Abstract
Directed differentiation of human embryonic stem cells (hESCs) has generated much interest in the field of regenerative medicine. Because of their ability to differentiate into any cell type in the body, hESCs offer a novel therapeutic paradigm for myocardial repair by furnishing a supply of cardiomyocytes (CMs) that would ultimately restore normal myocardial function when delivered to the damaged heart. Spontaneous CM differentiation of hESCs is an inefficient process that yields very low numbers of CMs. In addition, it is not clear that fully differentiated CMs provide the benefits sought from cell transplantation. The need for new methods of directed differentiation of hESCs into functional CMs and cardiac progenitors has led to an explosion of research utilizing chemical, genetic, epigenetic and lineage selection strategies to direct cardiac differentiation and enrich populations of cardiac cells for therapeutic use. Here, we review these approaches and highlight their increasingly important roles in stem cell biology and cardiac regenerative medicine.
Keywords: cardiac regeneration, cardiomyocyte, human embryonic stem cell, myocardial repair
Use of human embryonic stem cells for myocardial repair
Over 5 million people in the USA alone suffer with heart failure, resulting in approximately 60,000 deaths at a cost of US$37 billion/year [1]. Unlike some organs, the heart is unable to repair itself after injury. Heart transplantation remains the ultimate approach to treating endstage heart failure, but this therapy is invasive, costly and excludes some patients who are not candidates for transplantation given their comorbidities. Most importantly, there are not enough organs for transplanting the increasing number of patients with end-stage disease, and the infrastructure necessary to transplant organs is not available in all countries. New, accessible therapies are needed to treat the millions of patients with debilitating heart failure worldwide [2]. Stem cell transplantation may represent the first realistic strategy for reversing the deleterious effects of what has until now been considered terminal damage to the heart.
Human embryonic stem cells (hESCs) grow and divide indefinitely while maintaining the potential to develop into tissues derived of all three embryonic germ layers. As such, they provide an unprecedented opportunity to treat a variety of human diseases characterized by tissue loss or insufficiency. Under appropriate culture conditions, hESCs spontaneously differentiate into cardiomyocytes (CMs) with structural and functional properties characteristic of endogenous CMs [3]. To induce spontaneous differentiation, hESCs are cultured in suspension with serum for a period of 7–10 days to form 3D cell aggregates called human embryoid bodies (hEBs). hEBs are then allowed to adhere to gelatin-coated plates, where further cultivation results in the appearance of spontaneously contracting areas. This approach has been adopted by most laboratories as the standard for spontaneous CM differentiation from hESCs. Nonetheless, there are limitations to this protocol, most notably the small number of CMs produced. With this method, beating areas are visible in only 5–15% of hEBs, as reported by many groups [3-6].
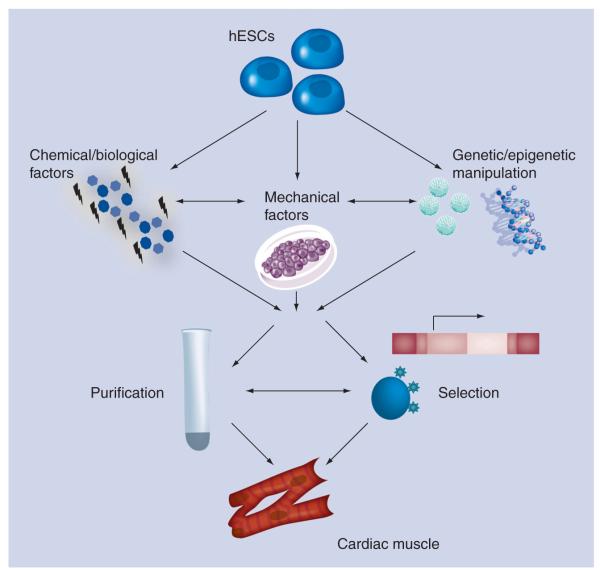
The low yield of CMs from spontaneous hESC differentiation, and the massive loss in cardiac cells from a typical myocardial infarction (~109 according to some studies, e.g., [7]) together present a critical issue in stem cell therapeutics. Clearly, the ability to generate sufficient numbers of hESC-derived CMs or cardiac progenitors will be required before cardiac regeneration through hESC transplantation can be realized. In addition, cardiac cell preparations will need to be of high purity in order to overcome the risk of local or distant teratoma formation. With these considerations in mind, much work has focused on directing the differentiation of hESCs into the cardiac lineage. Over the past decade, innovative enrichment, purification and selection strategies have been developed to guide cardiac differentiation to relatively pure homogeneity (Figure 1). These efforts have also provided new insight into human cardiac development, including the identification of multipotent cardiovascular progenitor cells. The prospect that cardiac progenitors will have a greater capacity to make functional connections with host myocardium as they differentiate in vivo after transplant has driven much of this work. Further investigations to delineate the human cardiac lineage tree will aid in the derivation of early cardiac progenitors from hESCs for utilization in myocardial therapies, and of specialized CM subtypes for specific myocardial applications.
Figure 1. Approaches to preparing human embryonic stem cell-derived cardiomyocytes for tissue repair.
Researchers are focusing on chemical (e.g., 5-azacytidine and p38 MAPK inhibitors) and biological (e.g., activin A, bone morphogenetic protein, basic FGF, VEGF and Dickkopf homolog 1) factors, genetic (e.g., miRNAs) and epigenetic (e.g., miRNAs and chromatin remodeling) manipulation, and mechanical factors (e.g., hydrodynamics and surface tension) to direct cardiomyocyte differentiation from hESCs. These approaches are complemented by purification methods that take advantage of the biochemical properties of human cardiomyocytes (e.g., Percoll density centrifugation and mitochondrial content), and selection strategies that rely on the expression of cardiac-specific genes (e.g., reporter lines and molecular beacons) and surface markers.
hESC: Human embryonic stem cell.
Chemical enrichment of CMs from hESCs
Defined culture media have been developed to direct human CM (hCM) differentiation from hESCs (Table 1). The cell-permeable small molecule, 5-azacytidine, which acts as a demethylating agent, was previously demonstrated to induce immortalized cells from a mouse cardiomyogenic cell line to become CMs [8]. Subsequently, others have shown that 5-azacytidine treatment at days 6–8 of hESC differentiation significantly increased cardiac α-MHC expression and enhanced CM differentiation, suggesting that DNA demethylation is a key factor in directing tissue-specific differentiation [4]. Similarly, exposure to SB203580, a small molecule inhibitor of p38 MAPK, has been shown to significantly improve CM differentiation of hESCs grown in medium conditioned by mouse END2 cells, supporting a role for p38 MAPK signaling in regulating hCM differentiation [9]. SB203580-treated hEBs displayed an increase in expression of both early mesoderm markers (Brachyury, Tbx6 and Mesp1) and cardiac α-MHC, as well as increased CM numbers. Our group has subsequently shown that p38 MAPK inhibition occurs in a dose- and stage-dependent manner, that it also causes the accelerated differentiation of hESC-derived CMs using the standard hEB formation method, and that it appears to act at the ectoderm/mesoendoderm branchpoint during hESC differentiation [10].
Table 1.
Chemical and biological medium supplements for directed differentiation of cardiomyocytes from embryonic stem cells.
| Factor/molecule | Concentration | Medium | Time added (days) | Ref. |
|---|---|---|---|---|
| 5-azacytidine | 10 μM | KO-DMEM/20% FBS | 6–8 | [4] |
| SB203580 | 5 μM | END2-CM | 0, 3, 6, 9 | [9,10] |
| PGI2 | 2 μM | Insulin-free, serum-free DMEM | 0, 3, 6, 9 | [13] |
| Activin A | 100 ng/ml | RPMI/B27 | 1 | [15] |
| BMP4 | 10 ng/ml | 2–5 | ||
| Activin A | 50 ng/ml | DMEM/F12/N2/B27 | 1–4 | [22] |
| BMP4 | 50 ng/ml | 1–4 | ||
| bFGF | 5 ng/ml | StemPro34 | 1–4, 8–14 | [23] |
| Activin A | 3 ng/ml | 1–4 | ||
| BMP4 | 10 ng/ml | 1–4 | ||
| VEGF | 10 ng/ml | 4–14 | ||
| DKK1 | 150 ng/ml | 4–14 |
bFGF: Basic FGF; BMP: Bone morphogenetic protein; DKK1: Dickkopf homolog 1; DMEM: Dulbecco’s modified Eagle’s medium; END2-CM: Serum-free DMEM conditioned by END2 cells; KO-DMEM: Knockout-DMEM; PGI2: Prostaglandin I2.
In the original study with SB203580, cells were subjected to an adapted differentiation system in which hESCs were differentiated in a suspension culture using serum-free medium conditioned by the mouse END2 cell line [11,12]. END2-conditioned medium alone exhibited CM-inducing activity during hESC differentiation [9], and biochemical as well as microarray analysis of END2-conditioned medium and END2 cells, respectively, identified prostaglandin (PG)I2, a product of prostaglandin synthase enzymes, as an inducing factor in hESC cardiac differentiation [13]. Two key enzymes involved in PGI2 synthesis were upregulated in END2 cells compared with control MES1 cells [14], which lack cardiogenic activity. PGI2 levels were between six- and ten-fold higher in END2-conditioned medium compared with control conditioned medium from MES1 cells. Moreover, insulin, a common supplement in media formulations, was discovered to be an inhibitor of hESC cardiac differentiation. END2-conditioned medium supplemented with increasing concentrations of insulin resulted in a dramatic decrease in hESC CM differentiation. Thus, addition of PGI2 in combination with insulin-free, unconditioned medium yielded effective cardiac induction similar to that produced by END2-conditioned medium. Cardiac differentiation was further augmented in the presence of SB203580. Taken together, these three components provide a basic, synthetic recipe for directing CM differentiation of hESCs.
Others have also undertaken the approach to chemically define medium conditions for controlling hESC differentiation. Another system utilizing sequential exposure of undifferentiated hESCs cultured on Matrigel™ to activin A followed by bone morphogenetic protein (BMP)4 within the first 5 days of differentiation proved to be 50-fold more efficient in generating CMs than the conventional serum induction of hEBs method [15]. Both factors were selected based on previous work showing that mesoderm formation and cardiogenesis are mediated by activin A and BMP4 [16-21]. Likewise, Yao et al. reported that hESCs seeded on Matrigel and treated with both activin A and BMP4 express specific CM markers (α-MHC, cardiac troponin I, Mef2, GATA4, Nkx2–5, atrial natriuretic factor [ANF]) [22].
This list of media supplements has grown to include basic FGF (bFGF), VEGF and the Wnt inhibitor, Dickkopf homolog (DKK)1. By mim-icking the signaling environment of the early mouse embryo, another group has established a three-stage protocol that supports cardiac development at high frequency in differentiating hESC cultures [23]. This protocol exposed hEBs to a combination of activin A, BMP4 and bFGF during the first 4 days of differentiation (stage 1) to induce primitive-streak formation, representing the onset of gastrulation. Between days 4 and 8 (stage 2), the differentiating hEBs were incubated in medium containing VEGF and DKK1 to induce cardiac mesoderm development and maturation. Previous studies had demonstrated that Wnt inhibition is required for cardiogenesis from mesodermal cells [24,25]. From day 8 to day 14 (stage 3), bFGF was added to VEGF and DKK1 to promote CM expansion. Gene expression analysis of stage 3 hEBs displayed expression of cardiac troponin T, atrial myosin light chain 2, and cardiac transcription factors Tbx5 and −20. By selecting particular exogenous factors known to play roles in early embryonic development and cardiac specification, and allocating them in the appropriate combinations and within an appropriate window of time, these studies have furnished base recipes for inducing hESCs toward cardiac lineages.
Effects of mechanical force on CM differentiation from hESCs
Since cardiac muscle is one of the few tissues that develops under the effects of dynamic force, it is not surprising that conditions generated by the force of fluids in motion can enhance CM differentiation. Sargent et al. discovered that supplying a constant rotary orbital motion for 7 days to suspension cultures of differentiating mouse EBs (mEBs) resulted in a significantly increased number of beating mEBs compared with mEBs cultured in static suspension [26]. Analysis of gene expression showed higher levels of mesodermal and cardiac proteins (Brachyury, GATA4, Nkx2–5, Mef2c, α-MHC and MLC2v) in rotary mEBs than in static mEBs. In addition, a greater proportion of rotary mEBs were positive for α-sarcomeric actin expression compared with static EBs. Morphologically, rotary orbital culture produced mEBs that were more uniform in size and higher in number than static suspension. Aggregation of individual mEBs was also inhibited with rotary culture compared with static culture. The enhanced CM differentiation was independent of rotary speed ranging from 25 to 55 rpm as determined by the expression of cardiomyogenic genes [27].
Domian et al. examined the effects of surface tension on cardiomyogenic differentiation of murine cardiac progenitors [28]. They cultured embryonic- and mouse embryonic stem cell (mESC)-derived progenitors on either fibronectin-coated slides or micropatterns of fibronectin alternating with a surfactant that blocks cell adhesion. They identified a population of cells that formed longitudinally aligned myocardial fibers specifically when grown on these micropatterned surfaces. In addition, culturing this population on micropatterned surfaces resulted in a statistically significant increase in the proportion of CMs, supporting a role for microenvironmental forces in CM differentiation.
Genetic & epigenetic manipulation of hESCs to enhance CM differentiation
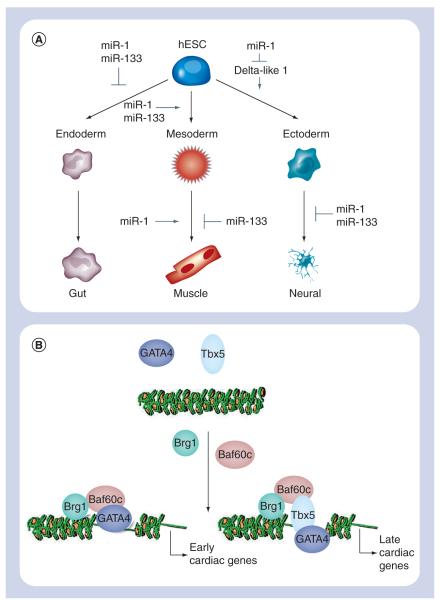
miRNAs are small, noncoding RNAs thought to regulate the expression of 30% of protein-coding genes [29]. Their biological importance in stem cell biology is underscored by recent studies demonstrating that mESCs lacking the miRNA processing enzyme Dicer display differentiation and proliferation defects [30-33]. miR-1 and miR-133 are specifically expressed in the mouse heart [34,35]. Targeted deletion or knockdown of these miRNAs results in dysregulation of cardiac morphogenesis, electrical conduction, cell-cycle and cardiac hypertrophy [34-37]. Recently, we have shown with our collaborators that miR-1 and miR-133 regulate the differentiation of mESCs and hESCs into the cardiac lineage [38]. Both miRNAs were enriched in mESC-derived CMs. Lentiviral introduction of either miR-1 or miR-133 into mESCs enhanced early mesoderm differentiation as evidenced by increased expression of Brachyury. miR-1 and miR-133 also reinforced mesoderm lineage decisions by repressing endoderm and neuroectoderm differentiation. When stimulated to differentiate into either endoderm or neuroectoderm lineages, mEBs expressing either miR-1 or miR-133 expressed lower levels of endodermal and neural markers compared with control mEBs. However, further differentiation revealed opposing roles of miR-1 and miR-133 (Figure 2a). miR-1 promoted differentiation of mesoderm into the cardiac and skeletal muscle lineages as determined by enhanced Nkx2–5 and myogenin expression, respectively, whereas miR-133 blocked induction of both markers. Importantly, the differentiation of hESCs in the presence of miR-1 behaved comparably to that of mESC differentiation. Overexpression of miR-1 in hESCs increased Nkx2–5 expression and yielded more than a threefold higher number of beating hEBs compared with wild-type controls.
Figure 2. Genetic and epigenetic manipulation to direct cardiomyocyte differentiation.
(A) miR-1 and -133 control the differentiation of embryonic stem cells into cardiac muscle, as well as ectoderm, as detailed in the text. In these studies, differentiation towards mesoderm was determined by expression of Nkx2–5, smooth muscle actin and myogenin, endoderm by α-fetoprotein and hepatocyte nuclear factor 4α expression, and ectoderm by expression of neural cell adhesion molecule 1, nestin and βIII tubulin. (B) Baf60c, a subunit of the human Swi–Snf (Brg–Brm) chromatin remodeling complex, is thought to be essential to the expression of GATA4-regulated early cardiac genes (e.g., Nkx2–5, Actc1, myosin light chain 7) and GATA4/Tbx5-regulated late cardiac genes (e.g., potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4).
hESC: Human embryonic stem cell.
While miRNAs direct cell lineage determination by controlling protein dosage, epigenetic regulation through chromatin remodeling has been shown to control cell fate as well (Figure 2b). Histone acetyltransferases and histone deacetylases promote chromatin unfolding and condensation to facilitate and silence active DNA transcription, respectively. Administration of the histone deacetylase inhibitor trichostatin A (TSA) during differentiation of mESCs has been reported to promote CM differentiation [39]. In a mESC line that expressed green fluorescent protein (GFP) under control of the cardiac-specific Nkx2–5 promoter, TSA exposure between days 7 and 8 of differentiation doubled the percentage of GFP+ cells, and boosted expression of Nkx2–5, β-MHC and ANF. While enhanced acetylation of histones 3 and 4 was seen in TSA-treated mEBs compared with untreated controls, there also was a concomitant increase in GATA4 acetylation and its DNA binding to the ANF promoter, suggesting that both epigenetic changes and post-translational modification of transcription factors contributed to CM differentiation.
Recently, Takeuchi et al. identified a minimal set of factors necessary to execute the cardiac transcriptional program [40]. Baf60c, a cardiac-enriched subunit of the Swi/Snf-like BAF chromatin remodeling complex, in combination with cardiac transcription factors GATA4 and Tbx5, was able to induce cardiac differentiation in mouse embryos when ectopically expressed. With this combination, 90% of the transfected embryos displayed expression of the early cardiac marker, cardiac α-actin (Actc1), and 50% of the transfected embryos exhibited beating tissue. GATA4 together with Baf60c was essential in initiating the cardiac gene program as assessed by expression of Actc1. None of the other transcription factors tested alone (Tbx5, Nkx2–5) or in concert with Baf60c were able to induce Actc1 expression. GATA4/Baf60c, however, was not sufficient for generating beating embryos: Tbx5 was required to achieve contracting CMs. Although the aforementioned factors have yet to be tested in hESCs, studies with Baf60c present defined, genetic platforms with which to alter cell fate decisions in hESCs.
Purification of hESC-derived CMs
The various differentiation protocols discussed have been developed with the aim of directing cardiac differentiation from pluripotent hESCs. While the efficiency of cardiac differentiation has markedly improved, differentiated hESC cultures may still consist of heterogeneous populations that include cell types derived from all three embryonic germ layers. If hESC-derived CMs are to be used in cell-based therapies, it is imperative that the final CM population is of high purity. Manual dissection of the beating areas has been one way to achieve high purity [3]. A less labor-intensive method utilizing Percoll gradient centrifugation has been described to purify hESC-derived CMs [4]. Differentiating hESCs were applied to a discontinuous Percoll gradient consisting of 40.5% Percoll layered over 58.5% Percoll. Following centrifugation, the majority of CMs resided within the 58.5% Percoll layer and expressed cardiac troponin I, sarcomeric MHC, α-MHC, β-MHC and N-cadherin. hCMs of 70% or less purity were obtained using this approach. It has been shown that Percoll-purified CMs can be further enriched by culturing the purified CM clusters in suspension for an additional week or longer [41]. These clusters, recultured for at least 8 days following purification, exhibited significant increases in cardiac α-MHC and β-MHC expression. Analysis by flow cytometry demonstrated that the cells comprising these clusters also expressed sarcomeric MHC, and that the percentage of sarcomeric MHC+ cells increased with time in culture.
A third purification strategy is based on the observation that CMs have high mitochondrial content compared with that of non-myocytes [42]. Using the fluorescent dye tetramethylrhodamine methyl ester perchlorate (TMRM) that freely diffuses into the mitochondrial matrix to label mitochondria, Hattori et al. found that TMRM fluorescence in embryonic rat hearts increases with developmental stage, suggesting that mitochondrial biogenesis is linked to cardiomyogenesis. In whole rat embryos, TMRM fluorescence in the heart was more robust than in other tissues, and when analyzed by flow cytometry, flow-sorted populations with the highest TMRM fluorescence were observed to express cardiac α-actinin. TMRM-labeled CMs derived from mESCs were positive for both Nkx2–5 and α-actinin. The CM content in cultured cells sorted from day 12 to day 25 mEBs was greater than 99% as determined by Nkx2–5 and α-actinin expression. Most notably, greater than 99% CM purity was also obtained in cultured cells sorted from differentiating hEBs.
Besides achieving a high degree of CM purity, these separation methods have the added advantage of not requiring genetic manipulation of hESCs. A disadvantage, however, is that none of these techniques allow purification of cardiac progenitor cells. Mechanical dissection can only be performed toward later stages of differentiation when a sufficient number of beating areas are visible. Percoll separation is less effective at earlier times of hESC differentiation [41]. hEBs used for TMRM purification experiments were between 50 and 90 days of differentiation [42]. Moreover, sorted TMRM-fluorescent cells from early mEBs failed to differentiate into CMs during subsequent culture.
Selection of hESC-derived cardiac progenitors
The use of genetic selection strategies has addressed both the issue of CM homogeneity and isolation of cardiac progenitors. Many laboratories, including our own, have developed transgenic/reporter hESC lines to derive pure CM populations. This approach relies on a cardiac-restricted promoter to drive the expression of a reporter gene or selectable marker. Huber et al. used lentiviral vectors to produce stable hESC lines in which enhanced GFP (eGFP) was expressed under control of the cardiacspecific human myosin light chain 2v promoter (MLC2v) [43]. Xu et al. generated stable hESC lines using a reporter plasmid consisting of the cardiac-specific mouse α-MHC promoter driving expression of the neomycin resistance gene [44]. Kita-Matsuo et al. designed a set of lentiviral vectors to generate multiple stable hESC lines with eGFP and mCherry reporters or with puromycin resistance downstream of the mouse α-MHC promoter [45]. In our lab, we have generated a cardiac-specific hESC reporter line using a lentiviral construct consisting of a fragment of the mouse α-MHC promoter upstream of eGFP. The specific promoter fragment used has allowed for the identification and analysis of early, multipotent cardiac progenitors expressing Nkx2–5, but before the onset of cardiac troponin T or chamber-specific myosin light chain expression [Wong S, Ritner C, King F, Bernstein HS, Pers. Comm.].
Collectively, fluorescence-activated cell sorting or antibiotic selection of these lines has yielded 85–99% pure CMs or cardiac progenitors that express cardiac-specific genes and exhibit action potentials characteristic of human embryonic CMs. eGFP-expressing cells derived from the MLC2v transgenic line formed stable intracardiac cell grafts following transplantation in rats [43]. Injection of neomycin resistance-selected hEBs into the hindlimb muscles of SCID mice resulted in no teratoma formation after 23 weeks [44]. Contractile forces in puromycin resistance-selected CMs were similar to those generated by rat neonatal ventricular CMs [45]. Whereas isolation of hESC-derived CMs from these transgenic/reporter lines were based on positive selection, Anderson et al. implemented a negative selection strategy to deplete undifferentiated, proliferating hESCs from cultures of hESC-derived CMs [46]. Their transgenic hESC line utilized a Herpes simplex thymidine kinase/ganciclovir (HSVtk/GCV) suicide gene system under the control of a constitutive phosphoglycerate kinase promoter. Following administration of the antiviral drug GCV, cells expressing HSVtk phosphorylate GCV, which then incorporates into nascent DNA chains of proliferating cells, causing chain termination and cell death. The increased number of α-actinin-positive cells after GCV treatment led to an almost sevenfold enrichment of CMs. An important caveat of this approach, however, is that other nonproliferating cell types would remain in the culture while proliferating hCMs would be depleted. The culture would still need to undergo a cardiac purification step and, as discussed below, the excluded proliferating hCMs and cardiac progenitors may be of greater benefit for transplantation than fully differentiated, nonproliferating CMs.
As an alternative to genetically modified hESC lines for tracking and isolating hCMs and cardiac progenitors, we have adapted dualfluorescence resonance energy transfer ‘molecular beacon’ technology for transient, real-time detection of gene expression during hESC differentiation (Figure 3). Molecular beacons are single-stranded oligonucleotide probes that have been employed to assay gene expression in vitro, as in real-time PCR, and in vivo using microscopy [47]. These consist of short sequences capable of forming stem–loop structures bearing a fluorescent reporter group at one end and a fluorescent quencher at the opposite end [47]. In the absence of a target sequence, the oligonucleotide self-anneals, forming a stem that brings the reporter and quencher in close proximity, thereby quenching fluorescence. In the presence of a target sequence, the oligonucleotide anneals to the target, separating the reporter and quencher, thereby allowing fluorescence. We have shown that appropriately designed, dual-fluorescence resonance energy transfer molecular beacon pairs can identify the expression of specific mRNAs by microscopy and flow cytometry, and facilitate the collection of specific hESC populations by fluorescence-activated cell sorting, while leaving the hESC genome intact [King F, Liszewski W, Ritner C, Bernstein HS, Pers. Comm.].
Figure 3. Dual-FRET molecular beacon technology for real-time analysis of stem cell differentiation.
Molecular beacons, consisting of short single-stranded oligonucleotides capable of forming stem–loop structures in solution, are designed complementary to adjacent sequences in a mRNA of interest. In the absence of the target mRNA, the stem–loop conformation juxtaposes the fluorescent dye and specific quencher molecule, extinguishing fluorescence. In the presence of an expressed target mRNA, binding of the molecular beacons relieves the quenching effect, allowing the donor dye to be excited by the FACS, emit at the excitation wavelength of the acceptor dye, and the acceptor dye to emit at a wavelength recognized by the sorting filter set of the FACS.
FRET: Fluorescence resonance energy transfer.
Researchers have exploited reporter ESC lines as research tools to identify cardiac progenitor cells with distinct molecular signatures. Wu et al. used a cardiac-specific mESC line expressing Nkx2–5:eGFP to isolate bipotent cardiac progenitors that express Nkx2–5 and the tyrosine kinase receptor, c-Kit [48]. When seeded as single cells and cultured for 13 days, the surviving Nkx2–5+/c-Kit+ single clones differentiated into both CMs and smooth muscles cells. Clonal progeny differentiated in three distinct patterns: beating CMs expressing Actc1 and ventricular myosin light chain, nonbeating cells expressing smooth muscle markers, SM22α and smooth muscle actin (SMA)-α, and a combination of beating CMs and vascular smooth muscle cells, implicating bipotential differentiation from a single cell.
At the same time, two groups have independently identified multipotent cardiovascular progenitors that give rise to all three major cardiovascular lineages. Previous work using a Brachyury:eGFP knock-in mESC line, in which an eGFP minigene was targeted to the Brachyury locus, showed that mEB-derived cells expressing Brachyury, but not fetal liver kinase (Flk)-1 had cardiac potential [49,50]. By following the maturation of this Bry+/Flk-1− cell subpopulation in culture, Kattman et al. discovered a second Flk-1+ population with high CM capacity that emerged following mEB reaggregation [51]. Differentiation of this second Flk-1+ population in methylcellulose followed by expansion in liquid culture resulted in spontaneous contracting colonies. These colonies expressed genes associated with cardiac (Nkx2–5, GATA4, Tbx5, Tbx20, MLC2a), endothelial (Flk-1, VE-cadherin, CD31) and vascular (SMA, calponin) differentiation, suggesting the presence of multilineage cardiovascular progenitors within this latent Bry+/Flk-1+ fraction.
In a separate study, genetic fate mapping and in vivo lineage tracing in Islet (Isl)1:IRES:Cre/R26R double heterozygous mice demonstrated that a population of Isl1+ precursors contributes to the generation of cardiac muscle, pacemaker, endothelial and smooth muscle cells in the heart [52]. To establish a source of cardiac Isl1+ precursors for further investigation, an Isl1:nlacZ knock-in mESC line was engineered in which a nuclear lacZ gene, followed by humanized Renilla GFP, was inserted into the Isl1 locus. Transcription profiling of cultured single cell-derived Isl1+ clones revealed that cells with the Isl1+/Nkx2–5+/Flk-1+ signature could give rise to cardiac, smooth muscle and endothelial cell derivatives under differentiation conditions. These clonal progenitors expressed markers of differentiated endothelial cells (VE-cadherin), CMs (cardiac troponin T) and smooth muscle cells (SM-MHC).
Significantly, both groups identified analogous multipotent cardiac progenitors derived from hESCs (Figure 4). Flow cytometric analysis of hEBs detected three distinct KDR+(Flk-1+)/c-Kit+ populations at day 6 of the three-stage differentiation protocol described earlier: KDRhigh/c-Kit+, KDRlow/c-Kit−, KDR−/c-Kit+ [23]. Of the three, the KDRlow/c-Kit− population contained cardiac progenitors that generated cells expressing markers of endothelial (CD31, CDH5, VE-cadherin, von Willebrand factor), vascular smooth muscle (calponin, SMA, sarcomeric MHC, caldesmon) and cardiac (Nkx2–5, Isl-1, Tbx5, Tbx20, cardiac troponin T, atrial myosin light chain 2) differentiation. To establish clonality, hESC lines expressing GFP or red fluorescent protein (RFP) were employed in methylcellulose colony assays. Mixing of KDRlow/c-Kit− populations isolated from both lines resulted in colonies expressing either GFP or RFP, but not both. Expression analysis of colonies from the mixed GFP/RFP cultures confirmed the presence of cardiac, endothelial and vascular smooth muscle lineages, suggesting that the three cell types arose from a single cell.
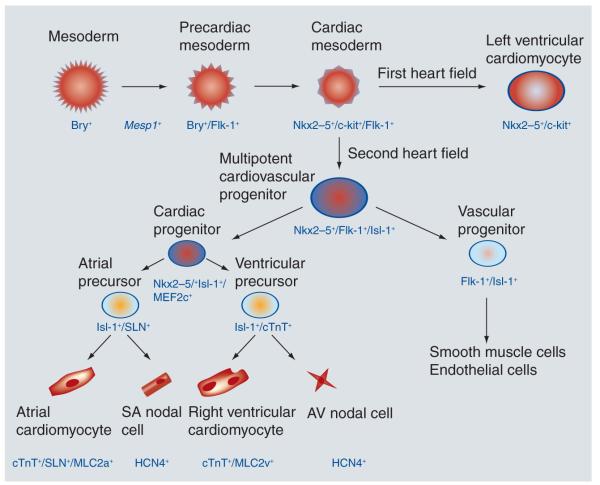
Figure 4. The cardiac lineage tree.
During cardiac myogenesis, the differentiation of progenitor cells into multiple heart cell lineages is under both temporal and spatial control. Cardiac mesoderm (Nkx2–5+/c-kit+/Flk-1+) gives rise to Nkx2–5+/c-kit+ cells that contribute to structures comprising the first heart field, and Nkx2–5+/Flk-1+/Isl-1+ progenitors that contribute to all three major cell lineages of the second heart field: cardiomyocytes, smooth muscle cells and endothelial cells. Cardiac progenitors (Nkx2–5+/Isl-1+/Mef2c+) specifically give rise to precursors that will develop into atrial cardiomyocytes, SA nodal cells, right ventricular cardiomyocytes and AV nodal cells.
AV: Atrioventricular; SA: Sinoatrial.
To track the fate of human Isl1+ cells and their progeny during hESC differentiation, Bu et al. used Isl1:cre hESCs transfected with a pCAG-flox-DsRed reporter plasmid to achieve irreversible DsRed expression in Isl1+ cells. In clonal assays of day 8 hEBs, approximately half of the DsRed+ (i.e., Isl1+) clones that were Nkx2–5+ expressed markers of all three major cardiac lineages: cardiac troponin T (CMs), PECAM1/CD31 (endothelial cells) and smooth muscle troponin (smooth muscle cells) [53]. Interestingly, KDR was not detected in DsRed+ cells from day 8 hEBs, but was present within 7 days after plating on mouse embryonic fibroblasts in the clonal assays, implying that Isl1+/Nkx2–5+/KDR+ cells may represent a more restricted downstream cardiac progenitor.
The specification of multipotent cardiovascular progenitors from early mesoderm has recently been shown to be governed by the basic helix–loop–helix transcription factor, Mesp1. In vivo cell lineage tracing, gene knockout and chimeric studies revealed that Mesp1 was expressed at the onset of gastrulation at E6.5 before that of Flk-1 or Nkx2–5, and gave rise to virtually all cells of the vascular system as well as cells of the myocardium and endocardium [54-57]. The results implicated Mesp1 as the earliest molecular marker of cardiovascular development identified to date. Subsequent work by others employing various Mesp1 mESC lines demonstrated that overexpression of Mesp1 directed the differentiation of mESCs toward cardiovascular lineages [58,59]. Compared with control mEBs, mEBs overexpressing human Mesp1 exhibited a five-fold enhancement in beating foci, and increased expression of Nkx2–5, GATA4, Mef2c, connexion 45 and 43, MLC2v, cardiac troponin I and ANF [60]. Bondue et al. showed that induction of Mesp1 resulted in upregulation of Isl1 expression, and accelerated mESC differentiation into all three main cardiac cell lineages (cardiac, vascular, smooth muscle), suggesting that Mesp1 specifies multipotent cardiovascular progenitors [58]. Lindsley et al. obtained similar findings, and also demonstrated that the activity of Mesp1 was dependent on Wnt [59]. In the hierarchical order of cardiac progenitors, these studies position Mesp1 as an early multipotent mesodermal progenitor that lies upstream of Isl1 and Flk-1.
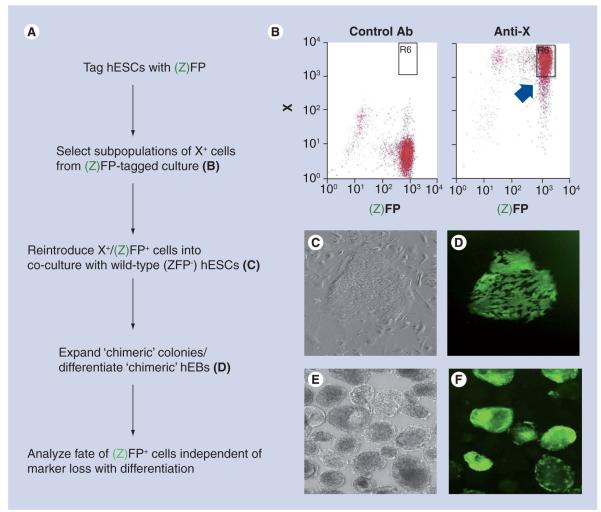
KDR/Flk-1 encodes a VEGF receptor and, as discussed above, has served as a cell surface marker to isolate a pool of cardiac progenitors during hESC differentiation [23]. Early studies of hESC lines demonstrated that subpopulations of hESCs characterized by the expression of specific surface markers exist within undifferentiated hESC cultures [61]. We recently developed a fluorescence-tagged co-culture system with which to track the fate of these subpopulations (Figure 5). Using this co-culture system, undifferentiated hESCs that constitutively expressed GFP from the ubiquitin C promoter were positively and negatively selected for CD133 or CD135 by fluorescence cell sorting. Sorted cells were added to dispersed colonies of undifferentiated, untagged hESCs and allowed to reaggregate to form chimeric colonies. Once reformed, cells were differentiated to determine their fates [62]. Under differentiation conditions, CD135+GFP+ and CD135−GFP+ cells were observed to differentiate into all three embryonic germ layers, suggesting that CD135 expression in undifferentiated hESCs did not track with a specific cell fate. By contrast, CD133+GFP+ cells gave rise almost exclusively to ectoderm as demonstrated by a threefold increase in nestin expression compared with CD135+GFP+ or CD135-GFP+ cells. CD133−GFP+ cells differentiated into endoderm and mesoderm as evidenced by more abundant α-fetoprotein (endodermal) and SMA (mesodermal) expression compared with CD133+GFP+ cells. Teratomas formed from co-cultures of CD133+GFP+ hESCs showed GFP-expressing cells exclusively within tissues of neuroectodermal origin. Subsequent to these studies with undifferentiated, CD133+ hESCs, we have found that 99% of CD133+ cells isolated from hEBs differentiated in culture for 8 days co-express Nkx2–5, suggesting a cardiac fate [Kwan HCK, Bernstein HS, Pers. Comm.]. Together, these experiments both validate the existence of subpopulations of undifferentiated hESCs that have a predetermined fate, and suggest that the relevance of surface marker expression may change with stage of differentiation. These also imply that other cell surface markers may exist with which to identify mesoderm- and CM-fated hESCs.
Figure 5. Fluorescence-tagged co-culture system for mapping stem cell fate.
(A) Overview of the scheme described in the text. (B) Example of fluorescence sorting of tagged cells co-expressing surface marker (X+). (C & D) Example of chimeric hESC colony containing fluorescently tagged, marker-expressing cells and untagged, nonselected cells. (C) Phase contrast image of colony; (D) immunofluorescence image of same colony. (E & F) Example of chimeric embryoid hEB derived from chimeric colony shown in (C & D). (E) Phase contrast image of hEBs; (F) immunofluorescence image of same hEBs.
Ab: Antibody; hEB: Human embryoid body; hESC: Human embryonic stem cell.
Adapted from [62].
Future perspective
Transplantation with hESC-derived CMs in animal models of myocardial injury has yielded promising, albeit modest, results. In nude rats, injection of hESC-derived CMs into uninjured hearts resulted in a 90% stable myocardial engraftment success rate [63]. However, injection of hCMs into rat hearts infarcted by permanent coronary artery ligation yielded only an 18% engraftment success rate due to poor survival of the transplanted cells [15]. This discrepancy prompted the investigators to deliver the CMs in the presence of a prosurvival cocktail targeting multiple cell death pathways [15]. Not only did the use of a prosurvival formulation improve graft survival in infarcted hearts, but the engrafted myocardium demonstrated improved ventricular function 4 weeks after transplantation, compared with controls. Comparable results have been obtained in a number of other studies evaluating the feasibility of transplanting hESC-derived CMs for myocardial repair in rodent models of myocardial infarction [64-69]. Although the methods for generating hCMs and monitoring engraftment, as well as the number of transplanted hCMs, varied between studies, hESC-derived CM transplantation led to measureable benefit.
Despite these encouraging results, challenges remain prior to clinical implementation. The cardiac-specific benefits determined from in vivo engraftment studies appear to be transient. Van Laake et al. reported that while cardiac function was improved 4 weeks after myocardial infarction, the functional benefit was no longer significant at 12 weeks, even after tripling the number of transplanted hCMs in a repeat study [64-66]. These results call into question the utility of fully differentiated hCMs for cardiac repair. The integration of hESC-derived CMs into existing muscle may be hampered by their limited developmental plasticity, whereas cardiac progenitors may retain the plasticity needed to enable extensive engraftment. Furthermore, as recent studies have shown, myocardial progenitors can diversify to become other cell types, including endothelial cells that would contribute to vascularization of the graft, thereby improving survival and integration of the transplanted cells.
The transient improvement seen in rodent models also argues for the use of preclinical animal models with hemodynamics that more closely resemble human physiology. Porcine models provide an opportunity to study the effects of cell transplantation in an animal with more relevant cardiovascular physiology [70,71]. One study used pigs as a large animal model of atrioventricular heart block. Injection of manually dissected, beating hEBs into the left ventricle of pig hearts with atrioventricular block resulted in successful pacing of the heart, manifested by the presence of a new ectopic ventricular rhythm as measured by body surface electrocardiography and electroanatomical mapping [72].
Transplanted hESC-derived cells will also need to evade immune rejection. hESCs appear to have a lower immunostimulatory potential compared with adult cells [73,74]. DNA microarray data of undifferentiated and differentiated hESCs indicate that almost half of the upregulated immunoregulatory genes in hematopoietic cells, lymphoid organs and other tissues are not similarly expressed in hESCs, implying that hESCs are immunologically immature [73,74]. These observations suggest that immuno suppressive regimens for hESC-based therapeutics may not need to be as rigorous as conventional organ transplantation. Nevertheless, transplanted hESC-derived CMs will be susceptible to immune rejection to some degree.
The next decade will usher in further advances in our understanding of the biology of pluripotent stem cells that will bring stem cell therapeutics closer to the clinic. These will likely include the establishment of a comprehensive tree for the human cardiac lineage; profiles of cell surface marker expression that define specific cardiac progenitor pools; nongenetic methods to derive and isolate cardiac progenitors and specialized CM subtypes to high purity and in sufficient quantities; techniques to ensure the absence of non-CM derivatives and undifferentiated hESCs to prevent tumor formation; strategies to circumvent immune rejection; and preclinical large animal models of heart failure for assessing cell engraftment, host immune response and myocardial function in both the short and long term.
Executive summary.
Use of human embryonic stem cells for myocardial repair
Efforts toward designing new molecular and cellular therapies to treat heart failure would address both an important source of human suffering as well as a significant healthcare expense.
Human embryonic stem cells (hESCs) offer an unlimited supply of human cardiomyocytes (hCMs) and cardiac progenitors for cell-based therapy because they divide indefinitely in culture and differentiate into hCMs.
Because spontaneous hCM differentiation is an inefficient process that yields low cell numbers, alternative strategies have been developed to enhance differentiation of hESCs into hCMs.
Chemical enrichment of hESC-derived CMs
Cell-permeable small molecules significantly improve hCM differentiation from hESCs.
Factors known to play a developmental role during embryonic cardiogenesis, including activin A, bone morphogenetic protein 4, VEGF and Dickkopf homolog 1, promote hESC cardiac differentiation.
Effects of mechanical force on CM differentiation from hESCs
Hydrodynamic forces provided by rotary orbital movement can enhance hCM differentiation.
Microenvironmental forces created by micropatterned surfaces containing adherent and nonadherent regions can promote CM differentiation.
Genetic & epigenetic manipulation of hESCs to enhance CM differentiation
hESC differentiation into the cardiac lineage is regulated by specific miRNAs that control Notch signaling.
Ectopic expression of a minimal set of chromatin remodeling and transcription factors Baf60c, GATA4 and Tbx5 is sufficient to generate contracting mouse CMs from mouse embryoid bodies.
Purification of hESC-derived CMs
Percoll gradient centrifugation is a simple way to separate hESC-derived hCMs from other hESC-derived cells.
Owing to the high mitochondrial content in hCMs, fluorescence-activated cell sorting based on mitochondrial content provides a novel way to purify hESC-derived hCMs.
Selection of hESC-derived cardiac progenitors
Reporter hESC lines allow genetic selection of hCMs and human cardiac progenitors based on expression of reporter genes or selectable markers under control of cardiac-specific promoters.
The use of reporter mouse ESC lines has facilitated the identification and selection of early cardiac progenitors with distinct molecular signatures.
Specific surface markers on subpopulations of undifferentiated hESCs may identify hESCs with predetermined mesodermal and cardiac fates.
Acknowledgements
The authors wish to thank members of the Bernstein Laboratory at University of California San Francisco for helpful discussion and communication of unpublished data.
SSY Wong is supported by a National Research Service Award from NHLBI (HL007544). HS Bernstein is supported by funds from the California Institute for Regenerative Medicine (RC1-00104) and NHLBI (HL085377).
Footnotes
Financial & competing interests disclosure The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Lloyd-Jones D, Adams R, Carnethon M, et al. Heart disease and stroke statistics – 2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119(3):480–486. doi: 10.1161/CIRCULATIONAHA.108.191259. [DOI] [PubMed] [Google Scholar]
- 2.Chien KR, Domian IJ, Parker KK. Cardiogenesis and the complex biology of regenerative cardiovascular medicine. Science. 2008;322(5907):1494–1497. doi: 10.1126/science.1163267. [DOI] [PubMed] [Google Scholar]
- 3.Kehat I, Kenyagin-Karsenti D, Snir M, et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J. Clin. Invest. 2001;108(3):407–414. doi: 10.1172/JCI12131. ▪First comprehensive characterization of human embryonic stem cell (hESC)-derived cardiomyocytes (CMs).
- 4.Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ. Res. 2002;91(6):501–508. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- 5.Itskovitz-Eldor J, Schuldiner M, Karsenti D, et al. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol. Med. 2000;6(2):88–95. [PMC free article] [PubMed] [Google Scholar]
- 6.He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ. Res. 2003;93(1):32–39. doi: 10.1161/01.RES.0000080317.92718.99. ▪ First characterization of electrophysiological heterogeneity among cultures of hESC-derived CMs.
- 7.Beltrami CA, Finato N, Rocco M, et al. Structural basis of end-stage failure in ischemic cardiomyopathy in humans. Circulation. 1994;89(1):151–163. doi: 10.1161/01.cir.89.1.151. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda K. Development of regenerative cardiomyocytes from mesenchymal stem cells for cardiovascular tissue engineering. Artif. Organs. 2001;25(3):187–193. doi: 10.1046/j.1525-1594.2001.025003187.x. [DOI] [PubMed] [Google Scholar]
- 9.Graichen R, Xu X, Braam SR, et al. Enhanced cardiomyogenesis of human embryonic stem cells by a small molecular inhibitor of p38 MAPK. Differentiation. 2008;76(4):357–370. doi: 10.1111/j.1432-0436.2007.00236.x. [DOI] [PubMed] [Google Scholar]
- 10.Gaur M, Ritner C, Sievers RE, et al. Timed inhibition of p38MAPK directs accelerated differentiation of human embryonic stem cells into cardiomyocytes. Cytotherapy. 2010 doi: 10.3109/14653249.2010.491821. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mummery CL, van Achterberg TA, van den Eijnden-van Raaij AJ, et al. Visceral-endoderm-like cell lines induce differentiation of murine P19 embryonal carcinoma cells. Differentiation. 1991;46(1):51–60. doi: 10.1111/j.1432-0436.1991.tb00865.x. [DOI] [PubMed] [Google Scholar]
- 12.Mummery C, Ward-van Oostwaard D, Doevendans P, et al. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107(21):2733–2740. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 13.Xu XQ, Graichen R, Soo SY, et al. Chemically defined medium supporting cardiomyocyte differentiation of human embryonic stem cells. Differentiation. 2008;76(9):958–970. doi: 10.1111/j.1432-0436.2008.00284.x. [DOI] [PubMed] [Google Scholar]
- 14.Mummery CL, Feijen A, Moolenaar WH, van den Brink CE, de Laat SW. Establishment of a differentiated mesodermal line from P19 EC cells expressing functional PDGF and EGF receptors. Exp. Cell Res. 1986;165(1):229–242. doi: 10.1016/0014-4827(86)90547-1. [DOI] [PubMed] [Google Scholar]
- 15.Laflamme MA, Chen KY, Naumova AV, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007;25(9):1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 16.Sugi Y, Lough J. Activin-A and FGF-2 mimic the inductive effects of anterior endoderm on terminal cardiac myogenesis in vitro. Dev. Biol. 1995;168(2):567–574. doi: 10.1006/dbio.1995.1102. [DOI] [PubMed] [Google Scholar]
- 17.Smith JC, Price BM, Van Nimmen K, Huylebroeck D. Identification of a potent Xenopus mesoderm-inducing factor as a homologue of activin A. Nature. 1990;345(6277):729–731. doi: 10.1038/345729a0. [DOI] [PubMed] [Google Scholar]
- 18.Nakajima Y, Yamagishi T, Ando K, Nakamura H. Significance of bone morphogenetic protein-4 function in the initial myofibrillogenesis of chick cardiogenesis. Dev. Biol. 2002;245(2):291–303. doi: 10.1006/dbio.2002.0637. [DOI] [PubMed] [Google Scholar]
- 19.Lough J, Barron M, Brogley M, Sugi Y, Bolender DL, Zhu X. Combined BMP-2 and FGF-4, but neither factor alone, induces cardiogenesis in non-precardiac embryonic mesoderm. Dev. Biol. 1996;178(1):198–202. doi: 10.1006/dbio.1996.0211. [DOI] [PubMed] [Google Scholar]
- 20.Ladd AN, Yatskievych TA, Antin PB. Regulation of avian cardiac myogenesis by activin/TGF-β and bone morphogenetic proteins. Dev. Biol. 1998;204(2):407–419. doi: 10.1006/dbio.1998.9094. [DOI] [PubMed] [Google Scholar]
- 21.Barron M, Gao M, Lough J. Requirement for BMP and FGF signaling during cardiogenic induction in non-precardiac mesoderm is specific, transient, and cooperative. Dev. Dyn. 2000;218(2):383–393. doi: 10.1002/(SICI)1097-0177(200006)218:2<383::AID-DVDY11>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 22.Yao S, Chen S, Clark J, et al. Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc. Natl Acad. Sci. USA. 2006;103(18):6907–6912. doi: 10.1073/pnas.0602280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L, Soonpaa MH, Adler ED, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453(7194):524–528. doi: 10.1038/nature06894. ▪ Identification of a human cardiac progenitor derived from hESCs.
- 24.Schneider VA, Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 2001;15(3):304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marvin MJ, Di Rocco G, Gardiner A, Bush SM, Lassar AB. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001;15(3):316–327. doi: 10.1101/gad.855501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sargent CY, Berguig GY, McDevitt TC. Cardiomyogenic differentiation of embryoid bodies is promoted by rotary orbital suspension culture. Tissue Eng. Part A. 2009;15(2):331–342. doi: 10.1089/ten.tea.2008.0145. [DOI] [PubMed] [Google Scholar]
- 27.Sargent CY, Berguig GY, Kinney MA, et al. Hydrodynamic modulation of embryonic stem cell differentiation by rotary orbital suspension culture. Biotechnol. Bioeng. 2010;105(3):611–626. doi: 10.1002/bit.22578. [DOI] [PubMed] [Google Scholar]
- 28.Domian IJ, Chiravuri M, van der Meer P, et al. Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science. 2009;326(5951):426–429. doi: 10.1126/science.1177350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39(3):380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl Acad. Sci. USA. 2005;102(34):12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanellopoulou C, Muljo SA, Kung AL, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat. Genet. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436(7048):214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 36.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ. Res. 2007;100(3):416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 37.Care A, Catalucci D, Felicetti F, et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13(5):613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 38.Ivey KN, Muth A, Arnold J, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2(3):219–229. doi: 10.1016/j.stem.2008.01.016. ▪▪ First demonstration that miRNAs control hESC differentiation.
- 39.Kawamura T, Ono K, Morimoto T, et al. Acetylation of GATA-4 is involved in the differentiation of embryonic stem cells into cardiac myocytes. J. Biol. Chem. 2005;280(20):19682–19688. doi: 10.1074/jbc.M412428200. [DOI] [PubMed] [Google Scholar]
- 40.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459(7247):708–711. doi: 10.1038/nature08039. ▪ Determined a role for chromatin remodeling in directing cardiac differentiation.
- 41.Xu C, Police S, Hassanipour M, Gold JD. Cardiac bodies: a novel culture method for enrichment of cardiomyocytes derived from human embryonic stem cells. Stem Cells Dev. 2006;15(5):631–639. doi: 10.1089/scd.2006.15.631. [DOI] [PubMed] [Google Scholar]
- 42.Hattori F, Chen H, Yamashita H, et al. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat. Methods. 2010;7(1):61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- 43.Huber I, Itzhaki I, Caspi O, et al. Identification and selection of cardiomyocytes during human embryonic stem cell differentiation. FASEB J. 2007;21(10):2551–2563. doi: 10.1096/fj.05-5711com. [DOI] [PubMed] [Google Scholar]
- 44.Xu XQ, Zweigerdt R, Soo SY, et al. Highly enriched cardiomyocytes from human embryonic stem cells. Cytotherapy. 2008;10(4):376–389. doi: 10.1080/14653240802105307. [DOI] [PubMed] [Google Scholar]
- 45.Kita-Matsuo H, Barcova M, Prigozhina N, et al. Lentiviral vectors and protocols for creation of stable hESC lines for fluorescent tracking and drug resistance selection of cardiomyocytes. PLoS ONE. 2009;4(4):e5046. doi: 10.1371/journal.pone.0005046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson D, Self T, Mellor IR, Goh G, Hill SJ, Denning C. Transgenic enrichment of cardiomyocytes from human embryonic stem cells. Mol. Ther. 2007;15(11):2027–2036. doi: 10.1038/sj.mt.6300303. [DOI] [PubMed] [Google Scholar]
- 47.Santangelo P, Nitin N, Bao G. Nanostructured probes for RNA detection in living cells. Ann. Biomed. Eng. 2006;34(1):39–50. doi: 10.1007/s10439-005-9003-6. [DOI] [PubMed] [Google Scholar]
- 48.Wu SM, Fujiwara Y, Cibulsky SM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127(6):1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 49.Kouskoff V, Lacaud G, Schwantz S, Fehling HJ, Keller G. Sequential development of hematopoietic and cardiac mesoderm during embryonic stem cell differentiation. Proc. Natl Acad. Sci. USA. 2005;102(37):13170–13175. doi: 10.1073/pnas.0501672102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fehling HJ, Lacaud G, Kubo A, et al. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130(17):4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- 51.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev. Cell. 2006;11(5):723–732. doi: 10.1016/j.devcel.2006.10.002. ▪ Among the first studies to demonstrate that major cardiac lineages may arise from a common progenitor.
- 52.Moretti A, Caron L, Nakano A, et al. Multipotent embryonic Isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127(6):1151–1165. doi: 10.1016/j.cell.2006.10.029. ▪ Among the first studies to demonstrate that major cardiac lineages may arise from a common progenitor.
- 53.Bu L, Jiang X, Martin-Puig S, et al. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460(7251):113–117. doi: 10.1038/nature08191. ▪ Identification of human cardiac progenitors derived from hESCs.
- 54.Kitajima S, Takagi A, Inoue T, Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127(15):3215–3226. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- 55.Saga Y, Hata N, Kobayashi S, Magnuson T, Seldin MF, Taketo MM. MesP1: a novel basic helix–loop–helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development. 1996;122(9):2769–2778. doi: 10.1242/dev.122.9.2769. [DOI] [PubMed] [Google Scholar]
- 56.Saga Y, Kitajima S, Miyagawa-Tomita S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc. Med. 2000;10(8):345–352. doi: 10.1016/s1050-1738(01)00069-x. [DOI] [PubMed] [Google Scholar]
- 57.Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126(15):3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 58.Bondue A, Lapouge G, Paulissen C, et al. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3(1):69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 59.Lindsley RC, Gill JG, Murphy TL, et al. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 2008;3(1):55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.David R, Brenner C, Stieber J, et al. MesP1 drives vertebrate cardiovascular differentiation through DKK-1-mediated blockade of Wnt-signalling. Nat. Cell Biol. 2008;10(3):338–345. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- 61.Carpenter MK, Rosler ES, Fisk GJ, et al. Properties of four human embryonic stem cell lines maintained in a feeder-free culture system. Dev. Dyn. 2004;229(2):243–258. doi: 10.1002/dvdy.10431. [DOI] [PubMed] [Google Scholar]
- 62.King FW, Ritner C, Liszewski W, et al. Subpopulations of human embryonic stem cells with distinct tissue-specific fates can be selected from pluripotent cultures. Stem Cells Dev. 2009;18(10):1441–1450. doi: 10.1089/scd.2009.0012. ▪▪ Determined that undifferentiated hESC cultures contain stem cell populations with predetermined fates.
- 63.Laflamme MA, Gold J, Xu C, et al. Formation of human myocardium in the rat heart from human embryonic stem cells. Am. J. Pathol. 2005;167(3):663–671. doi: 10.1016/S0002-9440(10)62041-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Laake LW, Passier R, Monshouwer-Kloots J, et al. Human embryonic stem cell-derived cardiomyocytes survive and mature in the mouse heart and transiently improve function after myocardial infarction. Stem Cell Res. 2007;1(1):9–24. doi: 10.1016/j.scr.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 65.van Laake LW, Passier R, Monshouwer-Kloots J, et al. Monitoring of cell therapy and assessment of cardiac function using magnetic resonance imaging in a mouse model of myocardial infarction. Nat. Protoc. 2007;2(10):2551–2567. doi: 10.1038/nprot.2007.371. [DOI] [PubMed] [Google Scholar]
- 66.van Laake LW, Passier R, Doevendans PA, Mummery CL. Human embryonic stem cell-derived cardiomyocytes and cardiac repair in rodents. Circ. Res. 2008;102(9):1008–1010. doi: 10.1161/CIRCRESAHA.108.175505. ▪ Extended the assessment of cardiac function in mice to 12 weeks following hESC-derived CM transplantation, underscoring the importance of long-term analysis and the challenges to using rodents as a preclinical model for myocardial therapy.
- 67.Leor J, Gerecht S, Cohen S, et al. Human embryonic stem cell transplantation to repair the infarcted myocardium. Heart. 2007;93(10):1278–1284. doi: 10.1136/hrt.2006.093161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kofidis T, Lebl DR, Swijnenburg RJ, Greeve JM, Klima U, Robbins RC. Allopurinol/uricase and ibuprofen enhance engraftment of cardiomyocyte-enriched human embryonic stem cells and improve cardiac function following myocardial injury. Eur. J. Cardiothorac. Surg. 2006;29(1):50–55. doi: 10.1016/j.ejcts.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 69.Caspi O, Huber I, Kehat I, et al. Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. J. Am. Coll. Cardiol. 2007;50(19):1884–1893. doi: 10.1016/j.jacc.2007.07.054. [DOI] [PubMed] [Google Scholar]
- 70.Angeli FS, Shapiro M, Amabile N, et al. Left ventricular remodeling after myocardial infarction: characterization of a swine model on β-blocker therapy. Comp. Med. 2009;59(3):272–279. [PMC free article] [PubMed] [Google Scholar]
- 71.Everett TH, 4th, Wilson EE, Foreman S, Olgin JE. Mechanisms of ventricular fibrillation in canine models of congestive heart failure and ischemia assessed by in vivo noncontact mapping. Circulation. 2005;112(11):1532–1541. doi: 10.1161/CIRCULATIONAHA.104.521351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kehat I, Khimovich L, Caspi O, et al. Electromechanical integration of cardiomyocytes derived from human embryonic stem cells. Nat. Biotechnol. 2004;22(10):1282–1289. doi: 10.1038/nbt1014. [DOI] [PubMed] [Google Scholar]
- 73.Drukker M, Katchman H, Katz G, et al. Human embryonic stem cells and their differentiated derivatives are less susceptible to immune rejection than adult cells. Stem Cells. 2006;24(2):221–229. doi: 10.1634/stemcells.2005-0188. [DOI] [PubMed] [Google Scholar]
- 74.Foldes G, Harding SE, Ali NN. Cardiomyocytes from embryonic stem cells: towards human therapy. Expert Opin. Biol. Ther. 2008;8(10):1473–1483. doi: 10.1517/14712598.8.10.1473. [DOI] [PubMed] [Google Scholar]
- 75.Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell. 2008;2(4):320–331. doi: 10.1016/j.stem.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 76.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132(4):661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]