals and Deviance residuals. Leave-one-out cross validation was used to predict the status of individual cases. Analysis of Covariance analysis was performed with the model below.
The variables were defined as follows. The race groups were Southern Chinese versus all the other groups. The stage classification was done according to the TNM-staging-systems of the UICC/AJCC (International Union Against Cancer/American Joint Committee on Cancer). Stage I and II were compared against stages III and IV. Age was analyzed as a continuous variable. Logistic regression analysis was done to explore the correlation between methylation and recurrence data
RESULTS
Determination of Appropriate Genes
Initially, 18 genes were evaluated for methylation by QMSP on 4 NPC cell lines (C-666, HONE, HNE, and CNE) and 5 normal microdissected epithelium samples from the nasopharynx. All of these genes were selected as TSG known to be methylated in cancer and as potential biomarkers in other cancers and NPC. Genes were chosen for additional analyses when genes were methylated in all NPC cell lines, and unmethylated or a methylation ratio of <10 for the normal nasopharynx tissues. A total of 11 genes met both criteria (APC, AIM1, CALCA, DCC, DLEC DLC1, ESR, FHIT, KIF1A, PGP9.5, and TIG1) and were then evaluated on additional tissue samples. Refer to Supplementary table 4 for methylation values.
Methylation in Nasopharyngeal tissues
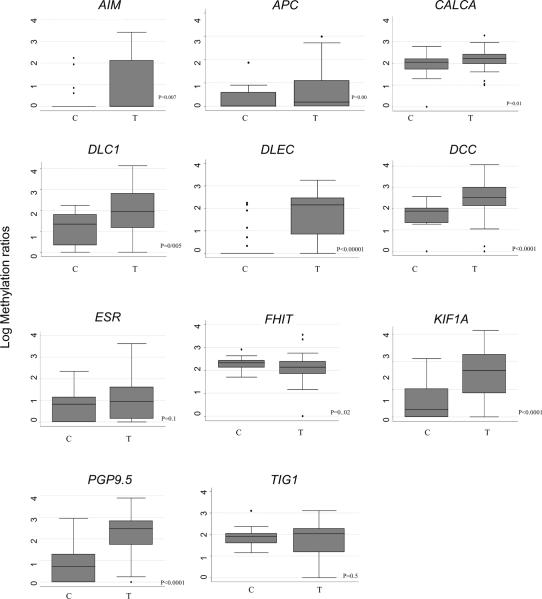
The resulting 11 genes were tested in a nasopharyngeal sample set that contained 50 NPC and 28 controls from benign nasopharyngeal biopsies. The controls included nasopharyngeal mucosa form 10 samples from patients with head and neck primaries outside of the nasopharynx. This subset of patients had considerable exposure to cigarette smoke and alcohol to their aerodigestive tract. The methylation values differ among NPC and control tissues. Box plots showing the methylation results by QMSP are shown in Figure 1. In general the methylation levels are higher in tumors compared to controls. Univariate logistic regression for the association between methylation and tumor status revealed a significant association between all the methylated genes except for TIG1 (p value<0.05).
Figure 1.
Promoter methylation levels in NPC cancer patients (T= tumor samples; n=50) and nasopharyngeal biopsies control (C= control samples; n=27). The quantity of each methylated gene promoter shown as the ratio of the PCR product of the gene of interest and the reference gene β-actin multiplied by 1,000. Boxplots show the middle 50% of data, the line is the median, and the bars extend 1.5 times the interquartile range. Mann-whitney test p values are shown.
The frequencies of methylation between nasopharyngeal carcinoma samples and controls as well as the cutoff values used are shown in Table 2. The sensitivity of the individual genes in tissue ranged from 26 to 66%. The specificity was above 92% for all genes except FHIT for which specificity was 21%. The combination of all the 11 genes displayed a sensitivity of 100% and a specificity of 57% (AUC of 0.78). Different combinations of methylation makers are shown on Table 3. Notably, the combination of PGP9.5, KIF1A, and DLEC had a sensitivity of 84% and a specificity of 92% (AUC 0.88). The combination of the two genes chosen for functional studies (see below), DLC1 and DCC, harbored a sensitivity of 66% and a specificity of 96% (AUC 0.81). A multigene predictive model was designed with the methylation values of 5 genes: PGP9.5, ESR, DCC, AIM1, and KIF1A. The methylation of the individual genes affects the probability of the samples being classified as a tumor or a control tissue. The model is shown bellow, where represent the gene methylations and regression coefficients, respectively.
Table 2.
Promoter methylation frequency for 11 genes analyzes in Nasopharyngeal carcinoma
| Gene | Frequency of positivity |
p | Cutoff | AUC (95% CI) | Sensitivity % | Specificity % | |
|---|---|---|---|---|---|---|---|
| Nasopharynegal carcinoma | Controls | ||||||
| AIM1 | 15/50 | 2/28 | 0.002 | 1.85 | 0.61 (0.53–0.69) | 30 | 92 |
| APC | 17/50 | 1/28 | 0.002 | 0.91 | 0.65 (0.57–0.72) | 34 | 96 |
| CALCA | 22/50 | 2/28 | 0.001 | 0.91 | 0.68 (0.59–0.76) | 40 | 92 |
| DCC | 23/46 | 1/27 | 0.01 | 2.49 | 0.77 (0.69–0.84) | 50 | 96 |
| DLEC | 29/48 | 1/28 | <0.0001 | 2.2 | 0.73 (0.65–0.81) | 60 | 96 |
| DLC1 | 21/48 | 0/27 | <0.0001 | 2.3 | 0.71 (0.64–0.77) | 43 | 100 |
| ESR | 13/50 | 1/28 | 0.01 | 1.61 | 0.61 (0.54–0.68) | 26 | 96 |
| FHIT | 22/50 | 6/28 | 0.05 | 2.1 | 0.38 (0.28–0.49) | 44 | 21 |
| KIF1A | 28/50 | 1/28 | <0.0001 | 1.61 | 0.76 (0.68–0.83) | 56 | 96 |
| PGP9.5 | 32/50 | 2/28 | <0.0001 | 2.3 | 0.78 (0.70–0.86) | 66 | 96 |
| TIG1 | 15/50 | 5/28 | 0.28 | 1.59 | 0.43 (0.43–0.53) | 26 | 92 |
| All genes | 50/50 | 12/28 | <0.0001 | 0.78 (0.69–0.87) | 100 | 57 | |
P values from two sided Fisher's exact test. AUC (area under the curve) and cutoffs derived from non parametric ROC curves
Table 3.
Sensitivity and specificity of different combinations of methylation markers
| Gene Combinations | AUC | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| PGP9.5, ESR, DCC, AIM1, and KIF1A | 0.87 (0.78–0.95) | 92 | 82 |
| PGP9.5, DLC1, DLEC | 0.84 (0.73–0.96) | 84 | 85 |
| PGP9.5, DLEC, DCC, KIF1A | 0.89 (0.86–0.96) | 90 | 89 |
| DLEC, DCC, KIF1A, AIM1 | 0.88 (0.81–0.96) | 92 | 85 |
| PGP9.5, KIF1A | 0.85 (0.77– 0.92) | 78 | 92 |
| PGP9.5, DLC1 | 0.85 (0.77– 0.92) | 78 | 92 |
| PGP9.5, DCC | 0.86 (0.78–0.96) | 84 | 89 |
| DLEC, KIF1A | 0.87 (0.80–0.93) | 78 | 96 |
| PGP9.5, KIF1A, DLEC | 0.88 (0.81–0.95) | 84 | 92 |
| AIM1, DCC | 0.79 (0.70–0.88) | 70 | 89 |
| DCC, DLC1 | 0.81 (0.73–0.88) | 66 | 96 |
| KIF1A, DCC, DLEC | 0.89 (0.82–0.96) | 86 | 92 |
AUC: area under the curve derived from nonparametric ROC curves
The coefficients in the model determine the influence a particular gene has in the classification. Given the methylation of other genes in the model, tumor occurs 7.3 (e1.99) times as often with a 10% increase in total methylation of PGP9.5. The model was able to accurately classify 40 of the 46 tumors (sensitivity of 87%) and discard 21 of the 26 controls (specificity of 81%). The goodness fit test for the model had a p value of 0.62 suggesting the model fits well. Six samples were excluded from the statistic model because we could not obtain methylation information for all 11 genes due to limited availability of DNA.
Additionally, we performed correlation analysis for methylation in all pairs of gene markers. Statistically significant correlations are shown in Supplementary Table 3. The strongest correlations were between DLEC and ESR (r>0.67), DCC and PGP9.5 (r>0.66), and DCC and DLC1 (r>0.6). Moderate correlations between CALCA and FHIT (r=0.65), KIF1A and TIG1 (r=0.48), and DLEC and KIF1A (r=0.56) were also found.
Correlation of methylation profile with clinicopathologic parameters
Several clinicopathologic and demographic variables were compared with the DNA methylation patterns in the tumor subset for which information was available. The ages were similar for the tumor and control groups in the study with a median age of 48 for the NPC patients and of 41 for the controls. The gender and race proportions were different between the groups. A smaller fraction of the NPC cases were females (20%) in comparison to the controls (70%). Also 60% of the NPC cases were Southern Chinese while only 20% of the control belonged to this racial group. All the tumors in the study were type II non-keratinizing carcinomas and all were EBV positive. No difference in methylation values was found between the 10 samples from the patients with other head and neck cancer and the rest of the control samples.
ANOVA analyses were done to determine whether the indentified methylation markers were associated with covariables based on the model described in materials and methods. A significant association with methylation and increased age was found for APC (p value 0.02). Patients from southern China were associated with an increase in methylation of CALCA, DLEC, ESR, KIF1A, and TIG1 (p value <0.05). Evaluating the methylation status in relationship to EBV status would give insights into the genetic profiles of the different NPC types. All of the NPC in our study tested positive for EBV status making comparison among methylation markers and EBV status impossible. We were unable to detect association between methylation and other clinicopathologic parameters.
In our sample set, two patients developed local and distal metastasis, two patients only distal metastasis, and another patient only local disease. The median follow up was 42.5 months with a range going from 6 to 154. We could not establish a correlation between recurrence and the methylation data.
Re-expression by treatment with Aza
We investigated if epigenetic inactivation could be reversed by the DNA methylation inhibitor Aza treatment. Five genes (DCC, DLC1, KIF1A, and PGP9.5) were randomly chosen to be evaluated in HNE and CNE cell lines. Aza treatment resulted in increased demethylation of the promoter regions of all chosen genes. We then analyzed the expression levels of the genes in the cell lines in Aza treated cell lines and controls. All Aza treated cell lines show dramatic re-expression in comparison to the controls (Figure 2). These results suggest that methylation contributes to the regulation of gene expression for the candidate genes we tested in NPC.
Figure 2.
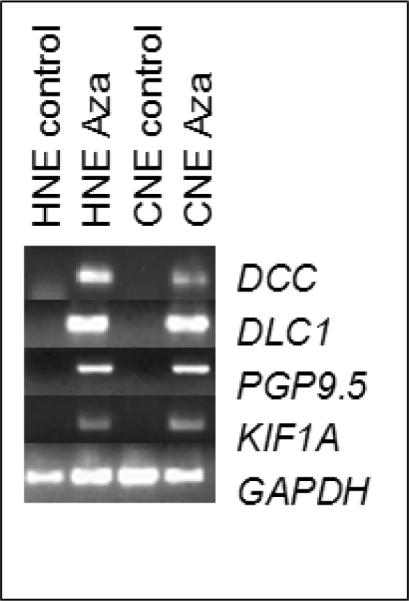
Re-expression of genes (DCC, DLC1, PGP9.5, and KIF1A) after treatment with Aza in NPC cell lines. HNE and CNE cell lines were treated for 5 days with Aza at a 5μM concentration. Side controls are shown. GAPDH was used as a normalizing loading control.
DCC and DLC1 functional studies
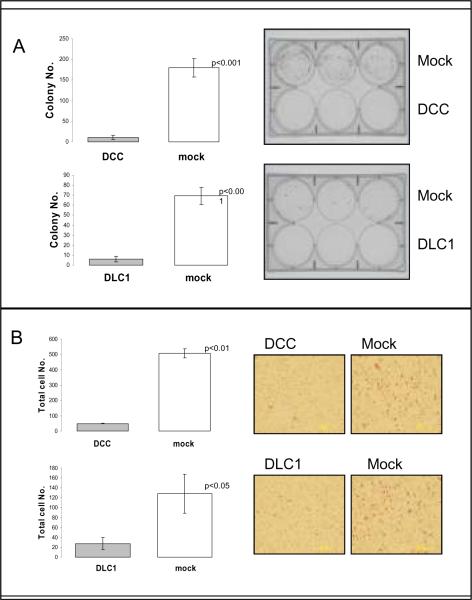
Two of these 5 genes, DCC and DLC1, were chosen to study further for tumor suppressor activity based on other tumor types for DCC29 and the high specificity for NPC of DLC1. Assays to evaluate cell growth and invasion properties were performed in transiently transfected HNE cell line. We performed colony focus assays after 2 weeks of selection in the presence of G418. Over-expression of DCC and DLC1 led to a notable decrease in the number of colonies in comparison to the control cells (vector only) (Figure 3A). Growth proliferation in control cells and over-expressing cells by MTT did not show a difference at 5 days. Invasion potential measured by the matrigel assay showed decreased invasion capability upon force expression of DCC and DLC1 in comparison to control cells (Figure 3B). These results suggest that both genes harbor basic tumor suppressor properties in NPC.
Figure 3.
DCC and DLC1 inhibit colony forming ability, and invasion in NPC cell lines. Panel A. Colony focus assays were performed in HNE cell lines the transiently transfected with DCC and DLC1. Colonies were counted under a microscope after 2 weeks of cell incubation in the presence of G418. Panel B. Matrigel invasion was performed on equal numbers of HNE cells transfected with the DCC or DLC1 and empty vector (mock). Cells were seeded to the upper wells of Matrigel-coated chambers in serum-free medium. After incubation for 48 hours, cells in the lower chambers were stained. All experiments were done in triplicate, and values indicate means ±s.d. Statistical significance was determined using Student's two-talied t test (p values<0.05).
DISCUSSION
Promoter methylation of tumor suppressor genes is an important mechanism of gene inactivation in human cancer. Because considerable difference exists between the methylation profiles of the distinct tumor types14, profiling the individual tumor types is a necessary task. The present study is a comprehensive attempt at profiling NPC based on a panel of methylation markers.
For the present study, the methylation profile was determined based on a large number of genes. The genes included in our study either had been widely reported as silenced by methylation in other tumor types or were identified recently by our unbiased pharmacologic unmasking strategy31, 32. Most of the genes in our panel (15) had never been evaluated in NPC. Our sample set contained 50 primary tumors as well as 28 controls. Obtaining appropriate controls for the study of NPC remains a challenge because of the difficultly in accessing the nasopharyngeal cavity. Published studies have a very limited number of normal nasopharyngeal samples (3 to 10 samples). For our study we found a moderate sized control group including a subset of samples that had other head and neck cancers to assess the use of these markers to differentiate field cancerization molecular alterations from cancer specific alteration. To our knowledge, this is the first study utilizing QMSP for the examination of NPC which gives us the advantage of quantitatively comparing the samples to more accurately segregate tumors from other samples. Even though several studies have looked at individual gene methylation status in NPC, this is one of the few studies looking at a panel of markers for this relatively rare tumor.
We report for the first time methylation of AIM1, APC, CALCA, DCC, ESR, FHIT, KIF1A, and PGP9.5 in a cancer-specific pattern for NPC (differential methylation in tumor than in normal). AIM1 and KIF1A are both newly discovered genes in cancer resulting from our pharmacologic unmasking study whose function is poorly understood. Our group has reported hypermethylation of AIM1 in bladder cancer and of KIF1A in a variety of human cancers including head and neck cancer, bladder, and breast cancer31, 32. AIM1 is involved melanoma tumorigenesis and in calcium binding, while KIF1A is in involved in anterograde transport along axonal microtubules33, 34. DCC is a tumor suppressor gene involved in epithelial differentiation and has been reported as hypermethylated in several cancers including colon, and head and neck29, 31. ESR stands for estrogen receptor alpha, which has growth suppression capabilities and is hypermethylated in breast cancer, colon cancer, and leukemias35, 36. PGP9.5 is a neuron specific protein that can function as a hydroxylase and a ligase and has been reported as hypemerthylated in gastric and esophageal squamous cell carcinomas37. APC, CALCA and FHIT have been reported as hypermethylated in a variety of malignancies such as colon cancer, breast cancer, and head and neck cancer14, 38. From a diagnostic point of view, a panel of only two gene markers could achieve sensitivity values close to 80% while still preserving high specificities (PGPG9.5 and KIF1A, PGP9.5 and DCC, PGP9.5 and DLC1, and KIF1A and DLEC).
We were also able to confirm a very high frequency of DLC1 and DLEC methylation for NPC. DLC1 is a tumor suppressor gene with loss of heterozygosity in liver cancer39, DLEC is a candidate tumor suppressor gene originally described in lung cancer20. The frequency of methylation in the tumors was similar between our study and previous studies. Considering any level of methylation as a positive as reported in non-quantitative assays, we found a tumor methylation frequency of 89% for DLC1 (previously 90%) and 85% for DLEC (previously 85%). The previous studies had examined mostly Asian samples, while our results show that both genes are methylated in EBV positive NPC from American patients as well. In our study, the levels of methylation for DLC1, DLEC, and TIG1 in the normal epitheliums were higher than that reported previously17, 18, 20. For TIG1 the levels of methylation in the controls could not be distinguished from the levels in the primary tumors. The discrepancy with the previous studies for the control samples could be attributed the content of the sample set and to methodology. The previous studies had only 3 or 4 normal nasopharyngeal epithelium samples while our study tested 28 samples. Additionally, the previous study used conventional MSP rather than QMSP.
Recent studies have reported that circulating EBV viral load correlates with tumor burden in NPC and can be used to detect recurrence10, 40. The use of EBV as a biomarker for early detection is limited by the rate of false positives, given that over 90% of the population worldwide is seropositive for the virus41. Methylation marker analysis in nasal scrapings or biopsies could be a more specific test for early detection than EBV viral load. Analysis of body fluids such as serum for methylation could also be further explored.
When analyzing the clinicopathologic and demographic variables, race and age were significantly associated with methylation. Race was significantly correlated with gene methylation of 5 genes. As mentioned before, the Southern Chinese population has an increased incidence of NPC. Methylation of CALCA, DLEC, ESR, KIF1A, and TIG1 was more likely to occur within this population and could be due to variability in genetic background or exposure to environmental carcinogens. Our study showed increased APC methylation with increased age and age-related methylation is a known phenomenon previously reported for APC42. A limitation of the present study is the low rate of recurrence which might have prevented us from detecting more prognostic correlations.
The functional studies showing growth suppressive effects for two commonly methylated genes in NPC lend support to the notion, that most if not all of the methylated genes specific to neoplastic epithelium, are true tumor suppressor genes. Pharmacologic demethylation experiments, confirm that methylation is an important regulatory mechanism for gene expression. Deleted in colorectal carcinomas (DCC) is located in 18q 21.3 and codes for a transmembrane protein similar to cell-adhesion proteins. DCC mediates apoptosis conditionally upon engaging with its ligand netrin-143. Carvalho et al. demonstrated that promoter methylation silenced expression of DCC by immunohistochemistry in head and neck squamous cell carcinomas29. They also showed that transfection of DCC lead to dramatic decrease in growth by CFA and this result was recreated in our NPC cell lines. Furthermore we showed a decrease in the invasion potential of the NPC cells upon DCC transfection. DCC has been implicated in poor prognosis for head and neck tumors and as a marker for local recurrence and increased risk of lymph node metastasis in esophageal carcinomas44.
Deleted in liver cancer 1 (DLC1) is a gene located in 8p22-21.3 that belong to the GTPase activiating proteins Rho family which binds to human tensins to suppress tumor growth by regulating actin adhesion45, 46. Seng et al. showed DLC1 was a downregulated by methylation in NPC and suppression of growth in the CNE cell line18. We confirmed the ability of DLC1 to decrease colony formation and decrease invasion capacity in another NPC cell line and showed decreased invasion properties. In breast cancer, DLC1 is downregulated in metastatic cell lines and the restoration of its expression leads to reduction of metastases in nude mice experiments47. Given the frequency of inactivation and their biological roles, it is possible that DCC and DLC1 could be important players in cell migration and particularly in NPC neck metastasis. Our results confirm that DCC and DLC1 act as tumor suppressor genes in NPC and that they are frequently inactivated by methylation.
The present study constitutes a comprehensive survey of methylated genes in NPC. Further studies looking at larger cohorts that validate these markers and their clinical applicability in NPC patients are required. Of particular interest will be studies to evaluate the use of these genes as diagnostic and prognostic biomarkers in body fluids that are easier to sample than the nasopharynx such as saliva and serum. For this purpose we are currently in the process of collecting serum samples from Southern Chinese NPC patients as well as matched controls.
Supplementary Material
Acknowledgments
We thank Dr. Maria Lung (Hong Kong University of Science and Technology) for the generous gift of the HONE and HNE cell lines.
Grant support: Specialized Program of Research Excellence (SPORE) grant P50-DE019032 from National Institute of Dental and Craniofacial Reseach (NIDCR) and National Institute of Health (NIH), as well as Early Detection Research Network (EDRN) grant U01-CA084986 from Cancer Institute (NCI) and NIH.
REFERENCES
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharyya N. The impact of race on survival in nasopharyngeal carcinoma: a matched analysis. Am J Otolaryngol. 2004;25:94–7. doi: 10.1016/j.amjoto.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Tao Q, Chan AT. Nasopharyngeal carcinoma: molecular pathogenesis and therapeutic developments. Expert Rev Mol Med. 2007;9:1–24. doi: 10.1017/S1462399407000312. [DOI] [PubMed] [Google Scholar]
- 4.zur Hausen H, Schulte-Holthausen H, Klein G, Henle W, Henle G, Clifford P, Santesson L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1056–8. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]
- 5.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–68. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 6.August M, Dodson TB, Nastri A, Chuang SK. Nasopharyngeal carcinoma: clinical assessment and review of 176 cases. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2001;91:205–14. doi: 10.1067/moe.2001.110698. [DOI] [PubMed] [Google Scholar]
- 7.Sham JS, Choy D, Wei WI. Nasopharyngeal carcinoma: orderly neck node spread. Int J Radiat Oncol Biol Phys. 1990;19:929–33. doi: 10.1016/0360-3016(90)90014-b. [DOI] [PubMed] [Google Scholar]
- 8.Altun M, Fandi A, Dupuis O, Cvitkovic E, Krajina Z, Eschwege F. Undifferentiated nasopharyngeal cancer (UCNT): current diagnostic and therapeutic aspects. Int J Radiat Oncol Biol Phys. 1995;32:859–77. doi: 10.1016/0360-3016(95)00516-2. [DOI] [PubMed] [Google Scholar]
- 9.Huang DP, Ho JH, Chan WK, Lau WH, Lui M. Cytogenetics of undifferentiated nasopharyngeal carcinoma xenografts from southern Chinese. Int J Cancer. 1989;43:936–9. doi: 10.1002/ijc.2910430535. [DOI] [PubMed] [Google Scholar]
- 10.Lo YM, Chan LY, Lo KW, Leung SF, Zhang J, Chan AT, Lee JC, Hjelm NM, Johnson PJ, Huang DP. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999;59:1188–91. [PubMed] [Google Scholar]
- 11.Fang Y, Guan X, Guo Y, Sham J, Deng M, Liang Q, Li H, Zhang H, Zhou H, Trent J. Analysis of genetic alterations in primary nasopharyngeal carcinoma by comparative genomic hybridization. Genes Chromosomes Cancer. 2001;30:254–60. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1086>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 12.Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–46. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 13.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 14.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–9. [PubMed] [Google Scholar]
- 15.Carvalho AL, Jeronimo C, Kim MM, Henrique R, Zhang Z, Hoque MO, Chang S, Brait M, Nayak CS, Jiang WW, Claybourne Q, Tokumaru Y, et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin Cancer Res. 2008;14:97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- 16.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTCNCIC trial. Lancet Oncol. 2009 doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 17.Kwong J, Lo KW, Chow LS, Chan FL, To KF, Huang DP. Silencing of the retinoid response gene TIG1 by promoter hypermethylation in nasopharyngeal carcinoma. Int J Cancer. 2005;113:386–92. doi: 10.1002/ijc.20593. [DOI] [PubMed] [Google Scholar]
- 18.Seng TJ, Low JS, Li H, Cui Y, Goh HK, Wong ML, Srivastava G, Sidransky D, Califano J, Steenbergen RD, Rha SY, Tan J, et al. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26:934–44. doi: 10.1038/sj.onc.1209839. [DOI] [PubMed] [Google Scholar]
- 19.Cui Y, Ying Y, van Hasselt A, Ng KM, Yu J, Zhang Q, Jin J, Liu D, Rhim JS, Rha SY, Loyo M, Chan AT, et al. OPCML is a broad tumor suppressor for multiple carcinomas and lymphomas with frequently epigenetic inactivation. PLoS ONE. 2008;3:e2990. doi: 10.1371/journal.pone.0002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong J, Chow LS, Wong AY, Hung WK, Chung GT, To KF, Chan FL, Daigo Y, Nakamura Y, Huang DP, Lo KW. Epigenetic inactivation of the deleted in lung and esophageal cancer 1 gene in nasopharyngeal carcinoma. Genes Chromosomes Cancer. 2007;46:171–80. doi: 10.1002/gcc.20398. [DOI] [PubMed] [Google Scholar]
- 21.Wong TS, Kwong DL, Sham JS, Wei WI, Kwong YL, Yuen AP. Quantitative plasma hypermethylated DNA markers of undifferentiated nasopharyngeal carcinoma. Clin Cancer Res. 2004;10:2401–6. doi: 10.1158/1078-0432.ccr-03-0139. [DOI] [PubMed] [Google Scholar]
- 22.Chang HW, Chan A, Kwong DL, Wei WI, Sham JS, Yuen AP. Detection of hypermethylated RIZ1 gene in primary tumor, mouth, and throat rinsing fluid, nasopharyngeal swab, and peripheral blood of nasopharyngeal carcinoma patient. Clin Cancer Res. 2003;9:1033–8. [PubMed] [Google Scholar]
- 23.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–8. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 24.Franklin WA, Gazdar AF, Haney J, Wistuba II, La Rosa FG, Kennedy T, Ritchey DM, Miller YE. Widely dispersed p53 mutation in respiratory epithelium. A novel mechanism for field carcinogenesis. J Clin Invest. 1997;100:2133–7. doi: 10.1172/JCI119748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li YH, Shao JY, Zhao MQ, Gao HY, Li LR, Guan ZZ, Zeng YX. Quantitative analysis of plasma Epstein-Barr virus (EBV) DNA for monitoring of recurrence and metastasis in nasopharyngeal carcinoma patients after radiotherapy. Ai Zheng. 2003;22:645–8. [PubMed] [Google Scholar]
- 26.Brait M, Begum S, Carvalho AL, Dasgupta S, Vettore AL, Czerniak B, Caballero OL, Westra WH, Sidransky D, Hoque MO. Aberrant promoter methylation of multiple genes during pathogenesis of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2786–94. doi: 10.1158/1055-9965.EPI-08-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandelker DL, Yamashita K, Tokumaru Y, Mimori K, Howard DL, Tanaka Y, Carvalho AL, Jiang WW, Park HL, Kim MS, Osada M, Mori M, et al. PGP9.5 promoter methylation is an independent prognostic factor for esophageal squamous cell carcinoma. Cancer Res. 2005;65:4963–8. doi: 10.1158/0008-5472.CAN-04-3923. [DOI] [PubMed] [Google Scholar]
- 28.Gallicchio L, Matanoski G, Tao XG, Chen L, Lam TK, Boyd K, Robinson KA, Balick L, Mickelson S, Caulfield LE, Herman JG, Guallar E, et al. Adulthood consumption of preserved and nonpreserved vegetables and the risk of nasopharyngeal carcinoma: a systematic review. Int J Cancer. 2006;119:1125–35. doi: 10.1002/ijc.21946. [DOI] [PubMed] [Google Scholar]
- 29.Carvalho AL, Chuang A, Jiang WW, Lee J, Begum S, Poeta L, Zhao M, Jeronimo C, Henrique R, Nayak CS, Park HL, Brait MR, et al. Deleted in colorectal cancer is a putative conditional tumor-suppressor gene inactivated by promoter hypermethylation in head and neck squamous cell carcinoma. Cancer Res. 2006;66:9401–7. doi: 10.1158/0008-5472.CAN-06-1073. [DOI] [PubMed] [Google Scholar]
- 30.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 31.Hoque MO, Kim MS, Ostrow KL, Liu J, Wisman GB, Park HL, Poeta ML, Jeronimo C, Henrique R, Lendvai A, Schuuring E, Begum S, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res. 2008;68:2661–70. doi: 10.1158/0008-5472.CAN-07-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ostrow KL, Park HL, Hoque MO, Kim MS, Liu J, Argani P, Westra W, Van Criekinge W, Sidransky D. Pharmacologic unmasking of epigenetically silenced genes in breast cancer. Clin Cancer Res. 2009;15:1184–91. doi: 10.1158/1078-0432.CCR-08-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray ME, Wistow G, Su YA, Meltzer PS, Trent JM. AIM1, a novel non-lens member of the betagamma-crystallin superfamily, is associated with the control of tumorigenicity in human malignant melanoma. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3229–34. doi: 10.1073/pnas.94.7.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yonekawa Y, Harada A, Okada Y, Funakoshi T, Kanai Y, Takei Y, Terada S, Noda T, Hirokawa N. Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein-deficient mice. The Journal of cell biology. 1998;141:431–41. doi: 10.1083/jcb.141.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horii J, Hiraoka S, Kato J, Saito S, Harada K, Fujita H, Kaji E, Yamamoto K. Methylation of estrogen receptor 1 in colorectal adenomas is not age-dependent, but is correlated with K-ras mutation. Cancer science. 2009 doi: 10.1111/j.1349-7006.2009.01140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Issa JP, Baylin SB, Belinsky SA. Methylation of the estrogen receptor CpG island in lung tumors is related to the specific type of carcinogen exposure. Cancer research. 1996;56:3655–8. [PubMed] [Google Scholar]
- 37.Tokumaru Y, Yamashita K, Kim MS, Park HL, Osada M, Mori M, Sidransky D. The role of PGP9.5 as a tumor suppressor gene in human cancer. International journal of cancer. 2008;123:753–9. doi: 10.1002/ijc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verri C, Roz L, Conte D, Liloglou T, Livio A, Vesin A, Fabbri A, Andriani F, Brambilla C, Tavecchio L, Calarco G, Calabro E, et al. Fragile histidine triad gene inactivation in lung cancer: the European Early Lung Cancer project. American journal of respiratory and critical care medicine. 2009;179:396–401. doi: 10.1164/rccm.200807-1153OC. [DOI] [PubMed] [Google Scholar]
- 39.Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer research. 1998;58:2196–9. [PubMed] [Google Scholar]
- 40.Lin JC, Wang WY, Chen KY, Wei YH, Liang WM, Jan JS, Jiang RS. Quantification of plasma Epstein-Barr virus DNA in patients with advanced nasopharyngeal carcinoma. The New England journal of medicine. 2004;350:2461–70. doi: 10.1056/NEJMoa032260. [DOI] [PubMed] [Google Scholar]
- 41.Yu KH, Lo YM, Tse GM, Chan KC, Chan AB, Chow KC, Ma TK, Vlantis AC, Leung SF, van Hasselt CA, Johnson PJ, Chan AT. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nonnasopharyngeal head and neck carcinomas. Clin Cancer Res. 2004;10:1726–32. doi: 10.1158/1078-0432.ccr-0991-3. [DOI] [PubMed] [Google Scholar]
- 42.Waki T, Tamura G, Sato M, Motoyama T. Age-related methylation of tumor suppressor and tumor-related genes: an analysis of autopsy samples. Oncogene. 2003;22:4128–33. doi: 10.1038/sj.onc.1206651. [DOI] [PubMed] [Google Scholar]
- 43.Mehlen P, Rabizadeh S, Snipas SJ, Assa-Munt N, Salvesen GS, Bredesen DE. The DCC gene product induces apoptosis by a mechanism requiring receptor proteolysis. Nature. 1998;395:801–4. doi: 10.1038/27441. [DOI] [PubMed] [Google Scholar]
- 44.Miyake S, Nagai K, Yoshino K, Oto M, Endo M, Yuasa Y. Point mutations and allelic deletion of tumor suppressor gene DCC in human esophageal squamous cell carcinomas and their relation to metastasis. Cancer research. 1994;54:3007–10. [PubMed] [Google Scholar]
- 45.Yuan BZ, Yang Y, Keck-Waggoner CL, Zimonjic DB, Thorgeirsson SS, Popescu NC. Assignment and cloning of mouse Arhgap7 to chromosome 8A4-B2, a conserved syntenic region of human chromosome 8p22-->p21. Cytogenetics and cell genetics. 1999;87:189–90. doi: 10.1159/000015462. [DOI] [PubMed] [Google Scholar]
- 46.Kawai K, Iwamae Y, Yamaga M, Kiyota M, Ishii H, Hirata H, Homma Y, Yagisawa H. Focal adhesion-localization of START-GAP1/DLC1 is essential for cell motility and morphology. Genes Cells. 2009;14:227–41. doi: 10.1111/j.1365-2443.2008.01265.x. [DOI] [PubMed] [Google Scholar]
- 47.Goodison S, Yuan J, Sloan D, Kim R, Li C, Popescu NC, Urquidi V. The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res. 2005;65:6042–53. doi: 10.1158/0008-5472.CAN-04-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.