Abstract
Histone deacetylase inhibitors (HDACi) are potent anti-cancer agents for variety of cancer types. Suberoylanilide hydroxamic acid (SAHA) has been approved as a drug to treat cutaneous T cell lymphoma, and the combination of HDACi and other agents have been actively tested in many clinical trials. Adenovirus 5 early region 1A (E1A) has been shown to exhibit high tumor suppressor activity, and gene therapy using E1A has been tested in clinical trials. Here, we showed that proapoptotic activity of HDACi was robustly enhanced by E1A in multiple cancer cells, but not in normal cells. Moreover, we showed that combination of E1A gene therapy and SAHA showed high therapeutic efficacy with low toxicity in vivo ovarian and breast xenograft models. SAHA downregulated Bcl-XL and upregulated proapoptotic BH3-only protein Bim, whose expression was further enhanced by E1A in cancer cells. These alterations of Bcl-2 family proteins were critical for apoptosis induced by the combination in cancer cells. SAHA enhanced acetylation of histone H3 in Bim promoter region, while E1A upregulated Egr-1, which was directly involved in Bim transactivation. Together, our results provide not only a novel insight into the mechanisms underlying anti-tumor activity of E1A, but also a rationale for the combined HDACi and E1A gene therapy in future clinical trials.
Keywords: HDAC inhibitors, E1A, apoptosis, Bim, gene therapy
Introduction
Cancer gene therapy is a developing therapeutic approach in which therapeutic cDNAs, antisense oligo DNAs or short interfering RNAs are systemically or locally administrated to patients to induce cell death or growth arrest of cancer cells that show an inadequate response to conventional chemotherapeutic drugs (Lo et al., 2005; Stoff-Khalili et al., 2006; Pirollo and Chang, 2008).
The human adenovirus type 5 early region 1A (E1A) associates with anti-cancer activities through multiple molecular mechanisms (Brader et al., 1997; Frisch, 2004; Lo et al., 2005; Liao et al., 2007). Studies using liposome or viral vector as a gene delivery vehicle have shown that the E1A gene inhibits tumor development effectively and prolongs survival in multiple orthotopic animal models (Yu et al., 1995; Ueno et al., 2002; Liao et al., 2004). On the basis of the safety study of E1A/liposome gene therapy, together with the high therapeutic efficacy in animal models (Xing et al., 1997, 1998), several clinical trials using E1A/liposome have been carried out in cancers of the breast, ovary and head and neck, showing the feasibility of E1A gene therapy in human (Hortobagyi et al., 2001; Yoo et al., 2001; Villaret et al., 2002; Madhusudan et al., 2004). E1A was also shown to sensitize cancer cells to chemotherapeutic drugs and enhance cell death (Lowe et al., 1993; Brader et al., 1997; Samuelson and Lowe, 1997; Cook and Routes, 2005; Liao et al., 2007). Particularly, based on our preclinical data that E1A increases the cytotoxicity and anti-tumor activity of paclitaxel in vitro and in vivo (Ueno et al., 1997, 2000; Liao et al., 2004), combined paclitaxel and E1A gene therapy is currently being investigated in a clinical trial for ovarian cancer patients.
As high expression of histone deacetylase (HDAC) has been reported in some types of cancer, HDAC is considered a promising target for cancer therapy (Yang and Seto, 2008). HDAC inhibitors (HDACi) are a novel class of anti-cancer agents that induce apoptosis more effectively in transformed cells than in normal cells (Burgess et al., 2004; Minucci and Pelicci, 2006; Xu et al., 2006; Marks and Breslow, 2007). Numerous HDACi and their combination with other chemotherapeutic drugs have been tested in clinical trials, and suberoylanilide hydroxamic acid (SAHA, vorinostat) has been approved as an anti-cancer drug to treat cutaneous T cell lymphoma (Marks and Breslow, 2007; Lane and Chabner, 2009).
HDACi induce hyperacetylation of core histone, resulting in modulation of gene expression through chromatin remodeling (Bolden et al., 2006). Anti-apoptotic Bcl-2 family proteins such as Bcl-2, Bcl-XL and Mcl-1 are downregulated, whereas pro-apoptotic proteins such as the death receptors DR5 and Fas and the pro-apoptotic Bcl-2 family proteins Bim and Bmf are upregulated in response to HDACi (Bolden et al., 2006).
In this study, we found that E1A efficiently enhanced cytotoxic effects of SAHA in a variety of human cancer cells. Combined treatment using E1A gene therapy and SAHA also showed highly effective anti-tumor activity in ovarian and breast cancer xenograft models, with lesser toxicity than that of E1A and paclitaxel. Moreover, we showed here, the underlying molecular mechanisms of HDACi-induced apoptosis enhanced by E1A. These results indicate that the combination therapy of E1A gene therapy and SAHA would be an attractive therapeutic strategy for treating human cancers.
Results
The combination of E1A plus SAHA effectively induces apoptosis in human cancer cells but not in normal cells
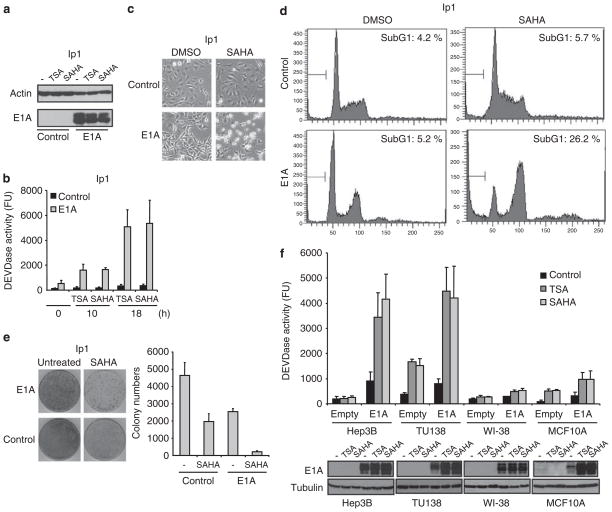
To obtain a general idea of which anti-cancer drugs are most effectively potentiated by E1A gene therapy in vitro, we transiently transfected human cancer cells with either E1A expression or empty plasmid, and then treated the cells with various anti-cancer drugs, including 5-fluorouracil, cisplatin, etoposide, paclitaxel and SAHA and assessed apoptosis by measuring the caspase activity (Supplementary Figure S1). Consistent with previous reports, we observed the sensitization effect of E1A on chemotherapeutic drugs. Interestingly, we found that the sensitizing effect of E1A on SAHA was stronger than the effect on the other chemotherapeutic drugs tested. The encouraging sensitization effect of E1A on SAHA prompted us to further investigate potential therapeutic effects of combination therapy using E1A gene therapy and SAHA. As a proof of concept, we compared the effects of SAHA and trichostatin A (TSA), another HDACi, on the ovarian cancer cell line SKOV3-ip1 and its E1A stable transfectant (Figures 1a and b) (Yu et al., 1993). HDACi strongly induced caspase activation in E1A-expressing cells, whereas control cells were relatively resistant to HDACi (Figures 1a and b). The sensitization effect of E1A in SAHA-induced apoptosis was also supported by the morphological change and the increase in the sub-G1 population (Figures 1c and d). Furthermore, the standard colony formation assay showed that the inhibitory effect of SAHA on colony formation of SKOV3-ip1 cells was much higher in E1A-expressing cells than the control cells, indicating that E1A affects the long-term survival after SAHA treatment (about 60 vs 90% reduction, Figure 1e). Similar results were obtained in the breast cancer cell line MDA-MB-231 and its E1A stable transfectant (Supplementary Figures S2a and b). To further confirm the E1A/SAHA sensitization effect, we also examined the effects of this combination in other ovarian cancer (2774-c10) and breast cancer (MDA-MB-468) cells, as well as in hepatocarcinoma (Hep3B) and head and neck cancer (TU138) cell lines (Figure 1f and Supplementary Figures S2c and d). In addition, to determine whether combination therapy using E1A and SAHA, is less toxic to normal cells, we also tested human normal fibroblasts (WI-38) and human normal mammary epithelial cells (MCF10A). We transiently transfected these cells with either E1A expression or empty plasmid, and determined caspase activity and E1A protein expression after HDACi treatment. The results showed that E1A robustly enhanced the caspase activation induced by HDACi in the cancer cell lines examined (Figure 1f). It is interesting to note that the pro-apoptotic effect of the combination of E1A and HDACi was much weaker in the two non-cancer cell lines than in cancer cells (Figure 1f). Together, these results show that E1A sensitizes SAHA to induce apoptosis in a variety of human cancer cells but not in normal cells.
Figure 1.
Adenovirus 5 early region 1A (E1A) enhances HDACi-induced cell death in human cancer cells. (a) E1A and actin expression in SKOV3-ip1 control or E1A cells was verified by immunoblot analysis. (b) SKOV3-ip1 control or E1A cells were treated with 250 nM of trichostatin A (TSA) or 5 μM of suberoylanilide hydroxamic acid (SAHA) for the indicated periods, and caspase-3 activity was determined by caspase assay, using a fluorescence substrate. (c) The cellular morphology of SKOV3-ip1 control or E1A cells treated with dimethyl sulfoxide (DMSO) or 5 μM of SAHA for 24 h. (d) SKOV3-ip1 control or E1A cells were treated with DMSO or 5 μM of SAHA for 20 h. The cells were then fixed and stained with propidium iodide, followed by flow cytometric analysis. The percentage of cells with a sub-G1 DNA content is shown within each box. (e) SKOV3-ip1 control or E1A cells were treated with 5 μM of SAHA for 24 h. Cellular sensitivity to SAHA was determined by using the clonogenic survival assay. The colony numbers were counted and shown in the bar graph (n=3). (f) Hep3B, TU138, WI-38 and MCF10A cells were transiently transfected with either empty or E1A expression plasmid and treated with 250 nM of TSA or 5 μM of SAHA for 16 h. Capase-3 activity was then determined by caspase assay using fluorescence substrate. E1A and tubulin expression in these cells was verified by immunoblot analysis.
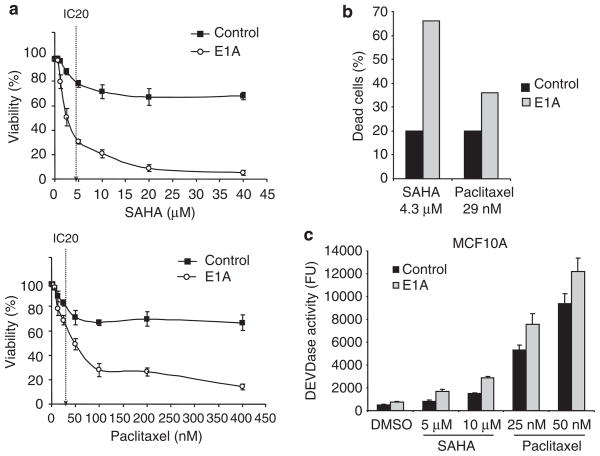
As the use of combination of E1A gene therapy and paclitaxel is under investigation in a clinical trial, we next compared the combined effects of E1A and SAHA with the combined effects of E1A and paclitaxel. We treated SKOV3-ip1 or MDA-MB-231 E1A stable and control cells with different concentrations of SAHA or paclitaxel and determined the cell viability with the trypan blue dye exclusion assay (Figure 2 and Supplementary Figure S3). In SKOV3-ip1 control cells, the drug concentrations that induced 20% cell death (IC20) were about 4.3 μM for SAHA and 29 nM for paclitaxel (Figure 2a). At these concentrations, SAHA killed 66% of SKOV3-ip1-E1A cells, whereas paclitaxel killed only 36% of the cells, indicating that the sensitization effect of E1A on SAHA is stronger than the effect on paclitaxel (Figure 2b). Similarly, E1A enhanced SAHA-induced cell death more effectively than paclitaxel-induced cell death in MDA-MB-231 cells (Supplementary Figure S3). To compare the toxicity of the combination of E1A and SAHA with that of the combination of E1A and paclitaxel in normal cells, we transiently transfected MCF10A cells with either E1A expression plasmid or empty vector and treated the cells with either SAHA or paclitaxel. Cytotoxicity was then evaluated by using the caspase assay (Figure 2c). Although SAHA induced more cell death than paclitaxel in SKOV3-ip1-E1A or MDA-MB-231-E1A stable cells at the drug concentrations used here (Figure 2c), the combination of paclitaxel and E1A showed more toxicity than the combination of E1A and SAHA did in MCF10A cells (Figure 2c). Together, these results indicate that the combination of E1A and SAHA has an high efficacy against the cancer cells, but less toxicity in normal cells than combined E1A and paclitaxel.
Figure 2.
Adenovirus 5 early region 1A (E1A) enhances suberoylanilide hydroxamic acid (SAHA)-induced cell death more effectively than paclitaxel-induced cell death in cancer cells but not in normal cells. (a) SKOV3-ip1 control or E1A stable cells were treated with the indicated concentrations of SAHA or paclitaxel for 48 h, and viability was determined by the trypan blue dye exclusion assay. IC20 in control cells was indicated as a dotted line. (b) Comparison of cell death of SKOV3 ip1-E1A cells at the drug concentrations that induced 20% cell death (4.3 μM for SAHA and 29 nM for paclitaxel) in SKOV3-ip1 control cells. (c) MCF10A cells were transiently transfected with either empty or E1A expression plasmid and treated with 5 or 10 μM of SAHA or 25 or 50 nM of paclitaxel for 24 h. Caspase-3 activity was then determined by caspase assay using fluorescence substrate.
The combinational effect of E1A gene therapy and SAHA in orthotopic xenograft model of ovarian and breast cancers
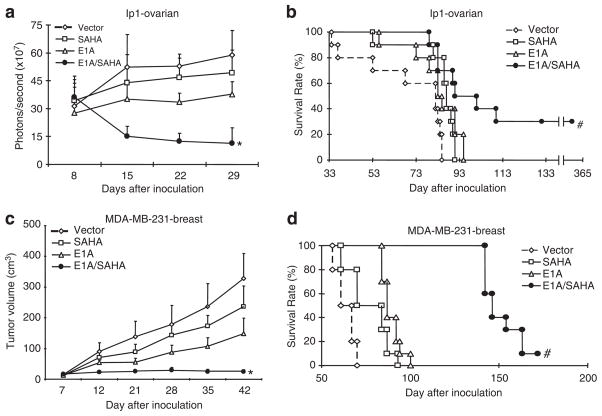
Next, to assess the effects of the combination of liposome/E1A gene therapy and SAHA in vivo, we determined tumor growth and survival in orthotopic xenograft models of human ovarian and breast cancers. In the ovarian cancer model, we intraperitoneally inoculated mice with SKOV3-ip1-luciferase stable cells; tumor formation, growth and reduction were monitored by a bioluminescent imaging system as described previously (Day et al., 2006; Xie et al., 2007). Under the conditions, in which each single treatment did not suppress tumor growth at the respective doses, tumor growth was readily suppressed by the combination treatment (Figure 3a). In addition, the mice treated with the combination survived significantly longer than mice treated with either E1A/liposome or SAHA alone, and 30% of treated mice live tumor-free for longer than 1 year (Figure 3b). We observed similar therapeutic effects in the MDA-MB-231 breast cancer mammary fat pad xenograft model (Figures 3c and d). Together, the in vivo results also support the sensitization effect of E1A gene therapy on SAHA.
Figure 3.
The combination of adenovirus 5 early region 1A (E1A) and suberoylanilide hydroxamic acid (SAHA) suppresses tumor growth in vivo. (a) Mice bearing SKOV3-ip1-luciferase tumors were treated with vector/liposome (control), E1A/liposome (E1A, 15 μg DNA/mouse), vector/liposome plus SAHA (SAHA 100 mg/kg) or E1A/liposome plus SAHA (E1A/SAHA), and luciferase signals in vivo were monitored. *P<0.01 vs single treatments. (b) Survival curves of animals used in (a). All treatments were terminated at day 31 after inoculation. #P<0.04 vs single treatments. (c) Mice bearing MDA-MB-231 breast tumors were treated with vector/liposome (control), E1A/liposome (E1A, 15 μg DNA/mouse), vector/liposome plus SAHA (SAHA, 100 mg/kg) or E1A/liposome plus SAHA (E1A/SAHA), and tumor sizes were measured. *P<0.01 vs single treatments. (d) Survival curves of animals used in (c). All treatments were terminated at day 31 after inoculation. and #P<0.01 vs single treatments.
To compare the toxicity of the combination therapy of E1A gene therapy and SAHA with that of E1A gene therapy and paclitaxel, we treated the mice with high doses of the drugs in combination. E1A expression plasmid/liposome concentration (50 μg DNA/mouse) was determined based on our previous reports (Xie et al., 2007, 2009). The concentrations of SAHA and paclitaxel used in clinic for human are around 400mg/m2 (daily) and 100 mg/m2 (once in 2–3 week), respectively. Thus, we tested SAHA: 130 mg/mouse (daily) and paclitaxel: 32 mg/mouse (once), that were calculated based on a standard conversion formula between human and mouse (Freireich et al., 1966; Reagan-Shaw et al., 2008). All the mice treated with E1A liposome plus paclitaxel died within 3 days, whereas no mice died after receiving E1A gene therapy plus SAHA (Supplementary Figure S4a). To evaluate liver toxicity in the mice treated with the clinical dose of E1A/liposome and SAHA, aspartate aminotransfrase level was determined in blood samples from the mice treated with E1A/liposome plus SAHA (Supplementary Figure S4b). The aspartate aminotransfrase level remained in the normal range, suggesting that the combination of E1A/liposome and SAHA does not cause much liver toxicity. Overall, the combination of E1A gene therapy and SAHA effectively reduced the tumor growth and prolonged animal survival with much better safety profile than the combination of E1A gene therapy and paclitaxel.
Bim and Bcl-XL are the critical mediators of apoptosis induced by E1A and SAHA
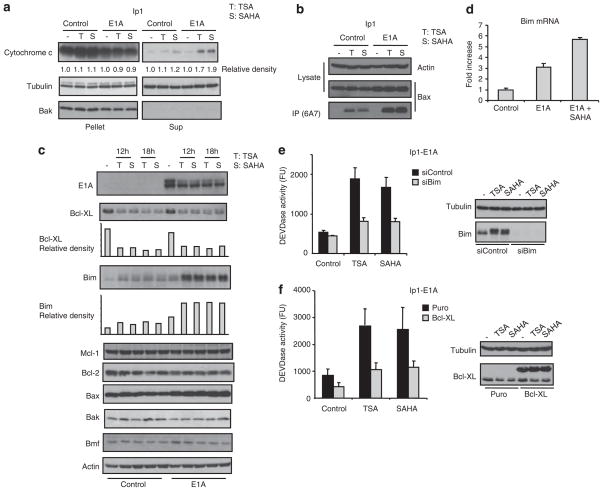
To determine the underlying mechanisms of apoptosis induced by the combination of E1A and SAHA, we first examined whether E1A might enhance cytochrome c release and Bax conformational change and therefore promote HDACi-induced apoptosis. Indeed, both HDACi-induced cytochrome c release (1.7–1.9 vs 1.1–1.2, Figure 4a) and Bax conformational change, which can be assessed by immunoprecipitation with a specific monoclonal antibody (6A7) that recognizes the active form of Bax, were promoted by E1A in SKOV3-ip1 (Figures 4a and b) and MDA-MB-231 cells (Supplementary Figures S5a and b). Next, to understand the molecular pathway regulating HDACi-induced Bax conformational change enhanced by E1A, we determined the expression of the Bcl-2 family proteins that have previously been shown to be involved in HDACi-induced apoptosis (Bolden et al., 2006). Immunoblot analysis in SKOV3-ip1 control and E1A cells treated or untreated with HDACi showed that Bcl-XL was downregulated and Bim was upregulated in response to HDACi (Figure 4c). In SKOV3-ip1 cells, E1A enhanced Bim expression but did not alter the Bcl-XL expression level (Figure 4c). RT–PCR analysis indicated that Bim upregulation and Bcl-XL downregulation occurred at the mRNA level (Figure 4d and Supplementary Figure S5c). Similar results were obtained when other cancer cell lines were tested by western blot analysis using Bim and Bcl-XL antibodies (Supplementary Figures S5d and e). Interestingly, in the normal MCF10A cells, E1A neither enhanced nor sensitized HDACi-induced Bim expression, which may explain, at least in part, why normal cells are relatively resistant to the combination treatment (Figure 1f and Supplementary Figure S5f). To determine the role of Bim and Bcl-XL in apoptosis induced by E1A and HDACi, we knocked down Bim or overexpressed Bcl-XL in SKOV3-ip1-E1A cells, and determined the effects of HDACi, by using the caspase assay. We found that caspase activation induced by HDACi was reduced by Bim knockdown or Bcl-XL overexpression in SKOV3-ip1-E1A cells (Figures 4e and f). Thus, these results suggest that both Bim and Bcl-XL have a critical role in apoptosis induced by the combination of E1A and SAHA. Apoptosis initiation is controlled by the balance between anti- and pro-apoptotic Bcl-2 family proteins (Youle and Strasser, 2008). To further confirm the role of Bim upregulation in apoptosis induced by the combination of SAHA and E1A, we transfected reduced amounts of short interfering RNA against Bim in SKOV3-ip1-E1A cells to knockdown Bim expression comparable to the level of Bim in the untreated cells (Supplementary Figure 6a). We found that reduction of Bim attenuated caspase activation induced by SAHA, indicating that induction of Bim might be critical for caspase activation induced by SAHA and E1A, and that E1A sensitizes SAHA-induced apoptosis partially by enhanced Bim expression. As we did not observe a complete inhibition of caspase activation when Bim was knocked down, it is likely that the loss of Bim expression raised the threshold, required for apoptosis and that other BH3-only Bcl-2 family proteins may also contribute to SAHA and E1A-induced apoptosis.
Figure 4.
The combination of adenovirus 5 early region 1A (E1A) and suberoylanilide hydroxamic acid (SAHA) activates the mitochondrial pathway of apoptosis though upregulating Bim and downregulating Bcl-XL. (a–d) SKOV3-ip1 control or E1A cells were treated with 250 nM of trichostatin A (TSA) or 5 μM of SAHA. (a) Sixteen hours after treatment, the cells were subjected to subcellular fractionation to separate the cytosolic and membrane fractions. Each fraction was analyzed by immunoblot analysis with the indicated antibodies. Bak is a mitochondria marker and could not be detected in the cytosolic fraction. (b) Sixteen hours after treatment, the Bax conformational change was determined by immunoprecipitation with anti-Bax 6A7 antibody that recognizes only the active form of Bax. (c) The expressions of the Bcl-2 family proteins were determined by immunoblot analysis with the indicated antibodies. (d) Six hours after treatment, the expressions of Bim mRNA were assessed by quantitative RT–PCR. (e) SKOV3-ip1 E1A cells were transiently transfected with siRNA oligo against Bim or control non-specific siRNA. Thirty-six hours after transfection, the cells were treated with 250 nM of TSA or 5 μM of SAHA for 12 h and subjected to caspase assay. Bim and tubulin expressions are shown in the right panel. (f) SKOV3-ip1 E1A stable cells were stably transfected with myc-tagged Bcl-XL or empty plasmid. The cells were then treated with 250 nM of TSA or 5 μM of SAHA for 14 h and subjected to caspase assay. Bcl-XL and tubulin expressions are shown in the right panel.
Bim is upregulated by E1A-induced Egr-1 and SAHA-induced chromatin remodeling
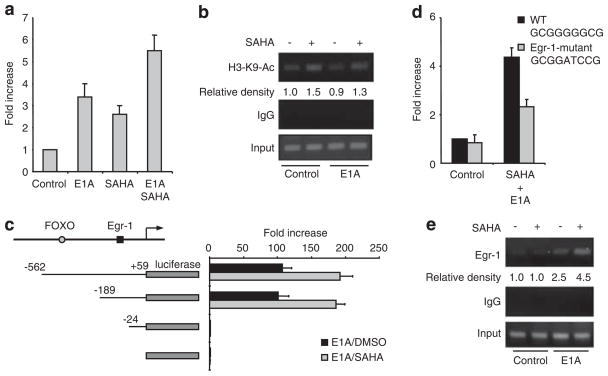
Next, we asked how Bim is upregulated by E1A and HDACi. We first examined the response of the Bim promoter region to E1A and SAHA by reporter assay (Figure 5a). The transactivation of Bim promoter was enhanced by SAHA, E1A and the combination well consistent with western blot and RT–PCR data (Figures 4c and d). HDACi induce hyperacetylation of core histones, resulting in the modification of chromatin structures and augmentation of transcriptional activity. We, therefore examined the acetylation level of histone H3 on the Bim promoter region by the chromatin immunoprecipitation assay using acetylated histone H3-K9 antibody. We found that the acetylation level of histone H3 around the Bim promoter was enhanced by SAHA, supporting that SAHA upregulates Bim through the hyperacetylation of histone on the Bim promoter (Figure 5b). However, interestingly, E1A does not affect the acetylation of Bim promoter regions, suggesting that other mechanisms exist for the combinational effect of E1A and SAHA on Bim expression. To determine the transcription factor(s) involved in Bim upregulation induced by E1A plus SAHA, we further analyzed the promoter region of Bim. Luciferase assay with the deletion mutants of Bim promoter narrowed down the potential transcription factor binding sites to the sequence between −24 and −189, which does not include FOXO3a binding site, previously identified (Figure 5c). We searched this sequence and found the Egr-1 binding sequence (GCGGGGGCG) in this region of both the human and mouse Bim promoters. Therefore, we tested the possible role of Egr-1 in Bim upregulation by E1A plus SAHA. We mutated the Egr-1 binding site in the Bim promoter and determined its response to E1A plus SAHA. Indeed, the SAHA/E1A-induced Bim promoter activity was significantly reduced in the Bim promoter containing the mutation in the Egr-1 binding site (Figure 5d). Similarly, dominant negative Egr-1 attenuated Bim promoter activation by SAHA in E1A-expressing cells (Supplementary Figure S6). Consistently, chromatin immunoprecipitation analysis using Egr-1 antibody showed that Egr-1 binds to the Bim promoter, which was enhanced by both SAHA and E1A (Figure 5e). These results suggest that Egr-1 is directly involved in Bim transcriptional regulation.
Figure 5.
Egr-1 binding site in Bim promoter is critical for Bim transactivaiton by the combination of E1A and HDACi. (a) SKOV3-ip1 cells were co-transfected with the Bim-promoter luciferase plasmid together with renilla luciferase expression plasmid (internal control) and E1A expression plasmid/empty vector. Twenty-four hours after transfection, the cells were treated with 5 μM of SAHA for an additional 6 h and subjected to the dual luciferase assay. The luciferase activity is shown relative to that of untreated control transfected with empty vector. (b) SKOV3-ip1 control or E1A cells were treated with SAHA for 6 h, and the chromatin immunoprecipitation (ChIP) assay was performed using control rabbit immunoglobulin G or anti-acetyl-histon H3-K9 antibody. Polymerase chain reaction was performed for Bim promoter. (c) Transactivation of the indicated deletion mutants of Bim promoter-luciferase plasmid in the presence of E1A was determined by dual luciferase activity as mentioned in (a). The luciferase activity is shown relative to that of pGL3 empty luciferase plasmid. (d) Transactivation of wild-type or Egr-1-binding site mutant Bim promoter were determined as mentioned in (a). (e) SKOV3-ip1 control and E1A stable cells were treated or untreated with SAHA for 12 h and subjected to the ChIP assay using Egr-1 antibody or control IgG.
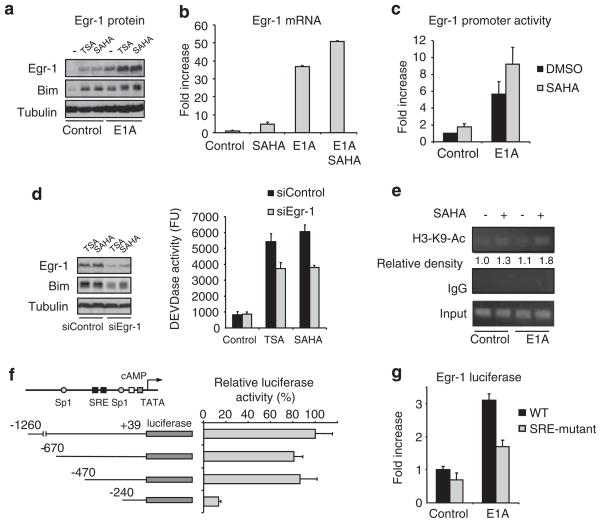
To further investigate how E1A and SAHA enhanced the Egr-1 activity, we found that E1A enhanced the expression of Egr-1 (Figure 6a), and Egr-1 expression was also enhanced by HDACi in SKOV3-ip1 cells, consistent with the previous reports (Pan et al., 2007; Lubieniecka et al., 2008) (Figure 6a). Quantitative RT–PCR analysis and reporter assay using Egr-1 promoter indicated that both E1A and SAHA upregulated transcription of Egr-1 (Figures 6b and c). When we knocked down Egr-1, Bim induction and caspase activation by HDACi were reduced in E1A-expressing cells (Figure 6d).
Figure 6.
Egr-1 is involved in Bim induction and apoptosis induced by the combination of adenovirus 5 early region 1A (E1A) and histone deacetylase inhibitors (HDACi). (a, b) SKOV3-ip1 control or E1A cells were treated with 250 nM of trichostatin A (TSA) or 5 μM of SAHA for 12 h; Egr-1, Bim and tubulin expressions were determined by immunoblot analysis (a). Bim mRNA expression was determined by quantitative RT–PCR (b). (c) SKOV3-ip1 cells were co-transfected with luciferase plasmid containing Egr-1 promoter together with renilla luciferase expression plasmid (internal control), E1A expression plasmid (E1A) or empty vector (control). Twenty-four hours after transfection, the cells were treated with 5 μM of SAHA for an additional 6 h and subjected to the dual luciferase assay. The luciferase activity is shown relative to that of untreated control transfected with empty vector. (d) SKOV3-ip1-E1A cells were transiently transfected with short interfering RNA oligo against Egr-1 or control non-specific siRNA. Thirty-six hours after transfection, the cells were treated with 250 nM of TSA or 5 μM of SAHA for 16 h and caspase activity was determined by caspase assay. Egr-1, Bim and tubulin expressions were determined by immunoblot analysis. (e) SKOV3-ip1 control or E1A cells were treated with SAHA for 6 h, and the ChIP assay was performed using control rabbit immunoglobulin G or anti-acetyl-H3-K9. Polymerase chain reaction was carried out for Egr-1 promoter. (f) Transactivation of the indicated deletion mutants of Egr-1 promoter-luciferase plasmid in the presence of E1A was determined by dual luciferase activity as mentioned in (c). (g) Transactivation of the wild-type or SRE mutant Egr-1 promoter were determined as mentioned in (c).
We next examined the acetylation level of histone H3 on the Egr-1 promoter region by the chromatin immunoprecipitation assay using acetylated histone H3-K9 antibody. We found that the acetylation level of histone H3 around the Egr-1 promoter was enhanced by SAHA, but not by E1A (Figure 6e), suggesting that SAHA induces the hyperacetylation of histone on Egr-1 promoter, similar to Bim promoter. To further investigate how E1A activates Egr-1 promoter activity, we searched for E1A-activating DNA elements that also reside in the Egr-1 promoter. Among the known DNA elements, serum responsible element (SRE) was identified. Interestingly, SRE was also shown to be responsible for serum-induced Egr-1 promoter activity (Thiel and Cibelli, 2002). Indeed, while testing with Egr-1 promoter and SRE-mutant Egr-1 promoter activity, we found that SRE of Egr-1 promoter was responsible for E1A-mediated Egr-1 activation (Figures 6f and g). Together, these results suggest that E1A enhances Bim expression through the upregulation of Egr-1 through its SRE.
Discussion
Together with mutation-activated Ras, E1A has been shown to transform primary rodent cells (Ruley, 1983; Byrd et al., 1988). As Ras oncoprotein also can transform only rodent cell lines but not primary rodent cells, and because immortalization is used to distinguish cell lines from primary cultured cells, the adenovirus type 5 E1A was considered an immortalization oncoprotein even though it does not associate with oncogenic activity (Frisch, 1991; Yu et al., 1991; Hung et al., 2000; Liao et al., 2007). Later, it was found that adenovirus E1A shows multiple anti-tumor activity (Yan et al., 1991; Yu et al., 1991; Frisch and Mymryk, 2002; Frisch, 2004; Liao et al., 2007). Overexpression of E1A induces growth arrest and apoptosis of human cancer cells in vitro, and it has been shown that apoptosis has a critical role in E1A-mediated anti-tumor activity (Frisch, 1991; Rao et al., 1992; Lowe et al., 1993; Deng et al., 1998). Moreover, liposome or adenovirus-mediated E1A gene transfer suppresses tumor growth and metastasis in animal models (Yu et al., 1995; Chang et al., 1997; Hubberstey et al., 2002; Ueno et al., 2002; Liao et al., 2004). On the basis of these preclinical studies, E1A gene therapy was tested in several clinical trials that show that this therapy is feasible in humans (Hortobagyi et al., 2001; Yoo et al., 2001; Villaret et al., 2002; Madhusudan et al., 2004). Moreover, a clinical trial of the combination of paclitaxel and E1A/liposome for ovarian cancer is currently ongoing. In addition to this particular combination therapy, we have been investigating more effective combinations of E1A gene therapy with other anti-cancer drugs. The present study shows that the combination of E1A gene therapy and SAHA is more potent in tumor suppression and safer than the combination of E1A gene therapy and paclitaxel, which justifies the combination of E1A gene therapy and SAHA in clinical trials in the near future.
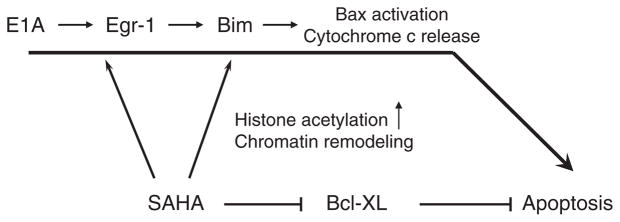
It has been shown that SAHA selectively kill transformed cells (Ungerstedt et al., 2005). In this study, we showed that the combination of E1A and SAHA also showed relatively low toxicity in normal cells. Our data also suggest that the selective alteration of Bcl-2 family protein expression might be involved in the killing effects of HDACi and E1A against transformed cells. In particular, Bim may be involved in the cancer cell killing activity of SAHA and E1A. Here, we showed that SAHA enhances the acetylation levels of histone H3 on the Bim promoter region (Figure 5b). This is likely to contribute to the transactivation of Bim induced by SAHA. However, we could not detect the additive effects on the acetylation of histone H3-K9 by the combination of E1A and SAHA (Figure 5b). Therefore, the combination of E1A and SAHA enhances Bim expression through a histone H3 acetylation-independent mechanism. Here, we showed that Egr-1, which functions as a tumor suppressor, is also involved in Bim upregulation and apoptosis in response to the combination of E1A and HDACi (summarized in Figure 7). Egr-1 expression was induced by either SAHA or E1A and further enhanced by the combination. Egr-1 was upregulated though SRE in its promoter by E1A, while SAHA enhances acetylation of histone H3 in Egr-1 promoter region (Figures 6e–g).
Figure 7.
The model of apoptosis signaling induced by the combination of adenovirus 5 early region 1A (E1A) and suberoylanilide hydroxamic acid (SAHA). E1A upregulates Bim expression through Egr-1 pathway, whereas SAHA upregulates Bim, as well as Egr-1 expression by enhancing its promoter acetylation. Bcl-XL is downregulated by SAHA. The alterations of Bim and Bcl-XL expression contribute to Bax activation, cytochrome c release and subsequent caspase activation.
In this study, we showed that the combination of E1A and SAHA induces cell death more effectively than E1A plus paclitaxel or etoposide, even though some drugs such as etoposide and paclitaxel are also known to induce Bim (Supplementary Figure S1 and Figure 2). It has been shown that etoposide and paclitaxel induce Bim through FOXO3a (Sunters et al., 2003; Liu et al., 2005). Our data showed that SAHA enhances Bim expression through chromatin remodeling, while E1A does through upregulation Egr-1 (Figures 5 and 6). In contrast, FOXO3a is not involved in Bim induction by the SAHA and E1A (Figure 5c). As SAHA also enhances Egr-1 expression though chromatin remodeling, SAHA enhances Bim expression not only by itself, but also enhancing the effects of E1A. These differences of mechanisms underlying Bim upregulaiton may explain, at least in part, why SAHA sensitize E1A more effectively than paclitaxel or etoposide.
In summary, we have shown that the combination of E1A gene therapy and SAHA shows high efficacy in vitro and in vivo with virtually no toxicity. In addition, we established a signal cascade, explaining the molecular mechanisms underlying apoptosis induced by the combination (Figure 7). Thus, this study provides us strong rationale to test the combination of E1A gene therapy and SAHA in future clinical trials.
Materials and methods
Reagents
Trichostatin A, paclitaxel, 5-fluorouracil, etoposide, anti-tubulin monoclonal antibody and actin-polyclonal antibodies were purchased from Sigma (St Louis, MO, USA). Anti-Bim and Bmf polyclonal antibodies were purchased from Calbio-chem (Gibbstown, NJ, USA). Anti-E1A, cytochrome c monoclonal antibodies and Mcl-1 polyclonal antibody were purchased from BD Biosciences (San Jose, CA, USA). Anti-Bax, Bcl-XL monoclonal antibodies and anti-Egr-1 polyclonal antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-Bak, Bax and Egr-1 antibodies were purchased from Cell Signaling (Danvers, MA, USA). Anti-acetylated histone H3 (K9) polyclonal antibody was purchased from Abcam (Cambridge, MA, USA). Control short interfering RNA and short interfering RNA against Bim and Egr-1 were purchased from Dharmacon (Lafayette, CO, USA). SAHA were synthesized in Department of Experimental Diagnostic Imaging at MD Anderson Cancer Center.
Plasmid
pUK21-CMV-E1A, which regulates E1A expression by CMV promoter, was used for transient transfection. Dominant negative Egr-1 was described previously (Zhang et al., 2003). Bim-luciferase plasmid was prepared by using PCR. Egr-1 luciferase plasmid was described previously (Baek et al., 2003). Egr-1luciferase deletion and SRE mutants were constructed by using PCR.
Cell culture and transfection
All the cell lines except MCF10A were maintained in Dulbecco’s modified Eagle’s medium/F12 medium supplemented with 10% fetal bovine serum. MCF10A cells were cultured in Dulbecco’s modified Eagle’s medium/F12 medium supplemented with 5% horse serum, 10 μg/ml insulin, 20 ng/ml EGF, 100 ng/ml cholera toxin and 500 ng/ml hydrocortisone. SKOV3-ip1 and MDA-MB-231 control and E1A stable cell lines were described previously (Ueno et al., 2000; Liao et al., 2004). Plasmid and short interfering RNA transfection was performed by using electroporation.
Immunoblot, subcellular fractionation, immunoprecipitation and quantitative–PCR
Immunoblot analysis was carried out by a standard protocol. To detect Bax conformational change, the cells were lysed in CHAPS lysis buffer (150mM NaCl, 10mM HEPES, pH 7.4, 1% CHAPS) containing protease and phosphatase inhibitors. Total proteins (500 μg) were subjected to immunoprecipitation using 1 μg of anti-Bax 6A7 monoclonal antibody and 15 μl of protein G agarose. Active form of Bax was detected by immunoblot analysis with anti-Bax polyclonal antibody. Subcellular fractionation was carried out as described previously (Uren et al., 2005). Quantitative RT–PCR was performed as described previously (Chou et al., 2009). The primers for Bim and Egr-1 are following; CCAGGCCTTCAA CCACTATC and TCTTGGGCGATCCATATCTC (Bim); TGAACAACGAGAAGGTGCTG and AGCGGCCAGTAT AGGTGATG (Egr-1).
Apoptosis assay
Caspase activity was measured as described previously (Yamaguchi et al., 2003). In brief, the cells were lysed in CHAPS lysis buffer, and 50 μg of total proteins was applied for the caspase assay using fluorescent caspase-3/7 substrate (Sigma). The fluorescence signal was read using a fluorescent plate reader following 1 h of incubation at room temperature. Results are presented as the mean plus standard error (n>3).
For analysis of sub-G1 populations by flow cytometry, cells treated or untreated with SAHA were harvested and fixed with methanol. In total, 106 cells were then incubated in 1ml of phosphate-buffered saline containing 100 μg/ml RNaseA and 40 μg/ml propidium iodide at 37 °C for 1 h. DNA contents were then analyzed by BD FACS Diva (BD Bioscience).
For the clonogenic survival assay, cells treated or untreated with SAHA were harvested and plated at 15 000 (SKOV3-ip1) or 20 000 (MDA-MB-231) cells per 100-mm dish without SAHA. After 12 days, the clones were visualized by crystal violet staining and counted.
Luciferase assay
Mouse Bim promoter regions were amplified by PCR and subcloned into pGL3-basic vector based on the previous report (Bouillet et al., 2001). Egr-1 binding site (GCGGGGGCG, from −37 to −27) was mutated (GCGGATCCG) by two-step PCR. SKOV3-ip1 cells were transiently transfected with the indicated plasmids, and the luciferase assay was performed by using the dual-luciferase system, according to the manufacturers’ protocol (Promega, Madison, WI, USA).
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation assay was carried out as described previously (Wang et al., 2006). SKOV3-ip1 control and E1A stable transfectants were treated with or without 5 μM SAHA for 12 h. The cells were then crosslinked with formaldehyde, lysed and sonicated. Soluble chromatin was incubated overnight with anti-histone H3 (acetyl-K9), anti-Egr-1 polyclonal antibody or rabbit immunoglobulin G, followed by incubation with protein A agarose by rotation for 2 h at 4 °C. Immune complexes were then reversed by protein–DNA crosslinking and digested with proteinase K and RNaseA. The purified DNA was dissolved with 20 μl of dH2O. The human Bim promoter sequences containing the Egr-1 binding site were amplified by PCR using the following primers: CAGGCAGAGTTACTCCGGTAAACACG and CAGAGCTCCAACAAACTGCAGACCAG. The primers used to determine the histone H3-K9 acetylation in were as follows: GAAGTGTACCCTAGCCTC and ACGGCCTCTG TCTCTTAG (Bim promoter); ACCCTTATTTGGGCAGC AC and TATGGGAAGCAGAAGCCCTA (Egr-1 promoter).
Animal study
In the ovarian cancer model, SKOV3-ip1-luciferase stable cells were inoculated intraperitoneally (3×106 cells/mouse). In the breast cancer model, MDA-MB-231 cells (3×106 cells/mouse) were injected into the mammary fat pad. One week after inoculation, the treatments were started. Liposome/E1A or empty vector (15 μg/mouse) was injected intravenously into the mice once weekly, followed by SAHA administration (100 mg/kg per mouse, intraperitoneally) four times per week. Each group contained 10 mice. The treatments were terminated 4 weeks after inoculation (at day 28 for gene therapy and day 31 for SAHA). Quantification of bioluminescence data and breast tumor volume was as described previously (Liao et al., 2004; Xie et al., 2007).
Supplementary Material
Acknowledgments
We thank Drs Jingwen Liu (Palo Alto Health Care System) and Thomas E Eling (National Institute of Health) for Egr-1 dominant negative expression plasmid and Egr-1 luciferase plasmid, respectively. We thank Dr Yongkun Wei for help in carrying out statistical analysis of xenograft experiments. We thank Drs Jennifer L. Hsu and Stephanie Miller for help in preparation of this paper. This work was supported by a SPORE Grant in ovarian cancer and breast cancer (P50 CA83639 and PO50 CA116199), the National Breast Cancer Foundation, MDACC China Medical University and Hospital Sister Institution Fund, Cancer Center Support Grant CA16672, DOH99-TD-C-111-005 Cancer Research Center of Excellence, Taiwan Department of Health, and the MARCUS Foundation. In memoriam, Mrs Serena Lin-Guo for her courageous fight against breast cancer.
Footnotes
Conflict of interest
The corresponding author, Dr Mien-Chie Hung, is an inventor on patents covering E1A as a therapeutic agent filed by the University of Texas MD Anderson Cancer. The remaining authors declare no conflict of interest.
References
- Baek SJ, Wilson LC, Hsi LC, Eling TE. Troglitazone, a peroxisome proliferator-activated receptor gamma (PPAR gamma) ligand, selectively induces the early growth response-1 gene independently of PPAR gamma. A novel mechanism for its anti-tumorigenic activity. J Biol Chem. 2003;278:5845–5853. doi: 10.1074/jbc.M208394200. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Zhang LC, Huang DC, Webb GC, Bottema CD, Shore P, et al. Gene structure alternative splicing, and chromosomal localization of pro-apoptotic Bcl-2 relative Bim. Mamm Genome. 2001;12:163–168. doi: 10.1007/s003350010242. [DOI] [PubMed] [Google Scholar]
- Brader KR, Wolf JK, Hung MC, Yu D, Crispens MA, van Golen KL, et al. Adenovirus E1A expression enhances the sensitivity of an ovarian cancer cell line to multiple cytotoxic agents through an apoptotic mechanism. Clin Cancer Res. 1997;3:2017–2024. [PubMed] [Google Scholar]
- Burgess A, Ruefli A, Beamish H, Warrener R, Saunders N, Johnstone R, et al. Histone deacetylase inhibitors specifically kill nonproliferating tumour cells. Oncogene. 2004;23:6693–6701. doi: 10.1038/sj.onc.1207893. [DOI] [PubMed] [Google Scholar]
- Byrd PJ, Grand RJ, Gallimore PH. Differential transformation of primary human embryo retinal cells by adenovirus E1 regions and combinations of E1A+ ras. Oncogene. 1988;2:477–484. [PubMed] [Google Scholar]
- Chang JY, Xia W, Shao R, Sorgi F, Hortobagyi GN, Huang L, et al. The tumor suppression activity of E1A in HER-2/neu-overexpressing breast cancer. Oncogene. 1997;14:561–568. doi: 10.1038/sj.onc.1200861. [DOI] [PubMed] [Google Scholar]
- Chou CK, Lee DF, Sun HL, Li LY, Lin CY, Huang WC, et al. The suppression of MAD1 by AKT-mediated phosphorylation activates MAD1 target genes transcription. Mol Carcinog. 2009;48:1048–1058. doi: 10.1002/mc.20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JL, Routes JM. Adenovirus E1A gene-induced tumor cell rejection through cellular sensitization to immune and nonimmune apoptotic injuries. Front Biosci. 2005;10:1396–1414. doi: 10.2741/1628. [DOI] [PubMed] [Google Scholar]
- Day CP, Rau KM, Qiu L, Liu CW, Kuo HP, Xie X, et al. Mutant Bik expression mediated by the enhanced minimal topoisomerase IIalpha promoter selectively suppressed breast tumors in an animal model. Cancer Gene Ther. 2006;13:706–719. doi: 10.1038/sj.cgt.7700945. [DOI] [PubMed] [Google Scholar]
- Deng J, Xia W, Hung MC. Adenovirus 5 E1A-mediated tumor suppression associated with E1A-mediated apoptosis in vivo. Oncogene. 1998;17:2167–2175. doi: 10.1038/sj.onc.1202148. [DOI] [PubMed] [Google Scholar]
- Freireich EJ, Gehan EA, Rall DP, Schmidt LH, Skipper HE. Quantitative comparison of toxicity of anticancer agents in mouse, rat, hamster, dog, monkey, and man. Cancer Chemother Rep. 1966;50:219–244. [PubMed] [Google Scholar]
- Frisch SM. Antioncogenic effect of adenovirus E1A in human tumor cells. Proc Natl Acad Sci USA. 1991;88:9077–9081. doi: 10.1073/pnas.88.20.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM. E1A as a tumor suppressor gene: commentary re S. Madhusudan et al. A multicenter Phase I gene therapy clinical trial involving intraperitoneal administration of E1A-lipid complex in patients with recurrent epithelial ovarian cancer overexpressing HER-2/neu oncogene. Clin Cancer Res. 2004;10:2905–2907. doi: 10.1158/1078-0432.ccr-04-0644. [DOI] [PubMed] [Google Scholar]
- Frisch SM, Mymryk JS. Adenovirus-5 E1A: paradox and paradigm. Nat Rev Mol Cell Biol. 2002;3:441–452. doi: 10.1038/nrm827. [DOI] [PubMed] [Google Scholar]
- Hortobagyi GN, Ueno NT, Xia W, Zhang S, Wolf JK, Putnam JB, et al. Cationic liposome-mediated E1A gene transfer to human breast and ovarian cancer cells and its biologic effects: a phase I clinical trial. J Clin Oncol. 2001;19:3422–3433. doi: 10.1200/JCO.2001.19.14.3422. [DOI] [PubMed] [Google Scholar]
- Hubberstey AV, Pavliv M, Parks RJ. Cancer therapy utilizing an adenoviral vector expressing only E1A. Cancer Gene Ther. 2002;9:321–329. doi: 10.1038/sj.cgt.7700436. [DOI] [PubMed] [Google Scholar]
- Hung MC, Hortobagyi GN, Ueno NT. Development of clinical trial of E1A gene therapy targeting HER-2/neu-overexpressing breast and ovarian cancer. Adv Exp Med Biol. 2000;465:171–180. doi: 10.1007/0-306-46817-4_16. [DOI] [PubMed] [Google Scholar]
- Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- Liao Y, Yu D, Hung MC. Novel approaches for chemosensitization of breast cancer cells: the E1A story. Adv Exp Med Biol. 2007;608:144–169. doi: 10.1007/978-0-387-74039-3_11. [DOI] [PubMed] [Google Scholar]
- Liao Y, Zou YY, Xia WY, Hung MC. Enhanced paclitaxel cytotoxicity and prolonged animal survival rate by a nonviral-mediated systemic delivery of E1A gene in orthotopic xenograft human breast cancer. Cancer Gene Ther. 2004;11:594–602. doi: 10.1038/sj.cgt.7700743. [DOI] [PubMed] [Google Scholar]
- Liu JW, Chandra D, Rudd MD, Butler AP, Pallotta V, Brown D, et al. Induction of prosurvival molecules by apoptotic stimuli: involvement of FOXO3a and ROS. Oncogene. 2005;24:2020–2031. doi: 10.1038/sj.onc.1208385. [DOI] [PubMed] [Google Scholar]
- Lo HW, Day CP, Hung MC. Cancer-specific gene therapy. Adv Genet. 2005;54:235–255. doi: 10.1016/S0065-2660(05)54010-0. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- Lubieniecka JM, de Bruijn DR, Su L, van Dijk AH, Subramanian S, van de Rijn M, et al. Histone deacetylase inhibitors reverse SS18-SSX-mediated polycomb silencing of the tumor suppressor early growth response 1 in synovial sarcoma. Cancer Res. 2008;68:4303–4310. doi: 10.1158/0008-5472.CAN-08-0092. [DOI] [PubMed] [Google Scholar]
- Madhusudan S, Tamir A, Bates N, Flanagan E, Gore ME, Barton DP, et al. A multicenter Phase I gene therapy clinical trial involving intraperitoneal administration of E1A-lipid complex in patients with recurrent epithelial ovarian cancer overexpressing HER-2/neu oncogene. Clin Cancer Res. 2004;10:2986–2996. doi: 10.1158/1078-0432.ccr-03-0291. [DOI] [PubMed] [Google Scholar]
- Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- Pan L, Lu J, Wang X, Han L, Zhang Y, Han S, et al. Histone deacetylase inhibitor trichostatin a potentiates doxorubicin-induced apoptosis by up-regulating PTEN expression. Cancer. 2007;109:1676–1688. doi: 10.1002/cncr.22585. [DOI] [PubMed] [Google Scholar]
- Pirollo KF, Chang EH. Targeted delivery of small interfering RNA: approaching effective cancer therapies. Cancer Res. 2008;68:1247–1250. doi: 10.1158/0008-5472.CAN-07-5810. [DOI] [PubMed] [Google Scholar]
- Rao L, Debbas M, Sabbatini P, Hockenbery D, Korsmeyer S, White E. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc Natl Acad Sci USA. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagan-Shaw S, Nihal M, Ahmad N. Dose translation from animal to human studies revisited. Faseb J. 2008;22:659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- Ruley HE. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983;304:602–606. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- Samuelson AV, Lowe SW. Selective induction of p53 and chemosensitivity in RB-deficient cells by E1A mutants unable to bind the RB-related proteins. Proc Natl Acad Sci USA. 1997;94:12094–12099. doi: 10.1073/pnas.94.22.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoff-Khalili MA, Dall P, Curiel DT. Gene therapy for carcinoma of the breast. Cancer Gene Ther. 2006;13:633–647. doi: 10.1038/sj.cgt.7700929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunters A, Fernandez de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA, et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem. 2003;278:49795–49805. doi: 10.1074/jbc.M309523200. [DOI] [PubMed] [Google Scholar]
- Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol. 2002;193:287–292. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- Ueno NT, Bartholomeusz C, Herrmann JL, Estrov Z, Shao R, Andreeff M, et al. E1A-mediated paclitaxel sensitization in HER-2/neu-overexpressing ovarian cancer SKOV3.ip1 through apoptosis involving the caspase-3 pathway. Clin Cancer Res. 2000;6:250–259. [PubMed] [Google Scholar]
- Ueno NT, Bartholomeusz C, Xia W, Anklesaria P, Bruckheimer EM, Mebel E, et al. Systemic gene therapy in human xenograft tumor models by liposomal delivery of the E1A gene. Cancer Res. 2002;62:6712–6716. [PubMed] [Google Scholar]
- Ueno NT, Yu D, Hung MC. Chemosensitization of HER-2/neu-overexpressing human breast cancer cells to paclitaxel (Taxol) by adenovirus type 5 E1A. Oncogene. 1997;15:953–960. doi: 10.1038/sj.onc.1201250. [DOI] [PubMed] [Google Scholar]
- Ungerstedt JS, Sowa Y, Xu WS, Shao Y, Dokmanovic M, Perez G, et al. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102:673–678. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren RT, Dewson G, Bonzon C, Lithgow T, Newmeyer DD, Kluck RM. Mitochondrial release of pro-apoptotic proteins: electrostatic interactions can hold cytochrome c but not Smac/DIABLO to mitochondrial membranes. J Biol Chem. 2005;280:2266–2274. doi: 10.1074/jbc.M411106200. [DOI] [PubMed] [Google Scholar]
- Villaret D, Glisson B, Kenady D, Hanna E, Carey M, Gleich L, et al. A multicenter phase II study of tgDCC-E1A for the intratumoral treatment of patients with recurrent head and neck squamous cell carcinoma. Head Neck. 2002;24:661–669. doi: 10.1002/hed.10107. [DOI] [PubMed] [Google Scholar]
- Wang YN, Chen YJ, Chang WC. Activation of extracellular signal-regulated kinase signaling by epidermal growth factor mediates c-Jun activation and p300 recruitment in keratin 16 gene expression. Mol Pharmacol. 2006;69:85–98. doi: 10.1124/mol.105.016220. [DOI] [PubMed] [Google Scholar]
- Xie X, Hsu JL, Choi MG, Xia W, Yamaguchi H, Chen CT, et al. A novel hTERT promoter-driven E1A therapeutic for ovarian cancer. Mol Cancer Ther. 2009;8:2375–2382. doi: 10.1158/1535-7163.MCT-09-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Xia W, Li Z, Kuo HP, Liu Y, Li Z, et al. Targeted expression of BikDD eradicates pancreatic tumors in noninvasive imaging models. Cancer Cell. 2007;12:52–65. doi: 10.1016/j.ccr.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Xing X, Liu V, Xia W, Stephens LC, Huang L, Lopez-Berestein G, et al. Safety studies of the intraperitoneal injection of E1A–liposome complex in mice. Gene Ther. 1997;4:238–243. doi: 10.1038/sj.gt.3300376. [DOI] [PubMed] [Google Scholar]
- Xing X, Zhang S, Chang JY, Tucker SD, Chen H, Huang L, et al. Safety study and characterization of E1A-liposome complex gene-delivery protocol in an ovarian cancer model. Gene Ther. 1998;5:1538–1544. doi: 10.1038/sj.gt.3300771. [DOI] [PubMed] [Google Scholar]
- Xu W, Ngo L, Perez G, Dokmanovic M, Marks PA. Intrinsic apoptotic and thioredoxin pathways in human prostate cancer cell response to histone deacetylase inhibitor. Proc Natl Acad Sci USA. 2006;103:15540–15545. doi: 10.1073/pnas.0607518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Bhalla K, Wang HG. Bax plays a pivotal role in thapsigargin-induced apoptosis of human colon cancer HCT116 cells by controlling Smac/Diablo and Omi/HtrA2 release from mitochondria. Cancer Res. 2003;63:1483–1489. [PubMed] [Google Scholar]
- Yan DH, Chang LS, Hung MC. Repressed expression of the HER-2/c-erbB-2 proto-oncogene by the adenovirus E1a gene products. Oncogene. 1991;6:343–345. [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo GH, Hung MC, Lopez-Berestein G, LaFollette S, Ensley JF, Carey M, et al. Phase I trial of intratumoral liposome E1A gene therapy in patients with recurrent breast and head and neck cancer. Clin Cancer Res. 2001;7:1237–1245. [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- Yu D, Matin A, Xia W, Sorgi F, Huang L, Hung MC. Liposome-mediated in vivo E1A gene transfer suppressed dissemination of ovarian cancer cells that overexpress HER-2/neu. Oncogene. 1995;11:1383–1388. [PubMed] [Google Scholar]
- Yu D, Wolf JK, Scanlon M, Price JE, Hung MC. Enhanced c-erbB-2/neu expression in human ovarian cancer cells correlates with more severe malignancy that can be suppressed by E1A. Cancer Res. 1993;53:891–898. [PubMed] [Google Scholar]
- Yu DH, Scorsone K, Hung MC. Adenovirus type 5 E1A gene products act as transformation suppressors of the neu oncogene. Mol Cell Biol. 1991;11:1745–1750. doi: 10.1128/mcb.11.3.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Lin M, Abidi P, Thiel G, Liu J. Specific interaction of Egr1 and c/EBPbeta leads to the transcriptional activation of the human low density lipoprotein receptor gene. J Biol Chem. 2003;278:44246–44254. doi: 10.1074/jbc.M305564200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.