Abstract
Sequencing-based DNA methylation profiling methods are comprehensive and, as accuracy and affordability improve, will increasingly supplant microarrays for genome-scale analyses. Here, four sequencing-based methodologies were applied to biological replicates of human embryonic stem cells to compare their CpG coverage genome-wide and in transposons, resolution, cost, concordance and its relationship with CpG density and genomic context. The two bisulfite methods reached concordance of 82% for CpG methylation levels and 99% for non-CpG cytosine methylation levels. Using binary methylation calls, two enrichment methods were 99% concordant, while regions assessed by all four methods were 97% concordant. To achieve comprehensive methylome coverage while reducing cost, an approach integrating two complementary methods was examined. The integrative methylome profile along with histone methylation, RNA, and SNP profiles derived from the sequence reads allowed genome-wide assessment of allele-specific epigenetic states, identifying most known imprinted regions and new loci with monoallelic epigenetic marks and monoallelic expression.
Keywords: DNA methylation, Sequencing, Bisulfite
DNA methylation plays a vital role in embryonic development, maintenance of pluripotency, X chromosome inactivation, and genomic imprinting through regulation of transcription, chromatin structure and chromosome stability1. DNA methylation occurs at the C5 position of cytosines within CpG dinucleo-tides2–4 and at non-CpG cytosines in plants and embryonic stem cells (ESC) in mammals. 5-Hydroxymethylation of cytosine also occurs in certain human and mouse cells5, 6 and is catalyzed by Tet proteins acting on methylated cytosine. Tet1 has a role in ESC maintenance and inner cell mass cell specification7. Several experimental methods detect methylation but not hydroxymethylation, while others detect both but cannot distinguish them.
Understanding the role of DNA methylation in development and disease requires knowing the distribution of these modifications in the genome. The availability of reference genome assemblies and massively parallel sequencing has led to methods which provide high-resolution genome-wide profiles of 5-methylcytosine8–16. In contrast to arrays, sequencing-based methods interrogate DNA methylation in repetitive sequences, and more readily allow epigenetic states to be assigned to specific alleles. Each method has unique characteristics leaving uncertainty about the optimal method for particular biological questions. DNA methylation maps are being produced by many laboratories worldwide, and their integration forms a basis for emerging international epigenome projects17. Thus, it is critical to determine the precision of each method, and how reliably they can be compared.
Here, we provide a detailed and quantitative comparison of four sequencing-based methods for genome-wide DNA methylation profiling. We focused on two methods that utilize bisulfite conversion (MethylC-seq8 and Reduced Representation Bisulfite Sequencing, or RRBS9), and two methods that utilize enrichment of methylated DNA (Methylated DNA Immunoprecipitation sequencing, or MeDIP-seq10, 11 and Methylated DNA Binding Domain sequencing, or MBD-seq12). We also developed an integrative methodology combining MeDIP-seq to detect methylated CpGs, with Methylation-sensitive Restriction Enzyme sequencing (MRE-seq)13, 14 to detect unmethylated CpGs. Unlike the enrichment methods alone, the integrative method can accurately identify regions of intermediate methylation which, in conjunction with SNP profiling from the sequencing data, allows for genome-wide identification of allele-specific epigenetic states.
RESULTS
Generation of DNA Methylation Profiles from Human Embryonic Stem Cells
Four individual sequencing-based methods and one integrative method were used to generate and compare DNA methylation profiles of three biological replicates of H1 ESCs. MethylC-seq8 involves shotgun sequencing of DNA treated with bisulfite, a chemical which converts unmethylated cytosines but not methylated cytosines to uracil. The second bisulfite-based method, RRBS9, reduces the portion of the genome analyzed through an MspI digestion and fragment size selection. MeDIP-seq10, 11 and MBD-seq12 involve enrichment of methylated regions followed by sequencing. In MeDIP-seq an anti-methylcytosine antibody is used to immunoprecipitate methylated single-stranded DNA fragments. MBD-seq utilizes the MBD2 protein methyl-CpG binding domain to enrich for methylated double-stranded DNA fragments. As a complementary approach for use in conjunction with methylated fragment enrichment methods, unmethylated CpGs are identified by sequencing size selected fragments from parallel DNA digestions with the methyl-sensitive restriction enzymes (MREs) HpaII (C^CGG), Hin6I (G^CGC) and AciI (C^CGC)(MRE-seq)13.
To reliably identify biological variation in methylation among samples from different individuals or biological states, it is important to determine the variation attributable to biological and technical replication. As an initial assessment of DNA methylation concordance among three H1 ESC biological replicates, the methylation status of 27,578 CpGs was assayed on the widely used bisulfite-based Infinium bead-array. The beta values, roughly representing CpG methylation levels, in the replicates were compared by calculating concordance correlation coefficients (CCC). The CCCs were very high, ranging from 0.992 to 0.996 (Supplementary Fig. 1). To assess technical variation, replicate #1 and replicate #2 were run a second time on the Infinium platform (data not shown). 98.9% of the total variation (technical and biological) was technical. Thus, platform comparisons using these replicates should be very informative.
As a second and more genome-wide analysis of variation in methylation calls, RRBS, covering approximately 1.6 million CpGs, was performed on all three biological replicates. The correlation between the biological replicates was high for RRBS (Supplementary Fig. 2) as it was for MeDIP-seq and MRE-seq (Supplementary Fig. 3). These results show that cell passage-related “biological variation” in methylation is present but minimal on a genome-scale. The rare biological variation in methylation levels was confirmed by pyrosequencing of selected loci (Supplementary Fig. 4, Supplementary Table 1).
Several algorithms are available for bisulfite-treated short read mapping, differences in which might alter local read density in a map, and ultimately impact methylation calls. Our assessment of overall concordance between aligners, including Bowtie18, BSMAP19, Pash20, RMAP21 and ZOOM22 applied to a subset of the MethylC-seq data9, indicated that, despite differences in speed and accuracy, aligner choice was unlikely to have a significant impact on the platform comparisons (Supplementary Table 2).
There are several important parameters in choosing an appropriate method for particular experimental goals, including the total number and local context of CpGs interrogated and the amount of sequencing required. To determine the impact of sequencing depth on coverage, we plotted CpG coverage genome-wide (Fig. 1c) and in CpG islands (CGIs) (Fig. 1d) as a function of read coverage threshold for CpGs. For MeDIP-seq and MBD-seq, the CpG coverage does not include CpGs for which a lack of methylation could be inferred from lack of reads (Fig. 1c–d). Thus, because CGIs are predominantly unmethylated, the CpG coverage in CGIs is lower than either RRBS or MethylC-seq. As an indicator of the cost efficiency for each method, we also graphed CpG coverage normalized to a single Gbp of sequence depth in the methylome maps (Supplementary Fig. 5). Enrichment methods had the lowest cost per CpG covered genome-wide, while RRBS had the lowest cost per CpG covered in CGIs. For the enrichment methods we examined the potential effect of CpG density on read coverage. While most of the genome is methylated and CpG poor, a small fraction is unmethylated and CpG rich (i.e. CGI). Consistent with this, MeDIP-seq and MBD-seq enrich primarily for low CpG density regions, along with a small subset of methylated CGIs. In contrast, MRE-seq interrogates higher CpG density regions because they have an abundance of unmethylated recognition sites for these enzymes. Therefore, the coverage of MRE-seq and enrichment methods is notably complementary (Supplementary Fig. 6).
Figure 1. Critical parameters in sequencing-based DNA methylation profiling.
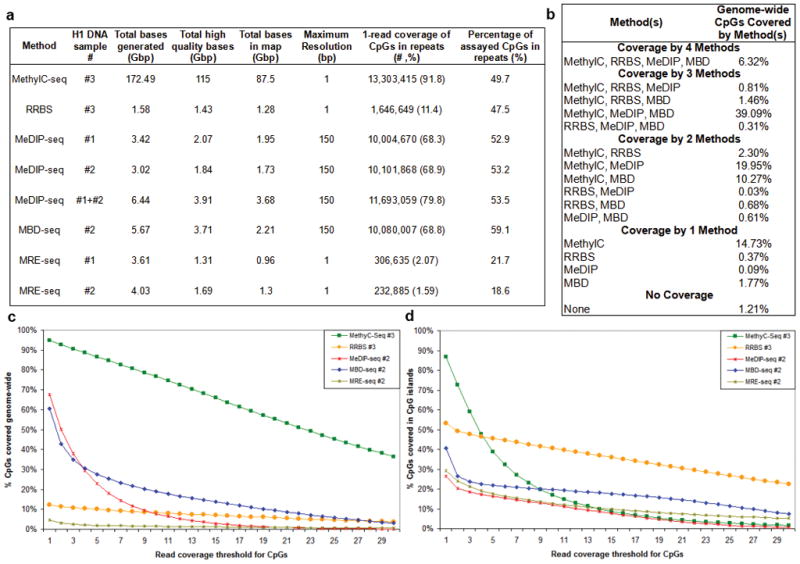
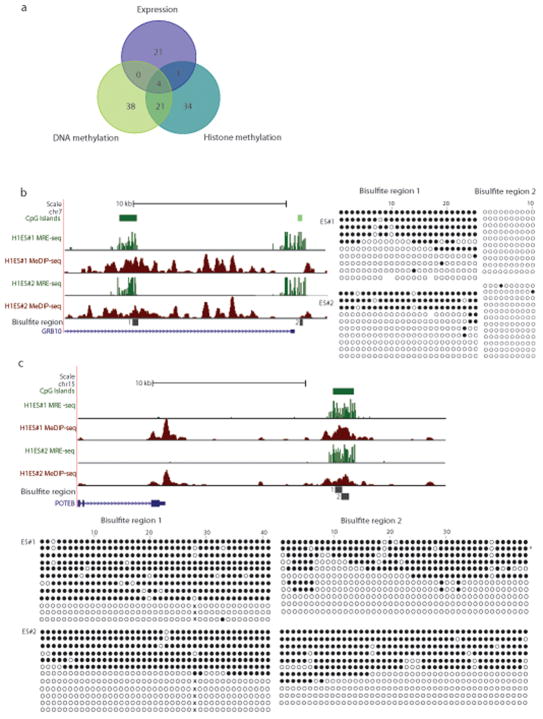
Sequencing statistics and CpG coverage are shown for MethylC-seq, (207 lanes, data analyzed here were from Lister, et al. 8.), RRBS (2 lanes), MeDIP-seq (4 lanes each), MBD-seq (3 lanes), and MRE-seq (3 lanes each). Since the amount of sequence produced per lane is increasing, we also provide “Gbp of sequence” as a measure of the relative cost of each method. The methods differ significantly in total bases generated by the Illumina sequencer, total high quality bases passing Illumina chastity filtering and mapping uniquely, and total bases used for generating methylome maps (high quality bases passing redundancy filters) (a) The H1 replicates assayed and the Gbp of sequence at successive processing stages by each method are shown. The bisulfite-based methods and MRE-seq resolve the methylation status of individual cytosines, whereas the MeDIP-seq and MBD-seq read mappings are extended to 150bp, resulting in a maximum resolution of 150bp. This extension is applied to calculations of CpG coverage but is not applied to the Gbp of sequence at the processing stages. Coverage information is shown for repeats (primarily transposon sequences) genome-wide. While maximum resolution of each method is reported, resolution can be assessed at various levels. As the level of resolution decreases, as a consequence of averaging of methylation scores over a window of larger size for example, imperfect coverage and limited accuracy become less limiting, provided that the average score is not affected by systematic biases in coverage and accuracy. Thus, methylome coverage and accuracy in methylation calls are a function of resolution. (b) The percentage of genome-wide CpGs (28,163,863) covered by multiple, single or no methods are shown. The percentage of CpGs covered genome-wide (c) or in CGI (d) are plotted as a function of read coverage threshold.
A major advantage of the sequencing-based methods over microarrays is their ability to interrogate CpGs in repetitive elements. Approximately 45% of the human genome is derived from transposable elements, a major driving force in the evolution of mammalian gene regulation23, 24, with nearly half of all CpGs falling within these repetitive regions. The extent to which different sequencing-based methods interrogate repeats is therefore of significant interest. In general, genome-wide CpG coverage was proportional to CpG coverage in repeats (Fig. 1a, c). The percent of interrogated CpGs in repeats was similar across all four methods, with MBD-seq capturing the highest fraction of repeat sequences (59.1%). Each of these methods is therefore useful for investigating this important and largely unexplored area. MRE-seq however, only minimally interrogates repeats, consistent with their dense methylation.
Only CpGs interrogated in common can be compared directly. The intersections of CpGs covered by the four methods were therefore determined (Fig. 1b). Overall, MethylC-seq provided the highest CpG coverage at 95% followed by MeDIP-seq at 67% and MBD-seq at 61%. RRBS covered the fewest CpGs genome-wide (12%), which drove the overlap of all methods to 6% of genome-wide CpGs.
For any given method, how deeply to sequence the library is an open question. As the sequencing depth increases, the number of unique reads covering a particular region approaches the total possible reads present in the library for each enriched region. This saturation occurs when further sequencing fails to discover additional regions above background. To understand the extent to which we sampled the regions represented in the RRBS, MeDIP-seq, and MBD-seq libraries, saturation analysis was performed. RRBS approaches but does not reach saturation at the current sequencing depth (Supplementary Fig. 7a). For MeDIP-seq and MBD-seq, saturation was observed when false discovery rate thresholds were applied, but not when unthresholded data were plotted (Supplementary Fig. 7b–c). Saturation was not observed for MRE-seq (Supplementary Fig. 7d–e), although the average restriction site was represented 13 times within each library, indicating that additional reads would mostly re-sample restriction sites already interrogated. Sequencing beyond saturation improves confidence in the observations and increases the CpG coverage, though at greater cost per CpG covered. Thus, sequencing below or up to saturation maximizes the number of samples that can be analyzed, while sequencing beyond saturation maximizes CpG coverage and improves confidence in methylation calls.
Comparison of Bisulfite-based Methods
Several observations from the CpG coverage analysis of MethylC-seq and RRBS are important to consider before assessing their concordance in methylation calls. First, RRBS provides substantial coverage of CpGs in CGIs, but low CpG coverage genome-wide (Fig. 1c–d). In contrast, MethylC-seq offers greater CpG coverage genome-wide. When coverage is normalized to one Gbp of sequence in the methylome map, RRBS shows higher coverage of CpGs in CGIs at all read depths (Supplementary Fig. 5). This difference points to RRBS as the method of choice if CGIs are the main focus of a study. However, at lower but still significant read thresholds, MethylC-seq samples far more CpGs in CGIs than RRBS.
A major advantage of bisulfite-based methods is that they allow quantitative comparisons of methylation levels at single base resolution. For MethylC-seq and RRBS, we calculated and compared the proportion of methylated reads at individual CpGs genome-wide. High concordance was observed using a simple method that makes methylation status calls at different minimum read depths and allows multiple methylation value cutoffs to be examined (Fig. 2a). The difference in methylation proportions (MethylC-seq - RRBS) at a minimum read depth of 5 was calculated for individual CpGs and concordance was declared if the difference did not exceed a given threshold (Fig. 2b). Of the CpGs compared between MethylC-seq and RRBS just 12.75% displayed identical methylation level, or a difference threshold of zero. If the difference threshold is relaxed to 0.1 or 0.25, the concordance increases to 53.85% or 81.82%, respectively. This analysis was also performed at minimum read depths of 2 and 10 (Supplementary Fig. 8a–b) which, for the 0.25 threshold, showed concordance of 80.28% and 83.89%, respectively, demonstrating that read depth has only a modest effect on concordance. We also performed this analysis on MethylC-seq on replicate #3 compared to RRBS on replicate #1 and #2, which showed a similar concordance (79.64% for #3 - #1; 82.95% for #3 - #2) (Supplementary Fig. 8c–f), while the concordance between MethylC-seq and RRBS both on replicate #3, (81.82%), falls between the concordances for different replicates. RRBS on replicate #1 and #2 was also compared (Supplementary Fig. 8g–h) and showed a higher concordance (91.54%) than any of the comparisons between MethylC-seq and RRBS, consistent with their high correlation coefficient (Supplementary Fig. 2). The RRBS and MethylC-seq discordant calls were not attributable to the local CpG density or genomic context of the individual CpGs (Fig. 2c–d). Taken together, these analyses suggest differences between replicates are attributable to technical or stochastic factors as well as modest biological variation.
Figure 2. Comparison of bisulfite-based methods.
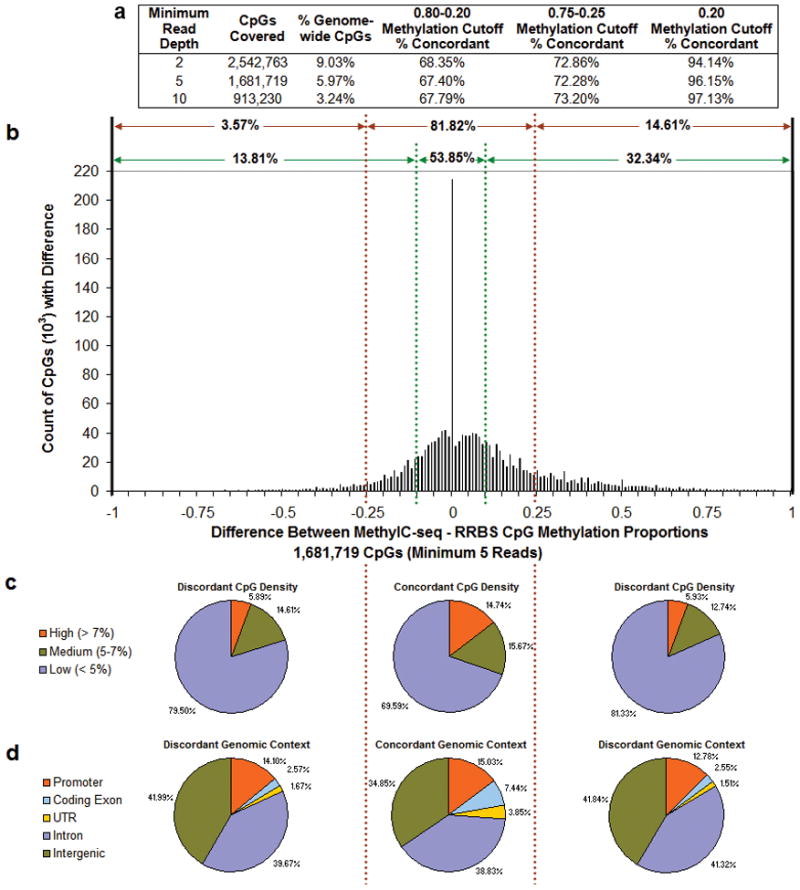
(a) Calls of highly/partially/weakly methylated (0.80-0.20 or 0.75-0.25 cutoff) or highly/weakly methylated (0.20 cutoff) were made for CpGs covered at several minimum read depths by MethylC-seq and by RRBS (both on replicate #3). The number and percent of genome-wide CpGs covered and the percent of concordant calls are shown for each minimum read depth and methylation call cutoff. (b) Differences (MethylC-seq - RRBS) in methylated proportions (methylated reads/(methylated reads + unmethylated reads)) for CpGs with a minimum coverage of 5 reads by both methods. Percentages of concordant and discordant methylation were determined at cutoffs of + and − 0.1 (green dashed lines) and 0.25 (red dashed lines). (c) CpG density in a 400bp window and (d) genomic context of concordant and discordant CpGs at the 0.25 cutoff.
Given the notable presence of non-CpG cytosine methylation in H1 ESC8, we also examined concordance between MethylC-seq and RRBS at CHH and CHG cytosines. Since CHH sites are asymmetric with respect to strand and 98% of CHG sites are hemi-methylated8, reads mapping to each strand were considered separately. When non-CpG cytosines were considered, either with (Supplementary Fig. 9) or without the zero (lack of methylation),methylation proportions (Supplementary Fig. 10), concordance was higher than concordance at CpGs. However, a lower degree of variation at non-CpG sites is expected because of the relatively narrow range of methylation levels for non-CpG sites.
For both CpG (Fig. 2b) and non-CpG cytosine (Supplementary Fig. 9 and 10) methylation, MethylC-seq showed slightly higher methylation proportions than RRBS on the same DNA, as demonstrated by the longer tail on the positive side of the graphs. This trend was also observed in comparisons of MethylC-seq to RRBS performed on replicate #1 and #2 (Supplementary Fig. 8c–f), suggest-ing that technical aspects are driving this difference.
Comparison of Methylated Cytosine Enrichment Methods
Concordance analyses for enrichment methods differ from bisulfite methods in two fundamental ways. First, binary methylation calls are used in enrichment methods, since methylation levels are not easily determined. Second, because of the lack of single CpG resolution inherent in enrichment methods, a windows based approach is used. The windows can include CpGs that are not directly covered by a read. Thus the percent of genome-wide CpGs contained in the compared windows is naturally higher than the percent of individual CpGs that overlap in the coverage comparison (Fig. 1b). We therefore assessed concordance between MeDIP-seq and MBD-seq by comparing binary highly methylated and weakly methylated calls from the average methylation across 1000bp and 200bp windows (see methods). For both window sizes, concordance was >90% at all read depths examined and improved with increasing minimum read depths (Fig. 3a). We confirmed the concordance between MeDIP-seq and MBD-seq at selected loci by bisulfite, PCR, cloning and sequencing (Supplementary Fig. 11, Supplementary Table 3). The substantially higher concordance relative to the bisulfite-based methods is in part related to the inference common to both enrichment methods that neighboring CpGs within a given window have similar methylation levels, and to the binary rather than quantitative methylation calls.
Figure 3. Comparison of methylated DNA enrichment methods.
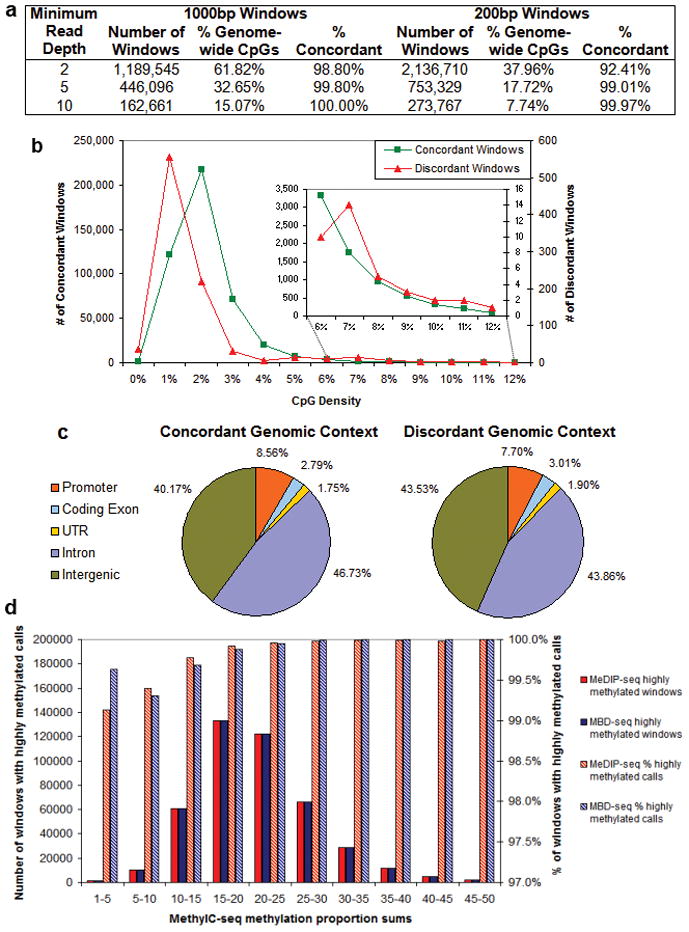
(a) Calls of highly/weakly methylated were made by averaging methylation scores for CpGs covered at varying minimum read depths by MeDIP-seq or MBD-seq in 1000bp and 200bp windows. The number of windows, percent of genome-wide CpGs covered, and the percent of concordant calls are shown for each minimum read depth and window size. For the 1000bp windows with a minimum read depth of 5, the (b) CpG density and (c) genomic context of the concordant and discordant windows are shown. The inset in (b) shows a close-up of the concordance/discordance of CpG densities consistent with CGIs. (d) For the 1000bp windows with a minimum read depth of 5, MethylC-seq methylation proportions for CpGs and non-CpG cytosines covered at a minimum read depth of 5, 444,590 windows, were summed and the windows were binned by the sum. For each of these bins, the number of windows called highly methylated by MeDIP-seq or MBD-seq is shown on the left y-axis and the percent of total windows with calls of highly methylated is shown on the right y-axis. Windows with a MethylC-seq methylation proportion sum greater than 15, representing 83% of all windows, were called highly methylated by MeDIP-seq and MBD-seq in 99.9% of cases. The windows with a methylation proportion sum from 1–15, representing 17% of all windows, were called highly methylated by MeDIP-seq and MBD-seq in at least 99.1% of cases.
When applied in the context of the enrichment methods, the minimum read depths limit the analysis to regions with at least a minimal methylation level. At sufficiently high sequencing depth however, greater confidence can be placed in the lack of methylation inferred from lack of reads. However, at lower sequencing depth, lack of methylation cannot be distinguished from lack of coverage due to the stochastic nature of read coverage. This is an important difference from the bisulfite-based methods which can identify unmethylated regions at a sequencing depth well below saturation.
The 1000bp windows covered at a minimum read depth of 5, representing 99.8% concordance, were examined for potential biases related to CpG density (Fig. 3b) and genomic context (Fig. 3c) on concordance between MeDIP-seq and MBD-seq calls. Concordant and discordant calls were similar in their genomic context, but discordant calls were shifted toward regions of lower CpG density compared to concordant calls. Thus, while these two methods differ in the extent of CpG coverage and read depth at sites covered (Fig. 1), in windows with even minimal coverage by both methods, the concordance is exceptionally high. To further examine the accuracy of the calls, we compared the methylation calls from MeDIP-seq to those from MethylC-seq. For regions with methylation detectable by MethylC-seq, MeDIP-seq and MBD-seq call highly methylated in nearly every case (Fig. 3d).
To examine the reliability of an enrichment based method specifically for inferring weakly methylated regions at different CpG densities, we compared MeDIP-seq to MethylC-seq (Supplementary text and Supplementary Fig. 4). These analyses and limited validation by pyrosequencing suggest that MeDIP-seq allows accurate inferences of lack of methylation/weak methylation in regions of high and medium CpG density, while accuracy is moderately reduced in low CpG density regions. Thus, increasing the sequencing depth of MeDIP-seq or using a complementary methodology targeting unmethylated CpGs may be useful.
Although MeDIP-seq and MBD-seq methylation calls are highly concordant, interesting differences exist between the regions each interrogates, and their sensitivity to detect non-CpG methylation. First, their rate of enrichment differs slightly with respect to local CpG density, with MeDIP-seq enriching more at regions with relatively low CpG density and MBD-seq enriching more at regions with slightly higher CpG density (Supplementary Fig. 6), which is also reflected in their moderate (46.33%) overlap in CpG coverage. Second, the ability of MeDIP-seq or MBD-seq to detect non-CpG methylation could be particularly important for evaluating the methylome of ESC which contains abundant non-CpG methylation9. To address this, read densities were examined in gene bodies with similar CpG methylation levels but different CHG methylation levels as measured by MethylC-seq. MeDIP-seq signal increased with increasing non-CpG cytosine methylation, while MBD-seq did not (Supplementary Fig. 12), suggesting a differential sensitivity in these two enrichment methods. However, the power to distinguish CpG methylation signal from CHG methylation signal is low, since non-CpG cytosine methylation is of-ten embedded within regions with high CpG methylation. As a negative control, regions in the genome that contain no CpGs were examined. MeDIP-seq and MBD-seq had only background level reads, consistent with the non-CpG cytosines being unmethylated in these regions (Supplementary Fig. 13).
Comparison of All Methods
To examine concordance of CpG methylation calls from the two bisulfite-based methods and the two methylation enrichment-based methods, a four-way comparison was performed. This can be viewed as combining the two previous pair-wise comparisons, but with three differences. First, to make the bisul-fite-based methods comparable to the highly/weakly methylated categorization of MeDIP-seq/MBD-seq scores, a binary calling scheme was applied with highly methylated defined as >0.20 methylation and weakly methylated defined as <= 0.20 methylation. When this calling scheme for individual CpGs was applied to bisulfite-based data alone, the concordance between methods was 94.14% for 2 reads, 96.15% for 5 reads and 97.13% for 10 reads. Second, in order to perform the comparison at the same level of resolution, the methylation proportions for individual CpGs in MethylC-seq and RRBS were averaged across windows. Third, in order to compare the bisulfite-based methods to the enrichment based methods without inferring an unmethylated state from complete absence of reads in enrichment methods, the comparison excluded regions lacking reads.
Methylation calls were made for 1000bp windows where all of the methods had at least one CpG covered by a minimum of 5 or 10 reads allowing for comparison of 199,438 or 87,363 windows respectively. Of all the windows covered by a minimum of 5 reads, 2.45% completely encompassed CGIs and 5.5% overlapped with CGIs. The four-way comparison revealed a high degree of concordance of methylation calls among all methods (Fig. 4a–b and Supplementary Table 4). To investigate the effect of applying different highly/weakly methylated cutoffs to MethylC-seq and RRBS, we performed the 4-way comparison at several cutoffs (Supplementary Fig. 14). Concordance remained above 90% up to a highly/weakly methylated cutoff of 0.55, suggesting the concordance results we report are applicable to a wide range of methylation call cutoffs. This result is congruent with the known partitioning of the genome into methylated and unmethylated zones.
Figure 4. Comparison of all methods.
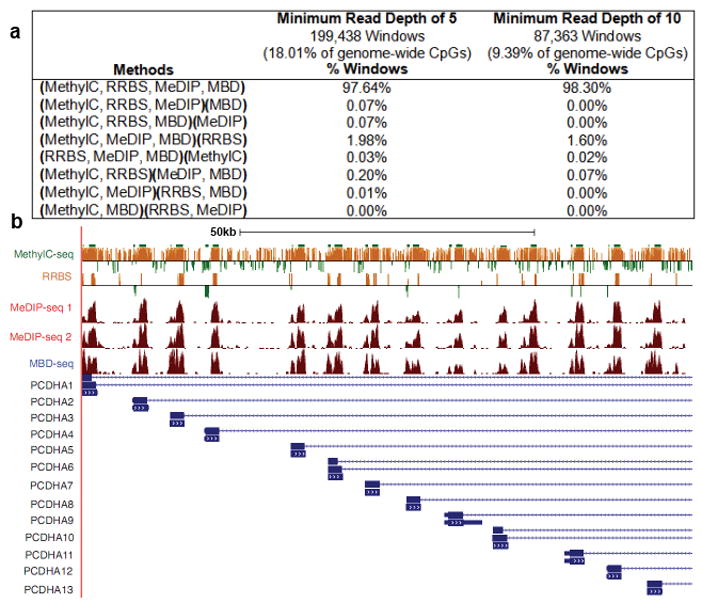
(a) The table shows the percentage of 1000bp windows with concordant and discordant MethylC-seq (replicate #3), RRBS (replicate #3), MeDIP-seq (replicate #2) and MBD-seq (replicate #2) calls at minimum read depths of 5 and 10. Methods making the same call are grouped together in parentheses. Calls were made for MethylC-seq and RRBS by averaging the methylation proportion of CpGs within the window that were covered at the minimum read depth and applying a highly/weakly methylated cutoff of 0.2. Calls were made for MeDIP-seq and MBD-seq by averaging the methylation score of CpGs within the window that were covered at the minimum read depth. (b) Genome browser view of the 100kb CpG rich Protocadherin alpha cluster (PCDHA) exemplifying the significant concordance in methylation status seen on a genome-wide level. For Methyl-seq and RRBS, the Y- axis displays methylation scores of individual CpGs. Scores range between -500 (unmethylated) and 500 (methylated), and the zero line is equivalent to 50% methylated. Negative scores are displayed as green bars and positive scores are displayed as orange bars. For MeDIP-seq 1, MeDIP-seq 2 and MBD-seq, the Y-axis indicates extended read density. Browsable genome-wide views of these datasets are available at http://www.genboree.org and http://genome.ucsc.edu/.
Since the limited coverage by RRBS constrained the number of windows that could be compared, a three-way comparison excluding RRBS was also performed. This allowed for the comparison of 444,494 1000bp windows, or 32% of CpGs genome wide compared to 18% in the 4-way comparison, which showed a three-way concordance of 99.69%. Using different minimum read depth and window sizes had little effect on concordance (Supplementary Table 5a and 5b).
To further evaluate the performance of the four methods, we compared them individually to the widely used Infinium bead-array. For the bisulfite-based methods, the differences in methylation for individual CpGs compared to beta values from the array assaying replicate #3 were calculated. At a difference threshold of 0.25, high concordance was observed between the array and MethylC-seq (96.41%; 20,885 CpGs) and between the array and RRBS (97.31%; 5,475 CpGs) (Supplementary Fig. 15). For the enrichment-based methods, the average methylation score was calculated for CpGs covered by a minimum of 5 reads in 200bp windows centered on CpGs assayed by the array, and used to make the binary methylation call. For the array assaying replicate #2, highly methylated was defined as >0.20 beta value and weakly methylated defined as <= 0.20 beta value. Both MeDIP-seq (96.19%; 4960 windows) and MBD-seq (90.80%; 4163 windows) calls showed high concordance with the array. This high degree of agreement between very different methods further supports the validity of comparing methylation profiles across platforms.
Integrative Method
To increase DNA methylome coverage while maintaining modest sequencing requirements, MeDIP-seq was integrated with MRE-seq13. The methylation scores from MRE-seq were inversely correlated with MeDIP-seq scores (Fig. 5a). The two methods combined assessed the DNA methylation status at 22 million CpGs (Fig. 5b). In regions where MRE-seq scores were high and MeDIP-seq scores were low, the MRE-seq reads corroborate the lack of methylation inferred from the absence of MeDIP-seq reads.
Figure 5. Integrative method increases methylome coverage and enables identification of a DMR.
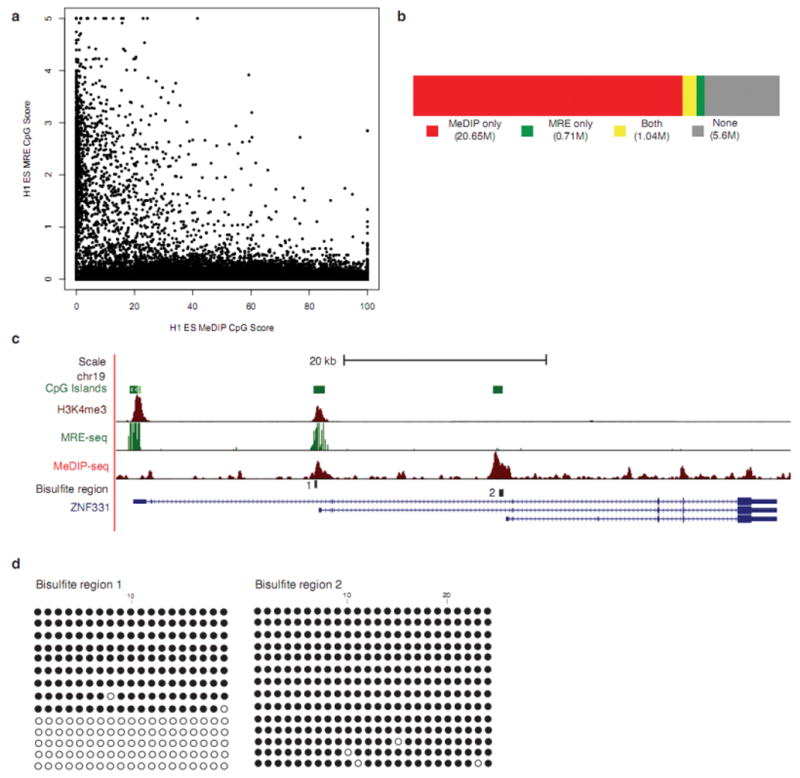
(a) MRE-seq involves parallel digests with methylation sensitive restriction enzymes (HpaII, AciI, and Hin6I), selection of cut fragments of approximately 50bp–300bp, pooling the digests, library construction, and sequencing. For every 600bp window along chromosome 21, MeDIP-seq scores were plotted against MRE-seq scores. The plot depicts the inverse relationship between MRE-seq and Me-DIP-seq signals. (b) Coverage of CpGs in the human genome by MeDIP-seq alone (red), MRE-seq alone (green), both (yellow), or neither method (no fill). Sequence from replicate #1 and #2 were used in these calculations (c) UCSC Genome Browser view of ZNF331in H1 ESC, showing overlap of Me-DIP-seq, MRE-seq and H3K4me3 (from ChIP-seq) signals at bisulfite region 1 and only MeDIP-seq signal at bisulfite region 2. (d) Clonal bisulfite sequencing results for specified regions in ESC from replicate #1. A filled circle represents a methylated CpG and an open circle indicates an unmethylated CpG.
Interestingly, there are a small but significant number of CGIs with overlapping MeDIP-seq and MRE-seq signals (Supplementary Table 6), indicating an intermediate methylation level. We tested two regions from one locus, ZNF331, by clonal bisulfite sequencing (Fig. 5c–d, Supplementary Table 7). Region 1 of ZNF331 showed overlap of signals from MeDIP-seq and MRE-seq, with bisulfite sequencing confirming intermediate and potentially monoallelic methylation. In contrast, region 2 exhibited MeDIP-seq signal only, and bisulfite sequencing confirmed nearly complete methylation. Interestingly, ZNF331 exhibits monoallelic expression in CEPH pedigrees25, 26, and allelic skewing in DNA methylation arrays27, supporting a provisional status of ZNF331 as a novel imprinted gene. Histone H3 lysine 4 trimethylation (H3K4me3), a mark enriched at promoters, overlapped with region 1 but not region 2 (Fig. 5c). A third CGI at the 5’ end of ZNF331 was fully unmethylated, and had an even stronger H3K4me3 peak. Thus, our integrative approach identified a differentially methylated region (DMR) in ZNF331 that may be a DNA methylation regulated promoter for one of the ZNF331 transcripts.
The analysis of ZNF331 suggested the possibility of using MeDIP-seq and MRE-seq to generate a list of candidate DMRs genome-wide (Supplementary Tables 6–7). Ultimately this could define all regions with an intermediate methylation level, encompassing DMRs of the human imprintome and sites of non-imprinted allelic epigenetic regulation. Consistently, our candidate list includes 16 of 19 DMRs of imprinted genes, including BLCAP, GRB10, H19, INPP5F, KCNQ1, MEST, SGCE, SNRPN, ZIM2, GNAS, GNASAS, DIRAS3, DLK1, NDN, PLAGL1, and TP73. Two of the known DMRs, in PEG3 and MEG3, appeared mostly methylated, potentially representing loss of imprint marks28. One of the 19 known DMRs (for NAP1L5) is not within a CGI but did in fact exhibit intermediate methylation (Supplementary Fig. 16). Thus, extension of this analysis to include CpG rich regions that are not strictly CGIs will be useful. The data indicate intermediate DNA methylation states that characterize DMRs within known imprinted regions and others are readily identifiable using an integrative approach.
Intersections of Monoallelic DNA Methylation, Histone Methylation, and Gene Expression
Sequencing-based methods present a unique opportunity to assign epigenetic marks and gene transcripts to specific alleles. We explored this possibility in the ESCs by identifying SNPs within sequence reads, focusing on the top 1000 CGI loci with extensive overlap between MRE-seq and MeDIP-seq signals (Fig. 6a, Supplementary Table 8–9). Of the 1000 loci examined, 203 contained an informative SNP and 63 of these exhibited monoallelic DNA methylation (Fig. 6a). The remaining 140 of the 203 loci with an informative SNP represent intermediate methylation states that may reflect heterogeneity in methylation across the cell population. In total, 119 of the 1000 loci exhibited evidence of monoallelic epigenetic modification and/or expression. Four DMRs were identified that were monoallelic in DNA methylation and histone methylation, and were associated with a gene exhibiting monoallelic expression (Supplementary Fig. 17). Strong corroborating evidence for monoallelic DNA methylation was obtained from similar analyses of the MethylC-seq data (Supplementary Fig. 18). These results demonstrate the excellent capabilities of sequencing-based epigenomic and transcriptome assays for identifying genes exhibiting monoallelic epigenetic marks and monoallelic expression.
Figure 6. Allelic DNA methylation, histone methylation, and gene expression in embryonic stem cells.
(a) Venn diagram summarizing the number of loci exhibiting monoallelic DNA methylation, histone methylation or monoallelic expression, and their overlap. Allelic methylation or expression was determined by comparison of genetic variation between pairs of assays. To further evaluate the top 1000 loci (average size of 2.9kb and encompassing a CGI) with potential allelic DNA methylation, the following pairs of assays were used: MRE-Seq and MeDIP-Seq for allelic DNA methylation within the loci, MethylC-seq and expression data for monoallelic expression of genes associated (+/−50kb) with the loci, MethylC-seq and histone modifications H3K4me3 and H3K9me3 for monoallelic histone methylation within 1kb from the loci. Evidence of genomic variation was found for 119 of 1000 loci; 4 loci in 3 genomic locations provided confirmation of monoallelic status from all three pairs of assays; an additional 26 loci were confirmed by two pairs of assays. (b–c) Validation of known and novel DMRs identified from MeDIP-seq and MRE-seq. DMRs are presented in a UCSC Genome Browser window with MeDIP-seq and MRE-seq signals in human H1 ESC, along with bisulfite sequencing results. The results from the biological replicates (#1 and #2) were very similar. (b) Imprinted gene GRB10 including a known DMR (Bisulfite region 1) and an upstream unmethylated CGI (Bisulfite region 2). (c) Novel DMR upstream of POTEB which exhibits allele specific DNA methylation. Open circle indicates an unmethy-lated CpG site. Filled circle represents a methylated CpG site. “x” indicates absence of a CpG site due to a heterozygous SNP which destroyed the 28th CpG. All clones without the CpG were unmethylated, while all the clones containing the CpG were methylated. Furthermore, the alleles could be distinguished in the sequence reads from MeDIP-seq (G, 9 of 9 reads) and MRE-seq (A, 30 of 30 reads).
To further assess the accuracy of methylation status predictions, 8 regions (total of 17 non-overlapping PCR products) which exhibited apparent monoallelic methylation from the MeDIP-seq and MRE-seq SNP analyses (Fig. 6a and Supplementary Table 8) were selected for clonal bisulfite sequencing. Adjacent CGI loci containing only MRE-seq reads were confirmed to be largely unmethylated (Fig. 6b), while loci containing only MeDIP-seq reads were heavily methylated (Supplementary Table 7). Individual bisulfite clones from two known imprinted genes INPP5F and GRB10 were either methylated or unmethylated at nearly all CpGs (Fig. 6b and Supplementary Table 7). GRB10 exhibited DNA methylation consistent with an isoform-specific imprint mark, as previously reported29. Seven (BCL8, FRG1, ZNF331, IAHI, MEFV, POTEB, ZFP3) of the eight putative DMRs showed evidence of differential methylation (Fig. 6c and Supplementary Table 7). Bisulfite analysis of a DMR upstream of POTEB at 15q11.2 provided direct evidence for allele-specific methylation (Fig. 6c, lower panel). The H3K9me3 signal at this locus is also monoallelic, as two nucleotides identified as heterozygous from the MethylC-seq reads both showed only a single allele in the H3K9me3 sequence reads (chr15:19346665, T in 4 of 4 reads; and chr15:19348112, C in 13 of 14 reads). In the 150 kb proximal to POTEB, three additional CGIs exhibit intermediate methylation levels, including one near the non-coding RNA, CXADRP2, and one encompassing the 5’ end of BCL8. The allelic pattern of DNA methylation of BCL8 was confirmed by bisulfite sequencing (Supplementary Table 7).
DISCUSSION
A quantitative comparison of four sequencing-based DNA methylation methods revealed that all four methods yield largely comparable methylation calls, but differ in CpG coverage, resolution, quantitative accuracy, efficiency and cost. The greater coverage provided by MethylC-seq comes at more than a 50 fold increase in cost compared to RRBS, MeDIP-seq and MBD-seq.
The methods also differ in their ability to detect methylation at non-CpG cytosines and to discriminate it from CpG methylation. However, the high degree of concordance, approaching 100% between MeDIP-seq and MBD-seq, suggests this differential ability to detect non-CpG methylation does not have a significant impact on the relative methylation levels within 1000 bp windows. This observation may be related to the low levels of methylation at non-CpG sites, and their presence in regions with high CpG methylation.
Our finding that MeDIP-seq enriches for regions with lower CpG density compared to MBD-seq is seemingly in contrast to the finding by Li et al.30 that MeDIP-seq was more sensitive to regions of high CpG density than MBD-seq. However, they also show that increasing eluent salt concentrations in MBD-seq enriches for increasingly higher CpG densities. Our comparison between MeDIP-seq and MBD-seq used a salt concentration of 1M compared to 700mM used by Li et al, which could account for the differences.
Variation in DNA methylation is a topic of wide interest. Variation may be between individuals, cell and tissue types, or within one cell type over time. Our biological replicates displayed variation that was similar in magnitude to variation from limited technical replicates, suggesting the concordance estimates may be marginally higher than what we report. Thus, to identify potentially rare variation in methylation between biological samples, the magnitude of technical variation should be considered.
There are numerous opportunities to increase methylome coverage. First, for RRBS or MRE-seq for example, selecting additional enzymes, increasing the size range of selected fragments, and increasing sequencing depth could dramatically increase CpG coverage. Second, increasing read length or employing paired-end sequencing could also positively impact each method. Third, integrative approaches could include MeDIP-seq or MBD-seq coupled with MRE-seq or RRBS, particularly for direct rather than inferred calling of unmethylated CpGs within high CpG density regions. Versatile methods such as Bisulfite Padlock Probes allow more targeted profiling, and could also complement the enrichment methods14.
Sequencing-based methods are unique in that they allow assessment of the methylation status of repetitive elements which encompass nearly half of all CpGs in the methylome. The epigenetic status of this entire genomic compartment has been inaccessible to microarrays, but is a critical component of epigenetic gene regulation, as many of the sequences have regulatory function 23, 31. Furthermore, the labile DNA methylation status of a particular transposon in the agouti locus influences phenotype in mice, including susceptibility to diabetes and cancer32, 33. These and other studies indicate that there is a great deal to be learned about the epigenetic regulation of these abundant but enigmatic elements.
Sequencing-based methylation analysis methods are also unique in that the sequence reads themselves can be used to construct a partial map of genetic variation, including common and rare variants. The comprehensiveness of the genetic map is a function of read coverage and whether reads contain 3 nucleotides (bisulfite methods) or 4 nucleotides (enrichment methods). The sites of genetic variation enable local epigenetic states to be associated with specific alleles. SNP microarrays have been similarly deployed for allelic DNA methylation analysis, but the detection of variants is confined to those present on the microarray34. Our combined epigenomic-genomic analyses identified all CGI with intermediate methylation states in H1 ESCs, many of which were confirmed as monoallelic. This represents an initial step towards characterizing the human imprintome and genome-wide monoallelic epigenetic states, a goal of basic biological and clinical importance in epigenomic research.
METHODS
Embryonic Stem Cells
H1 cells were grown in mTeSR1 medium35 on Matrigel (BD Biosciences, San Jose, California) for 10 passages on 10cm2 plates and harvested at passage 27. Cells were harvested by scraping prior to snap freezing for DNA isolation. Cells were also harvested from passage 30 and 32, and divided for isolation of DNA, RNA and chromatin.
Illumina Infinium Methylation Assay
Five hundred ng genomic DNA were used per sample for the Infinium methylation assay (Illumina, San Diego, CA), which measures methylation at 27,578 CpGs, with approximately 2 probes per gene (14,475 RefSeq genes). Bisulfite conversion was performed with the EZ DNA methylation kit (Zymo Research, Orange, CA) and each sample was eluted in 12 μl water. Amplification and hybridization to the Illumina HumanMethylation27 BeadChip were carried out according to manufacturer’s instructions at the UCSF Genomics Core Facility. Beta values, representing quantitative measurements of DNA methylation at individual CpGs, were generated with Illumina GenomeS-tudio software. Beta values were normalized to background and filtered to remove those with low signal intensity, and the filtered data were used for all subsequent analysis.
Shotgun bisulfite sequencing (MethylC-seq)
As described in Lister et al.8.
Reduced Representation Bisulfite Sequencing (RRBS)
RRBS analysis was performed as described previously36, 37, using approximately 30 ng of H1-derived DNA as input. The steps of the experimental protocol were as follows: (i) DNA digestion using the MspI restriction enzyme, which cuts DNA at its recognition site (CCGG) independent of the CpG methylation status. (ii) End repair and ligation of adapters for Illumina sequencing. (iii) Gel-based selection of DNA fragment sizes ranging from 40bp to 220bp. (iv) Two successive rounds of bisulfite treatment, after which we observed 98.4% converted cytosines outside of CpGs. Due to the presence of non-CpG methylation in ESC, this value is an underestimate of the actual bisulfite conversion rate. (v) PCR amplification of the bisulfite-converted library and sequencing on the Illumina Genome Analyzer II according to the manufacturer’s protocol.
A total of 2 lanes were sequenced, and the data were processed using Illumina’s standard pipeline for image analysis and base calling. The alignment was performed using custom software developed at the Broad Institute9. The non-RepeatMasked reference sequence is generated by size-selecting from an in-silico digest with the MspI restriction enzyme, and prior to the alignment all Cs in the reference sequence and in the aligned reads are converted into Ts. The alignment itself uses a straightforward seed-and-extension algorithm, identifying all perfect 12bp alignments and extending without gaps from either end of the seed. The best alignment is kept only in cases where the second-best alignment has at least three more mismatches, while all reads that match multiple times are discarded. The DNA methylation level of a specific CpG is calculated as the number of C-to-C matches between the unconverted reference sequence and the aligned read sequence divided by the sum of number of C-to-C matches and C-to-T mismatches.
Methylated DNA Binding Protein sequencing (MBD-seq)
3 μg of gDNA isolated as described above was sheared to ~300 bp using the Covaris E210 sonicator (Covaris Inc. Woburn, MA) and size separated by PAGE (8%). The 200–400bp DNA fraction was excised, eluted overnight at 4°C in 200μl of elution buffer (5:1, LoTE buffer (3 mM Tris-HCl, pH 7.5, 0.2 mM EDTA)-7.5 M ammonium acetate) and purified using a QIAquick purification kit (Qiagen, Mississauga, ON). The size selected DNA was end-repaired, A-tailed, and ligated to 2.5mMol of “paired-end” adapters (IDT Inc.) following the manufactures recommend protocol (Ilumina Inc.). The resulting product was purified on a Qiaquick MinElute column (Qiagen, Mississauga, Ontario) and assessed and quantified using an Agilent DNA 1000 series II assay and Qubit fluorometer (Invitrogen, Carlsbad, CA) respectively. 100ng of pre-adapted, size selected product was subjected to immunoprecipitation using the MethylMiner Methylated DNA Enrichment Kit (Invitrogen) following the manufactures recommended protocol. The bound fraction was eluted at 600mM, 1M and 2M NaCl and concentrated by the addition of 1μl (20ug/ul) mussel glycogen, 1/10th v/v 3M sodium acetate (pH5.2) and 2× v/v 100% ethanol. Samples were incubated at −80°C for 2 hours and subsequently centrifuged for 15 minutes at 14,000rpm at 4°C. Pellets were washed with 500μl cold 70% ethanol 2 times with 5 minute centrifugation at 14,000rpm at 4°C between washes and resus-pended in 60μl nuclease-free water. Following purification eluted products were subjected to PCR using Illumina paired-end adapters (Illumina Inc.) with 15 cycles of PCR amplification. PCR products were purified on Qiaquick MinElute columns (Qiagen, Mississauga, Ontario) and assessed and quantified using an Agilent DNA 1000 series II assay and size separated by PAGE (8%). The 320–520bp DNA fraction was excised and purified as described above. The products were assessed and quantified using an Agilent DNA 1000 series II assay and Qubit fluorometer (Invitrogen, Carlsbad, CA) respectively. A 1μl aliquot of each library was used as template in 2 independent PCR reactions to confirm enrichment for methylated (SNRPN promoter) and de-enrichment for unmethylated (CpG-less sequence on Chr15)(see Supplementary Materials for primer sequences). Cycling was 95° C for 30 s, 55° C for 30 s, and 72° C for 30 s with 30 cycles. PCR products were visualized by 1.8% agarose gel electrophoresis. Each library was diluted to 8nM for sequencing on an Illumina Genome Analyzer following the manufactures recommended protocol.
Methylated DNA Immunoprecipitation Sequencing (MeDIP-seq)
Two to five μg DNA isolated as described above was sonicated to ~100–500 bp with a Bioruptor sonicator (Diagenode). Sonicated DNA was end-repaired, A-tailed, and ligated to adapters following the standard Illumina protocol. After aga-rose size-selection to remove unligated adapters, adapter-ligated DNA was used for each immunopre-cipitation using a mouse monoclonal anti-methylcytidine antibody (1 mg/ml, Eurogentec, catalog # BI-MECY-0100). DNA was heat denatured at 95° C for 10 minutes, rapidly cooled on ice, and immunopre-cipitated with 1 μl primary antibody per microgram of DNA overnight at 4° C with rocking agitation in 500 μl IP buffer (10 mM sodium phosphate buffer, pH 7.0, 140 mM NaCl, 0.05% Triton X-100). To recover the immunoabsorbed DNA fragments, 1 μl of rabbit anti-mouse IgG secondary antibody (2.5 mg/ml, Jackson Immunoresearch) and 100 μl Protein A/G beads (Pierce Biotechnology) were added and incubated for an additional 2 hr at 4° C with agitation. After immunoprecipitation a total of 6 IP washes were performed with ice cold IP buffer. A nonspecific mouse IgG IP (Jackson Immunoresearch) was performed in parallel to methyl DNA IP as a negative control. Washed beads were resuspended in TE with 0.25% SDS and 0.25 mg/ml proteinase K for 2 hrs at 55° C and then allowed to cool to room temperature. MeDIP and supernatant DNA were purified using Qiagen MinElute columns and eluted in 16 μl EB (Qiagen, USA). Fifteen cycles of PCR were performed on 5 μl of the immunoprecipitated DNA using the single end Illumina PCR primers. The resulting reactions are purified over Qiagen MinElute columns, after which a final size selection (220–420 bp) was performed by electrophoresis in 2% agarose. Libraries were QC’d by spectrophotometry and Agilent DNA Bioanalyzer analysis, which indicated an average fragment size of 150bp. An aliquot of each library was diluted in EB to 5 ng/μl and 1 μl used as template in 4 independent PCR reactions to confirm enrichment for methylated and de-enrichment for unmethylated sequences, compared to 5 ng of input (sonicated DNA). Two positive controls (SNRPN and MAGEA1 promoters) and 2 negative controls (a CpG-less sequence on Chr15 and GAPDH promoter) were amplified (see Supplementary Materials for primer sequences). Cycling was 95° C for 30 s, 58° C for 30 s, 72° C for 30 s with 30 cycles. PCR products were visualized by 1.8% agarose gel electrophoresis.
Calculating MeDIP-seq and MBD-seq scores for single CpGs
MeDIP-seq and MBD-seq reads were mapped to the non-RepeatMasked human genome assembly (hg18) with MAQ. An algorithm was developed to calculate methylation scores for individual CpGs based on MeDIP-seq or MBD-seq data. Each uniquely mapped, non-redundant sequence read was extended to 150bp long, representing individual DNA fragments pulled down in the methylation enrichment experiment. The algorithm makes two assumptions: first, for a given fragment, this fragment is assigned to a CpG site that is covered by this fragment, and the probability of assigning it to a particular CpG, when there is more than 1 CpG is proportional to the level of methylation of the CpG site; the weighted sum of the probability of all CpGs covered by this fragment is always 1. Second, for a given CpG site, the number of fragments assigned to it is proportional to the level of methylation of this CpG site. The algorithm initiates by assigning a score of 1 to all CpGs, and then it iterates through two steps. In the first step, fragments are assigned to CpGs based on their scores. In the first round, since all CpGs have the same score of 1, an equal fraction of a fragment is assigned to each CpG that the fragment covers, and this is done for all fragments. In the second step, all the fractions of reads each CpG received in step 1 are added up, and this weighted sum is used as a methylation score for this CpG site. Then, the first step is repeated; only now individual CpGs may have a different prior for assigning reads. A fraction of a fragment is now assigned to CpGs that fragment covers based on methylation scores of the CpGs, i.e., the fraction assigned to each CpG is proportional to its methylation score. These updated fragment counts are summed again in step 2 and used as methylation score for individual CpGs. The algorithm iterates through these two steps until the methylation scores converge. These scores are in essence CpG density normalized read density.
Methylation sensitive restriction enzyme sequencing (MRE-seq)
Three parallel digests were performed (HpaII, AciI, and Hin6I; Fermentas), each with 1 μg of DNA. Five units of enzyme per microgram DNA were added and incubated at 37° C in Fermentas “Tango” buffer for 3 hrs. A second dose of enzyme was added (5 units of enzyme per microgram DNA) and the DNA was incubated for an additional 3 hrs. Digested DNA was precipitated with sodium acetate and ethanol, and 500 ng of each digest were combined into one tube. Combined DNA was size-selected by electrophoresis on a 1% agarose TBE gel. A 100–300 bp gel slice was excised using a sterile scalpel and gel-purified using Qiagen Qiaquick columns, eluting in 30 μl of Qiagen EB buffer. Library construction was performed using the Illumina Genomic DNA Sample Kit (Illumina Inc., USA) with single end adapters, following the manufacturer’s instructions with the following changes. For the end repair reaction, T4 DNA polymerase and T4 poly-nucleotide kinase were excluded and the Klenow DNA polymerase was diluted 1:5 in water and 1 μl used per reaction. For single end oligo adapter ligation, adapters were diluted 1:10 in water and 1 μl used per reaction. After the second size selection, DNA was eluted in 36 μl EB buffer using Qiagen Qiaquick columns, and 13 μl used as template for PCR, using Illumina reagents and cycling conditions with 18 cycles. After cleanup with Qiagen MinElute columns, each library is examined by spectrophotometry (Nanodrop, Thermo Scientific, USA) and Agilent DNA Bioanalyzer (Agilent, USA).
MRE scores
MRE-seq reads were mapped to the human genome assembly (hg18) with MAQ with an additional constraint that the 5’ end of a read must map to the CpG site within a MRE site. An MRE-score was defined for each CpG site as the number of MRE-reads that map to the site, regardless of the orientation, normalized by the number of million reads generated by the specific enzyme. An MRE-score for each genomic window (for example, any given 600bp window) was defined as the average MRE-score for all CpGs that have a score within the window.
RNA-seq
Polyadenylated RNA was purified from 20ug of DNAse1 (Invitrogen, Carlsbad, CA) treated total RNA using the MACS™ mRNA Isolation Kit (Miltenyi Biotec, Germany). Double-stranded cDNA was synthesized from the purified polyA+ RNA using Superscript™ Double-Stranded cDNA Synthesis kit (Invitrogen, Carlsbad, CA) and random hexamer primers (Invitrogen) at a concentration of 5μM. The resulting cDNA was sheared using a Sonic Dismembrator 550 (Fisher Scientific, Canada) and size separated by PAGE (8%). The 190–210bp DNA fraction was excised, eluted overnight at 4°C in 300 μl of elution buffer (5:1, LoTE buffer (3 mM Tris-HCl, pH 7.5, 0.2 mM EDTA)-7.5 M ammonium acetate) and purified using a QIAquick purification kit (Qiagen, Mississauga, ON). The sequencing library was prepared following the Illumina Genome Analyzer paired end library protocol (Illumina Inc., Hayward, CA) with 10 cycles of PCR amplification. PCR products were purified on Qiaquick MinElute columns (Qia-gen, Mississauga, Ontario) and assessed and quantified using an Agilent DNA 1000 series II assay and Qubit fluorometer (Invitrogen, Carlsbad, CA) respectively. The resulting libraries were sequenced on an Illumina Genome Analyzeriix following the manufacturer’s instructions. Image analysis and basecalling was performed by the GA pipeline v1.1 (Illumina Inc., Hayward, CA) using phasing and matrix values calculated from a control phiX174 library run on each flowcell.
ChIP-seq
Protocols for the chromatin immunoprecipitation assay and Illumina library construction are described in details elsewhere (O’Geen et al. in press). Briefly, crosslinked hESC were obtained from Cellular Dynamics, chromatin was extracted and sonicated to an average size of 500bp. Individual ChIP assays were performed using 50μg chromatin (equivalent to 5×10^6 cells) and 2ug of antibody were added to each ChIP reaction. The histone antibodies used in this study include H3AcK9 (Milli-pore#07-352), H3me3K4 (CST#9751S), H3me3K27 (CST#9733S), H3me3K9 (Abcam#ab8898), H3me3K36 (Abcam#ab9050), and H3me1K4 (Abcam#ab8895). ChIP libraries were created according to Robertson et al. using the entire purified ChIP sample. All ChIP samples except H3me1K4 were amplified using paired-end Illumina primers for a total of 18 cycles. Libraries were then run on a 2% aga-rose gel, and the 150–500-bp fraction of the library was extracted and purified. The H3me1K4 library was constructed by performing size selection of the 200–400bp library fragment prior to a15 cycle amplification. The libraries were quantified using a BioAnalyzer and sequenced. ChIP-seq peaks were called using the Sole-Search software38.
Bisulfite Pyrosequencing
Site-specific analysis of CpG methylation was performed by bisulfite pyro-sequencing. Genomic DNA (1.0 μg) was bisulfite modified and pyrosequencing was performed as previously described39. The quantitative performance of each pyrosequencing assay was verified by measuring methylation standards comprised of known proportions of unmethylated (whole genome-amplified) and fully methylated (SssI-treated) genomic DNA40.
Comparison was performed on three combinations of DNA methylome platforms: MethylC-seq versus reduced representation bisulfite sequencing (RRBS), and MethylC-seq versus methylated DNA immunoprecipitation sequencing (MeDIP-seq). H1 cell lines of different passage number were used in these experiments (Batch 3 for MethylC-seq, Batch 1 for RRBS and MeDIP). CpGs showing > 80% difference in methylation for the MethylC-seq RRBS comparison or > 80% difference between the methylated proportion and the methylation score for MethylC-seq and MeDIP comparisons were identified and regions with clusters of these sites were identified for pyrosequencing. Based on the distribution of target CpGs we looked for genomic regions with appropriate length (within range 50bp to 75bp), few or no non-CG cytosines, and 2 or many target CpGs. Pyrosequencing assays were designed and carried out in 16 regions selected for validation; 14 of these yielded reliable results. Genomic coordinates and primers used for pyrosequencing for the validated regions are listed in Supplementary Table 1.
Clonal Bisulfite Sequencing
Further validation of genome-wide data, particularly sites with apparent allelic DNA methylation, was performed by bisulfite sequencing. Total genomic DNA underwent bisulfite conversion following established protocol41 with a modified conversion conditions of: 95°C for 1 min, 50°C for 59 min for a total of 16 cycles. Bisulfite PCR primers (Supplementary Table 4) were utilized to amplify regions of interest and were subsequently cloned using pCR2.1/TOPO (Invitrogen). Single colony PCR and sequencing (QuintaraBio) provided contigs that were aligned for analysis.
Data Analyses
Comparison of CpG or non-CpG site methylation
Repeat masking of the reference genome assembly was not used in any of these analyses. For bisulfite-based methods, reads that mapped to the positive and negative strand were combined for CpG methylation calculations, but not for CHG and CHH methylation calculations due to the strand asymmetry of non-CpG methylation9. The methylated proportion was calculated for each CpG or non-CpG as (methylated reads/(methylated reads + unme-thylated reads)). Comparisons of methylation status calls were performed by imposing minimum requirements of 2, 5 or 10 reads covering a CpG or non-CpG site and applying varying methylated proportion cutoffs (0.80-0.20, 0.75-0.25, or 0.20) to make calls on the methylation status. Methylated proportion differences were calculated as (MethylC-seq proportion - RRBS proportion). Methylation proportion difference graphs were generated by counting the number of CpGs with a particular methylated proportion difference and plotting the count on the y axis. Concordance was then calculated as the percent of CpGs with a methylation proportion difference less than 0.1 or 0.25.
For enrichment based methods, methylation scores inferred for individual CpGs were averaged across CpGs covered by a varying minimum number of reads in 1000bp or 200bp windows. Methylation calls of highly methylated (methylation score >8) or weakly methylated (methylation score <= 8) were made based on the average methylation score for each window where at least one CpG was covered by the minimum number of reads.
Genomic Context of Concordant and Discordant CpGs
The overlap of concordant and discordant CpGs with annotated genes, as defined by the UCSC Genome Browser RefSeq Gene track (2010-01-24 version http://genome.ucsc.edu/cgi-bin/hgTrackUi?g=refGene), was identified. In order to deal with overlapping genes and multiple isoforms of genes, CpGs were classified into gene components based on the following prioritization order: Promoter (within 8000bp upstream of a transcription start site), Coding Exon, UTR, and Intron. CpGs that did not overlap with any of these gene components were identified as Intergenic.
Supplementary Material
Acknowledgments
We would like to thank the NIH Roadmap Epigenomics Program; sponsored by the National Institute on Drug Abuse (NIDA) and the National Institute of Environmental Health Sciences (NIEHS). JFC and MH are supported by NIH grant 5U01ES017154-02. AMi is supported by NIH grant 5U01DA025956-02. AMe and BEB are supported by NIH grant 6U01ES017155-02. JE and BR are supported by NIH grant 5U01ES017166-02. RPN was supported by NIH T32 CA108462-04 and F32CA141799. SLD was supported by CIRM TB1-01190. SDF was supported by NIH T32 CA108462-06. BEJ was supported by NIH T32 GM008568. M.A.M is a Terry Fox Young Investigator and a Michael Smith Senior Research Scholar. We thank Zefeng Zhang (Waterloo, ON, Canada) and Hao Lin (University of Waterloo) for analyses with the ZOOM algorithm.
Footnotes
Supplementary Information related to this paper is available at http://www.genboree.org/java-bin/project.jsp?projectName=Methylation%20Platform%20Comparison&isPublic=yes and hgwdev-remc.cse.ucsc.edu. Data used in this paper are available for download from the GEO NIH Roadmap Epigenomics Project Data Listings (http://www.ncbi.nlm.nih.gov/geo/roadmap/epigenomics/) and the Epigenomics Atlas (http://genboree.org/epigenomeatlas/edaccDataFreeze1.rhtml).
AUTHOR CONTRIBUTIONS
JFC, RAH, TW, MH, MAM, and AMi conceived and designed the experiments. RPN, CH, SLD, BEJ, SDF, YZ, and MH performed the MeDIP, MRE and bisulfite sequencing experiments. RAW and XZ designed and performed pyrosequencing and data analyses. HG, CB, AG, and AMe performed and analyzed RRBS. LE, HO, and PJF performed and analyzed Chip-seq experiments. RL, MP, and JRE analyzed MethylC-seq data and performed Bowtie aligner testing. RAH, TW, KF, JG, CC, MH, XZ, AD, and AO performed data analysis. TW, TB and DH developed MeDIP and MRE scoring algorithms and performed coverage analyses including repetitive sequence analyses. YX, WC, RL and WL compared bi-sulfite sequence aligners. JFC, RAH, MH, TW, RPN and RAW wrote the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 4.Gama-Sosa MA, Midgett RM, Slagel VA, Githens S, Kuo KC, Gehrke CW, et al. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983;740:212–9. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- 5.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010 doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacinto FV, Ballestar E, Esteller M. Methyl-DNA immunoprecipitation (MeDIP): hunting down the DNA methylome. BioTechniques. 2008:44, 35, 37, 39. doi: 10.2144/000112708. passim. [DOI] [PubMed] [Google Scholar]
- 11.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–85. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38:391–9. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–7. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ball MP, Li JB, Gao Y, Lee J, LeProust EM, Park I, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–8. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, et al. Shotgun bisul-phite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–9. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The American Association for Cancer Research Human Epigenome Task Force. European Union, Network of Excellence, Scientific Advisory Board Moving AHEAD with an international human epi-genome project. Nature. 2008;454:711–5. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coarfa C, Milosavljevic A. Pash 2.0: scaleable sequence anchoring for next-generation sequencing technologies. Pac Symp Biocomput. 2008:102–13. [PubMed] [Google Scholar]
- 21.Smith AD, Chung W, Hodges E, Kendall J, Hannon G, Hicks J, et al. Updates to the RMAP short-read mapping software. Bioinformatics. 2009;25:2841–2. doi: 10.1093/bioinformatics/btp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin H, Zhang Z, Zhang MQ, Ma B, Li M. ZOOM! Zillions of oligos mapped. Bioinformatics. 2008;24:2431–7. doi: 10.1093/bioinformatics/btn416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci USA. 2007;104:18613–8. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunarso G, Chia N, Jeyakani J, Hwang C, Lu X, Chan Y, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–4. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 25.Pant PVK, Tao H, Beilharz EJ, Ballinger DG, Cox DR, Frazer KA. Analysis of allelic differential expression in human white blood cells. Genome Res. 2006;16:331–9. doi: 10.1101/gr.4559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollard KS, Serre D, Wang X, Tao H, Grundberg E, Hudson TJ, et al. A genome-wide approach to identifying novel-imprinted genes. Hum Genet. 2008;122:625–34. doi: 10.1007/s00439-007-0440-1. [DOI] [PubMed] [Google Scholar]
- 27.Schalkwyk LC, Meaburn EL, Smith R, Dempster EL, Jeffries AR, Davies MN, et al. Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet. 2010;86:196. doi: 10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pick M, Stelzer Y, Bar-Nur O, Mayshar Y, Eden A, Benvenisty N. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–90. doi: 10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- 29.Arnaud P, Monk D, Hitchins M, Gordon E, Dean W, Beechey CV, et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum Mol Genet. 2003;12:1005–19. doi: 10.1093/hmg/ddg110. [DOI] [PubMed] [Google Scholar]
- 30.Li N, Ye M, Li Y, Yan Z, Butcher LM, Sun J, et al. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods. 2010 doi: 10.1016/j.ymeth.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Bourque G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev. 2009;19:607–12. doi: 10.1016/j.gde.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 32.Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nat Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- 33.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epige-netic gene regulation. Mol Cell Biol. 2003;23:5293–300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–3. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–46. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 36.Gu H, Bock C, Mikkelsen TS, Jager N, Smith ZD, Tomazou E, et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7:133–6. doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. High-throughput bisulfite sequencing in mammalian genomes. Methods. 2009;48:226–32. doi: 10.1016/j.ymeth.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blahnik KR, Dou L, O’Geen H, McPhillips T, Xu X, Cao AR, et al. Sole-Search: an integrated analysis program for peak detection and functional annotation using ChIP-seq data. Nucleic Acids Res. 2010;38:e13. doi: 10.1093/nar/gkp1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waterland RA, Lin J, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet. 2006;15:705–16. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- 40.Shen L, Guo Y, Chen X, Ahmed S, Issa JJ. Optimizing annealing temperature overcomes bias in bisulfite PCR methylation analysis. BioTechniques. 2007:42, 48, 50, 52. doi: 10.2144/000112312. passim. [DOI] [PubMed] [Google Scholar]
- 41.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65–5. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.