Abstract
Segmentation is a fundamental component of many medical image processing applications, and it has long been recognized as a challenging problem. In this paper, we report our research and development efforts on analyzing and extracting clinically meaningful regions from uterine cervix images in a large database created for the study of cervical cancer. In addition to proposing new algorithms, we also focus on developing open source tools which are in synchrony with the research objectives. These efforts have resulted in three Web-accessible tools which address three important and interrelated sub-topics in medical image segmentation, respectively: the BMT (Boundary Marking Tool), CST (Cervigram Segmentation Tool), and MOSES (Multi-Observer Segmentation Evaluation System). The BMT is for manual segmentation, typically to collect “ground truth” image regions from medical experts. The CST is for automatic segmentation, and MOSES is for segmentation evaluation. These tools are designed to be a unified set in which data can be conveniently exchanged. They have value not only for improving the reliability and accuracy of algorithms of uterine cervix image segmentation, but also promoting collaboration between biomedical experts and engineers which are crucial to medical image processing applications. Although the CST is designed for the unique characteristics of cervigrams, the BMT and MOSES are very general and extensible, and can be easily adapted to other biomedical image collections.
1. Introduction
Segmentation of anatomical structures or pathological regions plays an important role in medical image analysis. It is a critical component of numerous medical imaging applications because many image processing tasks, such as feature extraction, quantitative measurement, and object recognition, rely on previous segmentation results. Segmentation of regions of interest has been actively studied in a variety of medical domains with different image modalities and different organs, and many segmentation algorithms designed for specific applications have been proposed [1–6]. Even though it is an active research field with many published results, segmentation of medical images remains a very complicated and challenging task. In [1], four major difficult challenges are identified for this field. Two of them are: 1) the need to address the practical needs of its user community, i.e., biomedical scientists and physicians; and 2) the need to develop validation and evaluation approaches that are convincing and practical to carry out. In this paper, we report our research and development efforts to address these difficulties in the area of cervicographic image analysis. Our efforts include not only implementing and testing state-of-the-art cervicographic image processing algorithms, but also developing easy-to-use supporting tools to integrate these sophisticated algorithms within software testable by the biomedical community. The latter is not frequently emphasized by researchers in medical image analysis. These efforts are closely connected. Designing tools to reach more users and to be easily used by physicians may speed the refinement and improvement of the processing and analysis algorithms themselves. The performance and acceptance of the tools ultimately rely on the effectiveness and efficiency of the underlying algorithms. For cervicographic image segmentation, we have developed three tools, which compose a suite of complementary software. They address three strongly coupled, recurring tasks of image segmentation: “ground truth collection”, automated segmentation, and segmentation quality evaluation. The tools are implemented as three separate computer programs, unified at the data level through a consistent data exchange format. In the following, we first introduce the clinical significance of cervicographic images and then discuss these tools in detail.
1.1. Cervicographic images
Cervicographic images, also called “cervigrams”, are one commonly-used, cost-effective type of uterine cervix imaging, and are an important knowledge resource for cervical cancer diagnosis, study and training. Cervical cancer is the most common gynecological cancer and the leading cause of cancer deaths among women in many developing countries. This high prevalence is mainly attributed to the lack of widespread screening programs. Screening programs are very crucial for the early detection and treatment of precancer before the premalignant lesions progress into invasive cancer, a process generally being slow and spanning a decade. Cervicography is one low-cost visual technique for cervical cancer screening introduced in the 1970s [7]. It has been used as an alternative or adjunctive method to cytologic screening (i.e., Pap tests), which requires cell collection and examination, in areas with limited health care resources. In these areas, cytological screening is not viable due to its complexity and high cost. Like colposcopy, cervicography is based on acetowhitening, a transient phenomenon observed when a diluted acetic acid solution is applied to the surface of cervix. The tissue area where the color become whitish after acetic acid exposure are of high interest and are considered as one of the vital signs that may indicate abnormality. Cervicography requires the acquisition of cervicographic images, or “cervigrams”, which are low-magnification color photos of the cervix, taken during a short interval after treatment with acetic acid, when acetowhitening may occur. Cervicographic pictures are captured with a specially designed 35-mm camera, with a fixed focal lens (to obtain comparable pictures of cervices) and a mounted ring flash to provide the necessary illumination. The capture of cervigrams can be done by a physician or a nurse after a short period of training and does not require colposcopic expertise. The pictures are interpreted by experts, who have undergone specialized training in cervigram interpretation. The patients whose cervigrams are interpreted as being abnormal are then referred to colposcopy examination for further diagnosis.
Two examples of cervigrams are shown in Figure 1. The focal point of the cervigram is the cervix region, (or, more simply, the “cervix”) which is usually centered in the image and covers about half of the image. Outside the cervix region, there are vaginal walls and medical instruments. Text added during the image acquisition process may also appear in the image. Usually, the surface of the cervix is covered by two types of epithelium: squamous epithelium and columnar epithelium. The squamous epithelium consists of multilayer cells increasingly flatter toward the surface and normally appears pale pink. The columnar epithelium is composed of a single layer of tall cells and has a reddish and grainy appearance. Exposed to the acidic environment of the vagina, as a female ages, the columnar epithelium is gradually replaced by a newly formed squamous epithelium, a process called epithelium metaplasia [8]. The area on the cervix where epithelium metaplasia occurs is called the transformation zone. This zone is of great interest because almost all the cervical cancer cases arise in this area. The orifice in the center of the cervix is the external os, an important anatomical landmark. The effect of acetic acid, which is thought to cause abnormal squamous epithelium to turn whitish compared to the color of the surrounding normal squamous epithelium, can also be seen. Other clinically significant visual phenomena within the cervix region include areas of bleeding, mucus, cysts, and atypical vascular patterns.
Figure 1.
Examples of cervigrams
Cervigrams are a critical data component of two large-scale, multi-year cervical cancer projects funded and organized by the National Cancer Institute (NCI). One, the Guanacaste Project [9], was carried out in Guanacaste, a rural province of Costa Rica, where the incidence rate of cervical cancer is very high. The other is the ASCUS/LSIL Triage Study (ALTS) [10], which was conducted in the United States. The Guanacaste Project is the world’s largest prospective cohort study of HPV infection, the primary cause of cervical cancer and cervical neoplasia. It is based on enrollment and (up to) seven-year follow-up of over 10,000 women of 18+ years old randomly selected from Guanacaste. The field work included the fulfillment of a personal interview, cervical cytologic smears, cervicography, cervical swabs for HPV, and a blood specimen. The ALTS project goal is to study the clinical management of low-grade cervical cytologic abnormalities. It recruited about 5,000 women who had received a cytologic diagnosis of ASCUS (Atypia of Squamous Cells of Undetermined Significance) or LSIL (Low-Grade Squamous Intraepithelial Lesion) and followed them for two years at one of four American medical centers. The data collected includes cervical cell samples, a specimen for HPV testing, and cervicographic images. The National Library of Medicine (NLM), in collaboration with NCI, has digitized 100,000 cervigrams collected in these two projects, and has archived the images, as well as other associated clinical information, for research, and for training and education of colposcopic practitioners. NLM has been developing various software systems to search, disseminate, compress, and annotate these images, including the development of the Multimedia Database Tool (MDT) [11,12], for providing text-query access to the Guanacaste and ALTS databases, and is engaged in work to expand the search ability of these databases to include search by image contents. These efforts have resulted in development of a prototype Content-Based Image Retrieval (CBIR) system, CervigramFinder [13–15]. CervigramFinder (whose Web interface is shown in Figure 2) is a region-based search tool that retrieves medically-important regions in cervigrams based on their visual similarity to a query image. Region segmentation is of great interests because information extracted from these regions is valuable for supporting medical research for prevention of cervical cancer.
Figure 2.
Screenshot of CervigramFinder
1.2. Image segmentation triangle
Automated segmentation and analysis of cervigrams in such a large repository is a very challenging task. There are several factors that contribute to this. First, there is large variability in visual content of the images across the dataset. The shape and size of the cervix varies with the age, parity (child-bearing history), and hormonal status of patients. Also, the cervices of patients vary with respect to the visual phenomena which are present, and with respect to the sizes and spatial configurations of these phenomena, in particular, for the two types of epithelium and for the acetowhitened tissue. Second, there are irrelevant information and acquisition artifacts within the images which complicate and interfere with the detection of tissues of interest. For example, medical instruments, such as cotton swabs and the endocervical speculum, can be seen in the cervix region in some images. In addition, it is common to have small spots of intense brightness in various parts of the image that result from reflections from cervix fluids of the camera illumination. Lastly, boundary lines between different tissues may be obscured and the illumination of tissues is not uniform across images, complicating color comparisons from image to image. To address these complexities, a multi-step scheme has been proposed [16], where each step focuses on the sequential segmentation of a particular tissue type or anatomical feature. Evaluation of these algorithms, either subjectively or objectively, is critical. Both evaluation methods require the assistance of users, i.e. physicians or clinical experts in cervical cancer, and to achieve this, close collaboration between the biomedical and engineering communities is essential. An impediment to this collaboration has been the lack of medical image segmentation software that is convenient and easy-to-use. To address this problem, we have put significant effort into software development as well as algorithm research. Our work has resulted in three important tools for region segmentation and analysis of cervigrams: the Cervigram Segmentation Tool (CST), the Boundary Marking Tool (BMT), and Multi-Observer Segmentation Evaluation System (MOSES). The CST contains segmentation algorithms customized for automatic cervigram analysis, and allows a clinician to quickly obtain a segmented tissue result. The BMT provides capability for experts to manually draw region boundaries on the cervigram and to enter their interpretative findings. The data collected by the BMT can then be used as a “ground truth” dataset for “objective evaluation” of segmentation results. MOSES focuses on the problem of observer variability among image segmentations. It automatically generates a combined ground truth segmentation map from the segmentations created by each observer and computes the “performance” of each segmentation (i.e., it computes a sensitivity and specificity score for the observer who created the segmentation) It enables direct comparison of algorithm versus human observer performance in segmentation. As illustrated by Figure 3, the three tools each target one essential aspect of medical image segmentation: CST for automated image segmentation, BMT for ground truth data collection, and MOSES for segmentation quality evaluation. Even though each tool may function independently of the others, they are closely related. Specifically, the boundaries marked by multiple experts using BMT can be input to MOSES to generate a probabilistic estimate of the “true” segmentation; this result can then be used, again in MOSES, to assess the performance of CST for segmenting the same region automatically. All the tools are implemented to be Web-accessible and have a client/server architecture. Furthermore, a common data format is used across the tools, which is beneficial for data exchange, archive and extraction.
Figure 3.
The cervigram segmentation triangle
The BMT, CST, and MOSES are in varying stages of maturity. The most mature, the BMT, has been used in a number of data collection studies by the National Cancer Institute which have resulted in analytical results relating to uterine cervix cancer screening that have been published in the medical literature [17]. We envision that the CST will contribute to the collection of additional cervix region data by making this task less labor intensive, and that MOSES will be used to reconcile differences among boundary data collected from both human experts and automated methods (such as the CST). In the following sections, we will discuss the implementation of each tool in detail. Although our current focus is the analysis of cervigrams, many aspects of the system design and development of this software suite are general and can be straightforwardly applied to other types of medical image modalities.
2. BMT – Tool for Ground Truth Collection
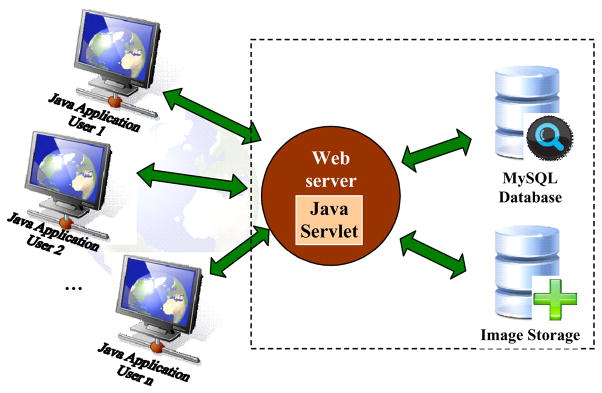
The Boundary Marking Tool is the most mature of the tools. Its development was motivated by the need to facilitate the visual study of cervigrams by gynecologists in their study of cervical neoplasia. In the development process, we have closely collaborated with biomedical experts at the National Cancer Institute, and the design has been shaped by their domain knowledge and usability requirements; have used an “early prototype” development model to expose our system concepts to the expert users at an early design stage; have put emphasis on design of a flexible and scalable system structure; and have taken as a goal quick turnaround on doctors’ suggestions for system changes and new functionalities to enhance the system. BMT is designed primarily for collecting image region data that is important to practicing and research physicians in uterine cervix oncology. To enable easy access and usability, BMT employs a client/server Web architecture as shown in Figure 4. It consists of four components: (1) a Java client application that provides a graphical user interface (GUI), (2) a Java servlet running on an Apache Tomcat server, (3) a database management system, and (4) an image storage server. The client application is written using Java Swing that provides a rich graphical interaction and supports a pluggable look and feel. The servlet is a Java server-side component used to extend the functionality of the Apache Web server. It coordinates communication between the client and other server-side components, including the management of user requests, authorization of user access, and interaction with the database. Client/servlet communication is by standard HTTP, and the servlet communicates with the database using Java database connectivity (JDBC). At startup, the client contacts the servlet to authenticate the username/password combination used for login. If the user had a previous session, his/her previously-recorded data for each image will be downloaded to the client. This is particularly useful for medical experts, since it allows them to resume unfinished work on their own schedules. Once logged in, the user selects the cervigram of interest, which will then be transmitted from the image server to the client for review and interpretation. The data collected by BMT is transmitted back to the server and stored in a MySQL database through the Java servlet. The database uses metadata tables that represent the system data at a high level of abstraction, with the goal of accommodating different image collections without or with minor code modifications.
Figure 4.
System architecture of BMT
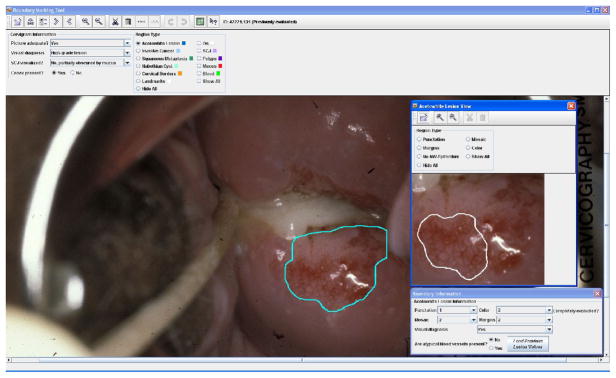
A screenshot of BMT’s user interface is shown in Figure 5. The interface allows users (in particular, gynecology professionals) to draw free-form boundaries interactively for regions of interest, and to enter annotations and interpretations for these regions. An “overall evaluation of the image” may be recorded, which includes an assessment of whether the image quality is satisfactory for visual examination, a final visual diagnosis categorized by six classes, a judgment about the visibility of the entire transformation zone, and an assessment confirming the presence of the cervix in the image. The central part of the interface is a canvas that displays the entire image downloaded from the server. The user can manually draw the boundary of specific types of regions on this canvas. Cervix regions that may be marked with boundaries and labeled include the acetowhite region, and regions with squamous metaplasia, invasive cancer, the external os, polyps, mucus, blood, cysts, and the transformation zone. Capabilities are also provided for drawing an “orientation landmark” (a 3–6–9–12 clockface that may be rotated to show the cervix orientation with respect to rotation) and the boundary that encloses the entire cervix region. Each region or landmark type is color-coded for easy identification. Depending on the type of region for which the user draws a boundary, pop-up or drop-down menus appear to record additional description information. For example, for acetowhite regions, a drop-down list of the Reid’s colposcopic index appears (when the user has finished drawing the boundary) for classifying the region’s margins and color characteristics. Since abnormal vascular patterns inside acetowhite regions are one of the crucial features associated with cervical neoplasia, the tool also provides an additional window with a detailed view of acetowhite regions and allows experts to delineate areas of vascular features by drawing sub-regions within the acetowhite regions. The user may also enter additional information about these sub-regions, such as the presence or absence of mosaicism (tile-like patterns) and punctuation (dot-like patterns) and the granularity classification (coarse or fine) of these patterns. Functions are provided for viewing detail and for editing the markings, including, zoom, erase, undo, and “auto-close” to force an open polygonal curve to become closed. Users may “export” their markings and recorded data to a local file, as well as save them to storage on the server.
Figure 5.
Screenshot of BMT
BMT has been actively used by NCI researchers for multiple studies [18, 19], including the assessment of how the findings of colposcopic examination correlate with high-grade disease, the analysis of the tropisms of HPV on the columnar and squamous epitheliums of the cervix, the evaluation of the effect of inter-observer and intra-observer variability on the accuracy of colposcopic diagnosis, and the comparison of the differences in cervix visual characteristics between patients with HPV-infections and those without infections The BMT is currently being used as a primary supporting tool in an NCI multi-year project to develop a new strategy for selection biopsy points on the cervix of women during colposcopy. In this study, BMT is used both “online” and “offline”. During the colposcopic examination, doctors draw lesion boundaries and mark multiple biopsy spots using BMT installed on a laptop computer that obtains an image of the cervix from a digital camera mounted on a colposcope. Later, in the offline mode, BMT is used to display the collected images, biopsy spots, and biopsy results (which are obtained by histology diagnosis of tissue collected at the spots marked by the BMT) during regular teleconferences by the study participants. BMT data is also incorporated into the MDT to allow the search and retrieval of this boundary-related information. Although the BMT was designed for the collection of uterine cervix data, its modularity and generality make it readily adaptable for collecting region-based data from other image modalities. For example, a modified version of BMT (shown in Figure 6) was employed in a dermatology study to measure the size of Kaposi sarcoma lesions during a clinical trial to determine whether the lesion sizes would regress when treated with nicotine patches. BMT is intended primarily as tool for clinical research. However, the region data collected by BMT may also play an important role in the evaluation and improvement of automatic image segmentation algorithms, as a source of multi-expert “ground truth” data. This is, in fact, what we have done to evaluate our own multi-step cervigram segmentation algorithm. The second-generation version of BMT has now been created, which works with the newly-created BMT Study Administration Tool (BSAT). With the BSAT, a privileged user may use a Web interface to completely define the configuration of the screens of the BMT, including which boundaries are to be collected, and what options the user will have for labeling the boundaries. The BSAT also allows the definition of which images will be used in a BMT data collection and which users (observers) will participate in the data collection, and allows the retrieval of the data collection results.
Figure 6.
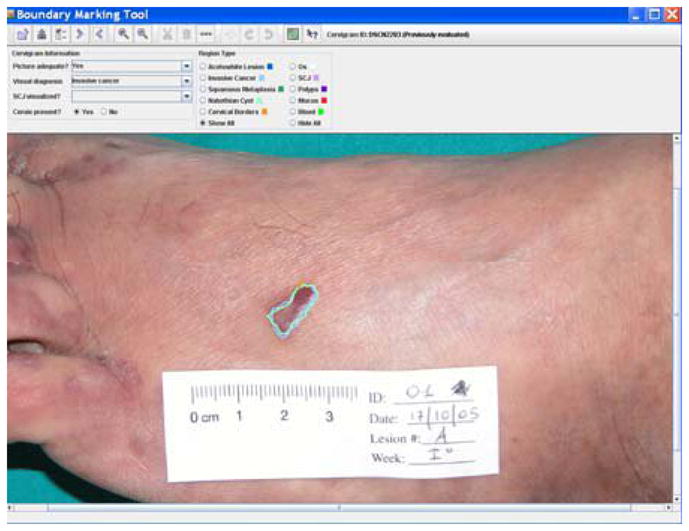
BMT for dermatology study
3. MOSES – Tool for Segmentation Quality Evaluation
Evaluation and validation of image segmentation quality are among the most prominent ongoing tasks in the image processing community, such as these articles in the field of dermoscopy [20, 21]. Effective evaluation methods should allow the comparison of different segmentation methods and facilitate the improvement of algorithms. Quantitative analysis and characterization of segmentation performance has long been recognized as a challenging problem, especially for medical images, due to the difficulty of obtaining a “true” segmentation as an evaluation standard. A typical approach is to compare the algorithmic segmentation results with manual segmentations by human experts. However, this approach is complicated by intra-observer and inter-observer variability in producing the expert segmentations. This variability is illustrated by an NCI study done with the BMT, using 939 cervigrams to study the correlation between cervical visual appearance and HPV types. In this collection, each image was manually marked and evaluated by two to twenty evaluators, who used the BMT at sites that were widely-geographically distributed. The twenty evaluators included twelve general gynecologists and eight gynecologist oncologists with varying years of experience. The evaluators were each assigned a set of images to evaluate and were asked to draw a boundary around the cervix and any acetowhite lesion they identified in each image. As shown in Figure 7, the variability among the experts’ segmentations of the same region is different across images. In some cases, the agreement in boundary marking is low, while in other cases, most of the experts have high agreement on the tissue boundaries. Using these 939 cervigrams, we have explored alternative approaches [22–24] aimed at 1) generating ground truth data with high consensus; 2) evaluating automated segmentation results; and 3) analyzing the inter- and intra-expert variability in spatial boundary marking. One result of this work has been the development of the MOSES system.
Figure 7.
Examples of cervigram regions marked by multiple medical experts.
MOSES was created to address the limitations of the STAPLE [25] system. STAPLE is a well-known expectation-maximization algorithm for multi-observer segmentation evaluation that generates a ground truth segmentation map from the observations of multiple experts and at the same time obtains a performance measure associated with each individual observation. Specifically, STAPLE estimates the sensitivity and specificity of each observer which maximizes the complete data log likelihood function:
| (1) |
where
D is an N×R matrix with element Dij indicates the binary decision made by observer j for pixel i. A value of 1 means presence (the pixel is in the segmentation) and 0, absence (the pixel is not in the segmentation). Suppose there are N pixels in the image whose segmentations are being evaluated by R observers.
T=(T1,T2,...,TN ) is an indicator vector of N elements and Ti represents the hidden binary “true segmentation” for pixel i.
p j= Pr( Dij=1 | Ti=1) represents the sensitivity of observer j (for any pixel i)
q j= Pr( Dij= 0 | Ti= 0) represents the specificity of observer j (for any pixel i)
MOSES explicitly takes into account both (1) the ground truth segmentation prior and (2) the observer performance prior, and balances the roles of the two priors to address different segmentation evaluation needs, by using Bayesian Decision Theory. Specifically, it considers the posterior probability distribution and applies the maximum a posteriori (MAP) optimization principle to obtain a probabilistic estimate of the “true” segmentation:
| (2) |
MOSES provides a more flexible approach for multi-observer evaluation by applying different methods for different scenarios depending on the availability and confidence of the estimation of the truth prior f (Ti=1) and the observer prior (p, q). These four scenarios are accounted for: 1) only the truth prior is known; 2) only the observer prior is known; 3) both priors are known, and (4) both priors are unknown. These scenarios can be addressed by incorporating initialization value for priors into equations 5 and 6. If a prior is unknown, it can be initialized either from a uniform probability distribution, or it may be based on observers’ previous segmentation data, as available knowledge allows. For example, in scenario two in which the sensitivity and specificity of each observer are known, we can differentiate observers into expert and non-expert categories, but we do not know the truth prior probability. If we assume there is no prior information available about the ground truth segmentation map, then we may initialize the truth prior under the assumption of equiprobability (i.e., we set according to a uniform probability distribution). If we assume the observers’ segmentation data reflects the prior distribution of the true segmentation, then we initialize it using this segmentation data. In this way MOSES handles more situations than STAPLE by integration of different priors for different application purposes. A detailed description of the MOSES algorithm can be found in [23, 24].
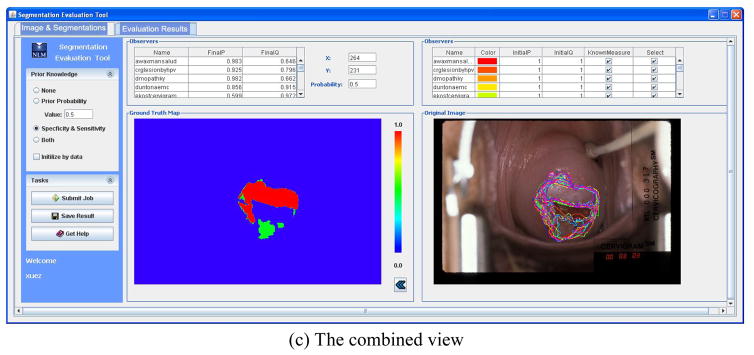
It is important to note that although MOSES is demonstrated using cervigrams, it can be used to analyze multiple observer segmentation applications of a wide variety of images. The system architecture of Web-based MOSES consists of only two major components: the client application and the servlet. The user interface had two tab panels. One is for loading and reviewing the image and segmentation information (Figure 8a). The segmentations of multiple observers are overlaid on the image and color-coded in an automatically-selected color sequence. The table lists the detailed information on segmentations including user names, colors of segmentations, and the initial observer performance values (p, q). The user may toggle the display of any segmentation on the image or exclude it from being used in the estimation of ground truth map. Segmentations in a new boundary file may be combined with existing segmentations, a handy feature when segmentations are being accumulated over time, for the same image. Therefore, the segmentations for one image can be saved in multiple files instead of one big file. The other tab is for displaying and exporting results (Figure 8b). This allows the user to select one of the scenarios with respect to the prior knowledge and set proper prior values. This information along with the segmentations and image size is then sent to the server. Please note that it is unnecessary to upload the image except its size and the segmentations are represented in a contour format. Since the transmitted data is not large, image compression is not an issue for MOSES. (Unlike CST, discussed in the next section, which does use image transmission.) The server-computed evaluation results include an estimated ground truth segmentation map and the final performance measures for each observer, which are displayed by the client on a panel and in a table, respectively. These results can be exported to the user’s local machine in a zip file which contains a grayscale image representing the ground truth map and a text file having the final (p, q) values. The original image with all observers’ boundaries and the resulting ground truth map can be displayed side-by-side for easy comparison as shown in Figure 8c. In addition to its role in assessing automatic segmentation algorithm performance relative to manual, multi-expert segmentations, MOSES is also expected to play a role in the study of observer manual segmentation variability among medical experts themselves.
Figure 8.
The user interface of MOSES
4. CST – Tool for Automated Segmentation
It is important to provide objective approaches for assessing the performance of automated segmentations. It is also important to provide domain experts with capability to directly experiment with and evaluate automatic algorithms within user-friendly software systems. These are the objectives that led to the development of CST, a Web-accessible tool for cervigram segmentation. Although the algorithms for segmenting important regions in cervigrams are still under development, a working prototype of CST has been built. One distinguishing aspect of CST is that it provides a solution to a problem facing many research groups who use Matlab to develop algorithms, namely that it combines the image-processing and algorithmic strengths of Matlab with the user interface and Web-friendly strengths of Java. The CST approach frees image processing researchers from rewriting existing algorithms coded in Matlab, while allowing various groups of collaborating researchers to evaluate the algorithms using their own images at geographically distributed locations without the installation of Matlab at each user site. The techniques presented here can be applied to various processing and analysis applications for medical images. In the following, we introduce the automatic cervigram segmentation algorithms, then discuss the practical implementation issues.
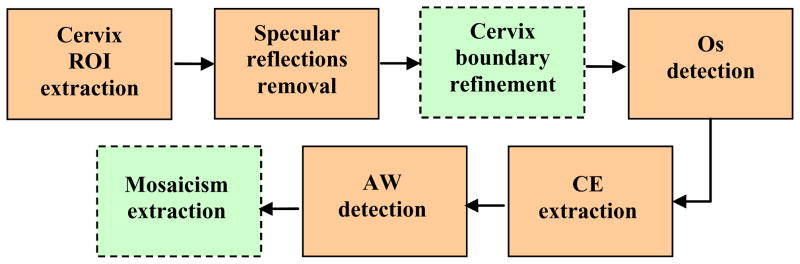
We attacked the complex problem of cervigram segmentation with a divide-and-conquer strategy that seeks to sequentially segment the various region types. As we target each individual region-of-interest, we identify corresponding, appropriate feature sets one-by-one. Note that this approach also permits research for each step to be conducted in parallel and, in principle, by different groups of collaborators. A block diagram of our multi-step segmentation scheme is given in Figure 9. The first step is to extract an approximate boundary of the cervix region to reduce the search space for the subsequent processing tasks (since all biomarkers of interest are inside this region). We used a method proposed in [26] which uses the Gaussian Mixture Modeling clustering technique with and the feature set being (1) the a channel of the pixel’s color in Lab color space, and (2) the pixel’s distance to the image center. This approach was motivated by the observation that the cervix region has a relatively pink color and is usually located in the central part of the image. An important factor that must be considered is that, on the cervix, there are often multiple Specular Reflection (SR) spots. These are caused by the bright illumination reflecting from fluid on the cervigram surface. The presence of these spots may interfere with the detection of the most significant visual diagnosis feature, the acetowhite regions, which have a color similar to that of the SR regions. Therefore, there is a need to identify or remove SR from the image. Two approaches for SR detection and filling were developed, and both were judged to be effective in tests that we performed. Both take into account the two representative characteristics of SRs: small area and high brightness. The first uses a morphological processing [27] method, the second, unsupervised clustering [28]. The third step, after removing the SR regions, is to obtain a more accurate cervix boundary by further refining the coarse cervix ROI that was extracted in step one. For this, we used a method of boundary energy minimization formulated as an active contour [29]. In this approach, the information characterizing the region, edge, and shape are all incorporated into the energy function; each term in this function is tailored to model the specific characteristics of the cervigrams.
Figure 9.
Block diagram of the multi-step segmentation scheme
After the refined cervix boundary is estimated, we proceed to segment the individual region types or significant boundaries within the cervix. These regions and boundaries are discussed in the following. The os, or opening into the uterus, is one important landmark on the cervix that is used by physicians as a reference for locating columnar epithelium or the transformation zone. It is detected using a concavity measure. This measure is motivated by the observation that, despite the large variation in size, color and shape of the os, it is always concave and the os center is near the center of the cervix [30]. Columnar epithelium (CE) tissues are extracted by using a texture feature and a color feature [31]. The texture feature is used to represent the coarseness of the tissue; in addition, a multi-scale texture-contrast feature is employed. The color feature is used to describe the redness/orange color of the tissue (the b channel of the Lab color space is used). The CE tissue pixels represented in this texture-color feature space are then identified using the GMM clustering technique. The detection of AW tissues is the most important, yet the most challenging segmentation task. Multiple methods have been explored and examined [32–35]. These methods include both supervised learning and unsupervised clustering approaches. Pixel-based, region-based and boundary-based have all been used. We have also investigated methods for analyzing vascular patterns within AW regions [36–39]. For example, an approach which utilizes Support Vector Machines and circumvents the need for actual vascular structure extraction was developed in [38]. We have evaluated these methods against segmentations/classifications made by medical experts. The performance of the segmentation algorithms that have been integrated into the CST, as shown by squares with solid boundaries in Figure 9, are summarized in Table 1. For detailed information about the algorithms and their performance, please refer to the corresponding references.
Table 1.
Performance of segmentation algorithms
| Step | Method | Performance |
|---|---|---|
| Cervix ROI extraction | Ref [26] | 120 cervigrams with manual segmentations of cervix available were used for testing. Criteria for judging an acceptable ROI were that it should include the cervix region in its entirety while excluding most of the irrelevant surroundings. These requirements were largely satisfied in all of the testing images. |
| SR removal | Ref [28] | A subjective quality grading was conducted with the cooperation of a medical expert on 120 images (no ground truth SR-free images were available). Out of 120 images, the expert judged the algorithm detection of SR to be “very high” in 85% of the cases, using a 5-point scale with “very high” the best agreement that the expert could choose. |
| Os detection | Ref [16] Ref [30] |
A quantitative evaluation of the os detection was performed using a dataset of 120 images (set 1) and a dataset of 158 images (set 2), for which a manual delineation of the os region by a medical expert was available. The minimal distance between the automatically-detected os marker and the manually- segmented os contour was calculated. The distances were less than 10 pixels (the approximate width of the os region as marked by the expert) in 82% of the cases in set 1 and in 79% of the cases in set 2. |
| CE extraction | Ref [31] | The performance of the algorithm was demonstrated by several cervigram examples. The detected CE regions were consistent with the assumed CE model: a rough and red region located around the cervix center. The detected regions were in general close to the manually marked CE regions. Miss- detection of the CE regions was apparent in the cases that were different from the assumed CE model. |
| AW detection | Ref [32] | A comparison with respect to the color space representations, the granularity level, the kernel function for SVM classifier in terms of training time and memory usage was carried out. Qualitative segmentation results on a 939 cervigram images with multiple expert markings were obtained. |
We have implemented most of these algorithms in Matlab to take advantage of its functionality for numerical computations, matrix manipulations, and image analysis and to quickly develop algorithmic implementations for experimentation. However, there are practical challenges in deploying systems developed using Matlab to multiple sites. To execute Matlab programs, either an installation of the Matlab environment or the operating system-specific software libraries are required along with the developed applications. The former requires the purchase of licenses, and the latter may be cumbersome for inexperienced users. In contrast, Java has clear advantages with respect to easy dissemination, platform independence, system updates, and code integrity. However, it is undesirable to rewrite the Matlab codes using Java when the algorithms are under constant improvement and are not finalized. To allow clinical experts, who might have little or no knowledge of Matlab programming, to test or validate the segmentation algorithms using their own images with ease of use, we designed the CST architecture to combine the merits of both languages. Our three-tier system framework is intended to circumvent the need to re-coding the sophisticated MATLAB algorithms, while simultaneously permitting easy access and deployment. The system architecture (Figure 11) consists of three main components: (1) the Java application on the user machine, (2) the Java servlet on the Web server, and (3) Matlab on a server-side Windows machine. Similarly to MOSES and BMT, the CST Java application provides a graphical user interface for collecting the end user’s input and also for presenting back to the user the segmentation results. The servlet serves as a mediator between the Java application and the Matlab machine. Through the servlet, the information collected by the Java application is sent to the segmentation algorithms running on the server-side Matlab machine, and the segmentation results obtained by the Matlab programs are then returned to the Java application for display. The servlet communicates with the Matlab machine via Java sockets (which are supported by the Matlab Java interface). Since the Matlab Java interface does not support the passing of user-defined structures but only the passing of strings, the servlet also handles the necessary reformatting of the data transferred back and forth. The server-side Matlab machine runs a specially-written Matlab function, which uses an infinite loop to wait for requests from the servlet and to handle socket calls from the servlet. It accepts the image and other related data forwarded by the servlet and then calls appropriate region/landmark segmentation algorithms to process the cervigram image. It also returns the segmentation results, such as the coordinates of the boundaries of the segmented regions, back to the servlet via the Java socket. The CST is a research application, and the automated cervigram segmentation algorithms are still in the early stage, thus frequent changes in the code for both the user interface and the Matlab segmentation algorithms are expected. This architecture has several advantages for minimizing the work involved in system updates. First, it decouples the user interface from the Matlab segmentation algorithms. Second, the client application can be conveniently deployed to end users from the designated project webpage and the deployment of the most current version of the tool is guaranteed. And last, the Matlab segmentation functions are organized in a modular structure for easy identification and replacement of algorithm components. Besides the system architecture shown in Figure 11, we also investigated two other options for Matlab and Java integration using the technique of Matlab Builder JA recently made available by the MathWorks, Inc. Matlab Builder JA is a Java compiler for Matlab. It is useful for deploying Matlab functions into Java classes and these classes can then be used in a Java application. One option is to bundle the JAR file created by Matlab Builder JA with the user interface together and made them available for downloading to client sites. This architecture does not require the Matlab runtime environment on the client sites. However it is not platform independent and continues to pose a challenge to inexperienced users or to those with limited administrative access to their computers. Another possibility is to deploy the JAR file on the server. This maintains the proposed client/server architecture, but can be a burden on the Web server if heavy computational demands are posed on it. It also puts an extra workload to the developer and is not convenient for code debugging. As stated previously, the cervigram specified by the user is required to be transferred to the server for being processed by algorithms coded by Matlab. The size of the cervigram to be uploaded could be large. Therefore, an image compression method needs to be applied for fast transmission and efficient memory usage. To address this issue, we incorporated a novel image codec designed and implemented by our collaborators [40] into the system. The codec has several very nice features, such as extremely low memory usage, high image quality at very high compression ratios, and fast coding speed. The image is encoded on the client side using the codec if its size is larger than a threshold. The compressed image is then decoded back on the server and moved to the Matlab machine to be segmented. Since it is a lossy codec, the effect of compression on segmentation results was examined. Preliminary tests indicate that the compression does not degrade the performance of segmentation to a noticeable level.
Figure 11.
The user interface of CST
One design objective for CST interface was to be clear and simple; the current implementation is shown in Figure 10. There are three main panels: a thumbnail panel, a property panel and an image view panel. The thumbnail panel provides an overall view of the image, at reduced spatial resolution, and can be used to navigate the full-size image. The image view panel shows a rectangular view of part of the cervigram at its original spatial resolution. The property panel displays the image information and allows users to toggle the view of the boundaries on the image. The part of the image shown in the image view panel is kept in sync with the part of the image marked by the blue rectangle in the thumbnail panel. Therefore, users can see the details of different areas of the cervigram by moving either the scroll bars of the image view panel or the blue rectangle. A progress bar for informing users that the segmentation is ongoing and a button for aborting the segmentation process are also provided. The system has incorporated the functionality of segmenting the regions of cervix ROI, SR, os, CE, and AW (the steps enclosed in solid squares in Figure 9). With this GUI, there are only a few user interactions required to obtain the segmentation of interest. First, the user provides a cervigram. Then, the user clicks the appropriate button for each desired segmentation. The cervigram, along with related parameters, is then submitted to the server. The server executes the request and sends the segmented boundaries back to the client. The boundaries are then displayed on the image. A working prototype of CST that includes fundamental functionalities has been created, and more work is planned for segmentation algorithm development and interface usability. (For example, the current version does not allow users to modify or edit the automatic segmentation results.) The modification of the user interface, the additions of new algorithms, and the frequent update of segmentation algorithms has become relatively easy with the flexibility offered by the system. Most importantly, it provides a convenient way to make the Matlab coded algorithms executable over the Web and relieves inexperienced users from the work involved in Matlab installation and programming. It also saves researchers effort and time in re-programming the complex MATLAB segmentation algorithms in Java.
Figure 10.
The system architecture of CST
5. Integration of Tools
Jointly, BMT, MOSES and CST compose a set of complementary tools which are important for uterine cervix image segmentation. They each address one crucial subtopic in the image segmentation domain. These three subtopics are strongly interconnected. BMT collects region data from medical experts. The data acquired by BMT can be used to validate the automated segmentation algorithms, to compare multiple segmentation methods, to train a supervised segmentation approach, and to assist in the analysis of observer variability. It also allows image processing researchers to gain more knowledge of the visual appearance of cervix tissues from clinical experts’ graphical markings and text annotations. MOSES quantitatively measures the variability among multiple observers and specifically addresses various situations with different prior knowledge scenarios. It provides an automatic approach for objective evaluation of multiple observer performance on the data collected by BMT. CST incorporates automatic segmentation algorithms and allows a direct visual examination of the automated segmentation results by domain experts. The accuracy of the automatic segmentation results output by CST can also be evaluated by MOSES. Although each tool runs independently and has different characteristics, it is desirable to unify them since all of them are involved in creating or processing region data which can be represented in the format of contours. It is very convenient to be able to seamlessly exchange data among these tools. For example, region data output by BMT and CST can be loaded into MOSES, and the data from BMT can be loaded into CST, directly, and without any reformatting. This data interchange capability is achieved by representing the region data of all three tools in Extensible Markup Language (XML) format. The XML standard and the corresponding set of techniques for generating, validating, formatting, transforming and extracting XML documents is now widely used and offers many advantages, such as flexibility and portability, with respect to information interchange. For exchange of the XML region data files among the three tools, the data elements and their relationships used in both CST and MOSES are designed to be consistent to that of BMT. The XML syntax defined is simple and straightforward and contains elements for describing the image and elements for describing the boundary information. Through the use of this consistent data format, BMT, MOSES, and CST are integrated into a suite of software for cervigram segmentation and analysis.
6. Discussion
The development of automatic segmentation requires effective evaluation. Objective evaluation methods require reliable “ground truth”. Subjective evaluation methods involve the visual examination and judgments of domain experts. We put effort in both approaches: BMT and MOSES can be used to obtain ground truth data, and CST is for visual inspection. Both evaluation methods rely on the involvement of medical experts. Therefore, it is very important to foster an active and close collaboration between the engineering and medical expert communities. These three tools represent our efforts towards this objective. They are developed with an “expert-centered” philosophy that aims to increase the satisfaction and productivity of medical experts using engineering tools, and to better leverage their expertise in advancing and refining the engineering tool development. Major considerations are to make these tools easy to access and use by medical experts. With these considerations in mind, the tools have been created with Web-accessible clients and with high emphasis placed on friendliness of the interfaces to the medical community. In addition, we have addressed several other practical issues, such as the need for convenient exchange of data among our tools, the need for compression and transmitting large images, and the integration of Java and Matlab capabilities. Although this set of tools was developed for the analysis of uterine cervix images, they are not limited to this image type and may be extended to other domains which require region-based data analysis. The generality of BMT has been demonstrated by its extension to be used in dermatology study. MOSES can be applied to any image modality. The CST is more cervigram-oriented, but several aspects of its implementation and design, such as how to conveniently deploy Matlab-coded algorithms to multiple sites, can be adopted by many image processing research systems.
7. Conclusions
Automatic segmentation of uterine cervix images is important for indexing and analyzing the content of the images. It is a very challenging task and it is still in its early stages. This paper represents our significant research and development work in this area. It describes the design and implementation of three important systems, BMT, CST, and MOSES, which we developed for analyzing uterine cervix images. It covers three crucial aspects of medical image segmentation: the development of segmentation algorithms, the collection of ground truth data, and the evaluation of multiple observers’ segmentation. To the best of our knowledge, this is the first paper in the literature which addresses these three topics in a unified manner. For image analysis researchers and engineers, a key issue is to enhance the effectiveness of collaboration with physicians and biomedical researchers, which is crucial for algorithm improvement and the eventual adoption of the applications by the end users. Toward this goal, in addition to developing image processing algorithms, we also focus on designing and developing associated tools which are easy-to-use and convenient from the standpoint of biomedical collaborators. Although these tools have their own unique characteristics and are customized for uterine cervix image analysis, they may be extended to other image analysis applications.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM), and Lister Hill National Center for Biomedical Communications (LHNCBC).
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors do not hold any conflicts of interest that could inappropriately influence this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duncan JS, Ayache N. Medical image analysis: progress over two decades and the challenges ahead. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2000 Jan;22(1):85–106. [Google Scholar]
- 2.Maulik U. Medical image segmentation using genetic algorithms. IEEE Transactions on Information Technology. 2009;13(2):166–173. doi: 10.1109/TITB.2008.2007301. [DOI] [PubMed] [Google Scholar]
- 3.Yao W, Abolmaesumi P, Greenspan M, Ellis RE. An estimation/correction algorithm for detecting bone edges in CT images. IEEE Transactions on Medical Imaging. 2005;24(8):997–1010. doi: 10.1109/TMI.2005.850541. [DOI] [PubMed] [Google Scholar]
- 4.Ray N, Acton ST, Altes T, de Lange EE, Brookeman JR. Merging parametric active contours within homogeneous image regions for MRI-based lung segmentation. IEEE Transactions on Medical Imaging. 2003;22(2):189–199. doi: 10.1109/TMI.2002.808354. [DOI] [PubMed] [Google Scholar]
- 5.Parker BJ, Feng D. Graph-based Mumford-Shah segmentation of dynamic PET with application to input function estimation. IEEE Transactions on Nuclear Science. 2005;52(1):79–89. [Google Scholar]
- 6.Falcao AX, Bergo FPG. Interactive volume segmentation with differential image foresting transforms. IEEE Transactions on Medical Imaging. 2004;23(9):1100–1108. doi: 10.1109/TMI.2004.829335. [DOI] [PubMed] [Google Scholar]
- 7.Steiner C. Cervicography: a new method for cervical cancer detection. American Journal of Obstetrics and Gynecology. 1989;139(7):815–825. doi: 10.1016/0002-9378(81)90549-4. [DOI] [PubMed] [Google Scholar]
- 8.Sellors JW, Sankaranarayanan R. In: Colposcopy and Treatment of Cervical Intraepithelial Neoplasia - A Beginner’s Manual. Sellors JW, Sankaranarayanan R, editors. International Agency for Research on Cancer; France: 2003. [Google Scholar]
- 9.Bratti MC, Rodriguez AC, Schiffman M, Hildesheim A, Morales J, Alfaro M, Guillen D, Hutchinson M, Sherman ME, Eklund C, Schussler J, Buckland J, Morera LA, Cardenas F, Barrantes M, Perez E, Cox TJ, Burk RD, Herrero R. Description of a seven-year prospective study of human papillomavirus infection and cervical neoplasia among 10, 000 women in Guanacaste, Costa Rica. Revista Panamericana de Salud Pública/Pan American Journal of Public Health. 2004;15(2):75–89. doi: 10.1590/s1020-49892004000200002. [DOI] [PubMed] [Google Scholar]
- 10.Schiffman M, Adrianza ME. ASCUS-LSIL Triage Study: Design, methods and characteristics of trial participants. Acta Cytologica. 2000;44(5):726–742. doi: 10.1159/000328554. [DOI] [PubMed] [Google Scholar]
- 11.Long LR, Antani S, Thoma GR. Image informatics at a national research center. Computerized Medical Imaging and Graphics. 2005 February;29(2):171–193. doi: 10.1016/j.compmedimag.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Bopf M, Coleman T, Long LR, Antani S, Thoma GR. An architecture for streamlining the implementation of biomedical text/image databases on the web. Proceedings of the 17th IEEE Symposium on Computer-Based Medical Systems; June 2004.pp. 563–568. [Google Scholar]
- 13.Xue Z, Long LR, Antani S, Thoma GR. A Web-accessible content-based cervicographic image retrieval system. Proceedings of SPIE Medical Imaging Conference; April 2008.pp. 07-1–9. [Google Scholar]
- 14.Xue Z, Antani S, Long LR, Jeronimo J, Schiffman M, Thoma G. CervigramFinder: a tool for uterine cervix cancer research. Proceedings of the AMIA Annual Fall Symposium; 2009. [Google Scholar]
- 15.Xue Z, Long LR, Antani S, Thoma GR. A system for searching uterine cervix images by visual attributes. Proceedings of the 22nd IEEE International Symposium on Computer Based Medical Systems; August, 2009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenspan H, Gordon S, Zimmerman G, Lotenberg S, Jeronimo J, Antani S, Long R. Automatic detection of anatomical landmarks in uterine cervix images. IEEE Transactions on Medical Imaging. 2009;28(3):454–468. doi: 10.1109/TMI.2008.2007823. [DOI] [PubMed] [Google Scholar]
- 17. [last accessed on April 14th, 2010]; http://archive.nlm.nih.gov/proj/impactOfTools.html.
- 18.Jeronimo J, Long R, Neve L, Michael B, Antani S, Schiffman M. Digital tools for collecting data from cervigrams for research and training in colposcopy. Journal of Lower Genital Tract Disease. 2006 January;10(1):16–25. doi: 10.1097/01.lgt.0000194057.20485.5a. [DOI] [PubMed] [Google Scholar]
- 19.Jeronimo J, Schiffman M, Long LR, Neve L, Antani S. A tool for collection of region based data from uterine cervix images for correlation of visual and clinical variables related to cervical neoplasia. Proceedings of the 17th IEEE Symposium on Computer-Based Medical Systems; 2004. pp. 558–562. [Google Scholar]
- 20.Celebi ME, Schaefer G, Iyatomi H, Stoecker WV, Malters JM, Grichnik JM. An improved objective evaluation measure for border detection in dermoscopy images. Skin Res Technol. 2009;15(4):444–450. doi: 10.1111/j.1600-0846.2009.00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joel G, Schmid-Saugeon P, Guggisberg D, Cerottini JP, Braun R, Krischer J, Saurat JH, Murat K. Validation of segmentation techniques for digital dermoscopy. Skin Res Technol. 2002;8(4):240–249. doi: 10.1034/j.1600-0846.2002.00334.x. [DOI] [PubMed] [Google Scholar]
- 22.Gordon S, Lotenberg S, Long LR, Antani S, Jeronimo J, Greenspan H. Evaluation of uterine cervix segmentations using ground truth from multiple experts. Computerized Medical Imaging and Graphics. 2009 February;33(3):205–216. doi: 10.1016/j.compmedimag.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Y, Wang W, Huang X, Lopresti D, Long LR, Antani S, Xue Z, Thoma G. Balancing the role of priors in multi-observer segmentation evaluation. Journal of Signal Processing Systems. 2008 May;55(1–3):158–207. doi: 10.1007/s11265-008-0215-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Y, Huang X, Lopresti D, Long LR, Antani S, Xue Z, Thoma G. Web-based multi-observer segmentation evaluation tool. Proceedings of the 21st IEEE International Symposium on Computer-Based Medical Systems; June 2008.pp. 167–169. [Google Scholar]
- 25.Warfield SK, Zou HK, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Transactions on Medical Imaging. 2004;23(7):903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon S, Zimmerman G, Long R, Antani S, Jeronimo J, Greenspan H. Content analysis of uterine cervix images: initial steps towards content based indexing and retrieval of cervigrams. Proceedings of SPIE Medical Imaging; 2006. pp. 1549–1556. [Google Scholar]
- 27.Srinivasan Y. Internal Progress Report on Contract Number 476-MZ-501319, Computer Vision and Image Analysis Lab (CVIAL) Texas Tech University; 2006. NLM cervigram segmentation. [Google Scholar]
- 28.Zimmerman G, Greenspan H. Automatic detection of specular reflections in uterine cervix images. Proceedings of SPIE Medical Imaging; 2006. pp. 2037–2045. [Google Scholar]
- 29.Lotenberg S, Gordon S, Greenspan H. Shape priors for segmentation of the cervix region within uterine cervix images. Journal of Digital Imaging. 2009;22(3):286–296. doi: 10.1007/s10278-008-9134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmerman G, Gordon S, Greenspan H. Automatic landmark detection in uterine cervix images for indexing in a content-retrieval system. Proceedings of IEEE International Symposium on Biomedical Imaging; 2006. pp. 1348–1351. [Google Scholar]
- 31.Zimmerman G, Gordon S, Greenspan H. Content-based indexing and retrieval of uterine cervix images. Proceedings of the 23rd IEEE Convention of Electrical and Electronics Engineers; Israel. 2004. pp. 181–185. [Google Scholar]
- 32.Huang X, Wang W, Xue Z, Antani S, Long LR, Jeronimo J. Tissue classification using cluster features for lesion detection in digital cervigrams. Proceedings of SPIE Medical Imaging; 2008. p. 69141Z. [Google Scholar]
- 33.Gordon S, Greenspan H. Segmentation of non-convex regions within uterine cervix images. Proceedings of IEEE International Symposium on Biomedical Imaging; 2007. pp. 312–315. [Google Scholar]
- 34.Alush A, Greenspan H, Goldberger J. Lesion detection and segmentation in uterine cervix images using an arc-level MRF. Proceedings of IEEE International Symposium on Biomedical Imaging; 2009. [Google Scholar]
- 35.Artan Y, Huang X. Combining multiple 2ν-SVM classifiers for tissue segmentation. Proceedings of IEEE International Symposium on Biomedical Imaging; 2008. pp. 488–491. [Google Scholar]
- 36.Tulpule B, Hernes DL, Srinivasan Y, Mitra S, Sriraja Y, Nutter BS, Phillips B, Long RL, Ferris DG. A probabilistic approach to segmentation and classification of neoplasia in uterine cervix images using color and geometric features. Proceedings of SPIE Medical Imaging; Feb, 2005. pp. 995–1003. [Google Scholar]
- 37.Srinivasan Y, Nutter BS, Mitra S, Phillips B, Sinzinger E. Classification of cervix lesions using filter bank-based texture mode. Proceedings of the 19th IEEE Symposium on Computer-Based Medical; 2006. pp. 832–840. [Google Scholar]
- 38.Xue Z, Long LR, Antani S, Jeronimo J, Thoma GR. Segmentation of mosaicism in cervicographic images using support vector machines. Proceedings of SPIE Medical Imaging; 2009. p. 72594X-72594X-10. [Google Scholar]
- 39.Srinivasan Y, Gao F, Tulpule B, Yang S, Mitra S, Nutter B. Segmentation and classification of cervix lesions by pattern and texture analysis. International Journal of Intelligent Systems Technologies and Applications. 2006;1(3):234–246. [Google Scholar]
- 40.Guo J, Hughes B, Mitra S, Nutter B. Ultra high resolution image coding and ROI viewing using line-based backward coding of wavelet trees (L-BCWT). 27th Picture Coding Symposium; May 6–8; Chicago, Illinois. 2009. [Google Scholar]