Abstract
Natural tools for recombinant protein production show technological limitations. Available natural promoters for gene expression in Pichia pastoris are either constitutive, weak or require the use of undesirable substances or procedures for induction. Here we show the application of deletion variants based on the well known methanol inducible AOX1 promoter and small synthetic promoters, where cis-acting elements were fused to core promoter fragments. They enable differently regulated target protein expression and at the same time to replace methanol induction by a glucose or glycerol feeding strategy. Trypsinogen, the precursor of the serine protease trypsin, was expressed using these different promoters. Depending on the applied promoter the production window (i.e. the time of increasing product concentration) changed significantly. In fedbatch processes trypsinogen yields before induction with methanol were up to 10 times higher if variants of the AOX1 promoter were applied. In addition, the starting point of autoproteolytic product degradation can be predetermined by the promoter choice.
Keywords: Pichia pastoris, AOX1 promoter variants, Porcine trypsinogen, Production window
Introduction
The applicability of simple genetic techniques, strong expression of foreign genes and the feasibility of high cell density cultures made the methylotrophic yeast Pichia pastoris a favourable host system for heterologous protein production (Cregg et al. 1985, 1993, 2000). High levels of foreign proteins have been expressed in P. pastoris under the control of the strong, methanol inducible AOX1 promoter (Hasslacher et al. 1997; Werten et al. 1999; Xiong et al. 2006). A benchmark was set in 1997 by the intracellular production of an industrially applied (S)-hydroxynitrile lyase with a titer of 22 g l−1 protein (Hasslacher et al. 1997). Generally, the Pichia pastoris system is a faster, more simple and less expensive alternative to higher eukaryotic expression systems, such as mammalian cell cultures, still able to perform typical eukaryotic posttranslational modifications like glycosylation or disulfide bridge formation (Cregg et al. 1985, 1993). Although Pichia pastoris has often been used successfully in recombinant protein production still little is known about the regulation of the AOX1 promoter on a molecular level (Hartner et al. 2008). Briefly, transcription can be regulated by positively and negatively acting sequences in the promoter (Verdier 1990). Glucose and other repressing sugars can affect the rate of transcription by interfering with activators or repressors directly or by regulating the expression of such transcription factors. Inan et al. (2004) first described two cis-acting elements, A and C, within the AOX1 promoter (PAOX1) binding to yet unknown DNA-binding proteins. In addition the positive acting transcription factor Mxr1p interacting with the AOX1 promoter and responsible for the activation of many genes in response to methanol was identified by Lin-Cereghino et al. (2006). Kranthi et al. (2009) identified 6 Mxr1p binding sites within the AOX1 promoter. In addition, a comprehensive study of Hartner et al. (2008) described the influence of mutations, deletions and multiplications of putative transcription factor binding sites on the regulatory properties of the AOX1 promoter. Indicating Mxr1p as an important regulator of the methanol utilization in P. pastoris, deletion of 5 of the 6 Mxr1p sites resulted in reduced activity on methanol. In agreement also Xuan et al. (2009) showed the AOX1 promoter region −638 to −510 to be upstream activating.
Additionally to mechanistic information, the developed library of Hartner et al. (2008) allows adjusting the level of transcription according to the individual expressed genes. Further, first AOX1 promoter variants became available, which allow glucose regulated expression based on a simple repression-derepression principle.
In this study we show the effect of different synthetic promoter variants on heterologous gene expression in Pichia pastoris. A major interest was put in the question how differently regulated promoters influence production window and product quality under different environmental conditions, here in presence of methanol, glucose or glycerol. For these comparative studies our new Escherichia coli/Pichia pastoris shuttle vector (pPpT2) was employed. As model target porcine trypsinogen was selected. Trypsinogen (zymogen) is the inactive precursor of the serine protease trypsin which is secreted in the pancreas (Hanquier et al. 2003; Stroud et al. 1977). In the small intestine it is further activated to trypsin through proteolytic cleavage by enterokinase. Enterokinase cleaves after the lysine of the (Asp)4-Lys sequence in the propeptide and releases active protease (Hanquier et al. 2003). Active trypsin can cleave its own zymogen by cleaving at the carboxyl-terminal ends of lysine and arginine starting an autocatalytic cascade. Trypsin is used in industrial biotechnology for the processing of enzymes and pharmaceutical proteins (e.g. insulin production). Thus its precursor trypsinogen also became an interesting model target for protein expression studies (Chance et al. 1997; Frank et al. 1995; Hanquier et al. 2003).
Materials and methods
Strains and plasmids
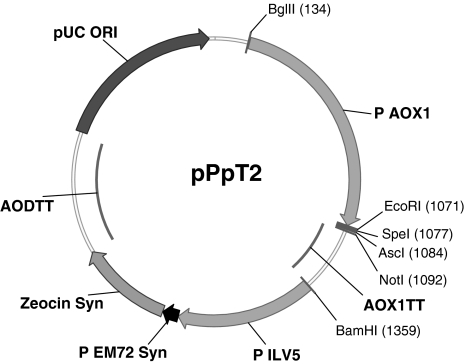
Porcine trypsinogen (GenBank accession no. P00761) was codon optimized applying a Pichia pastoris high methanol codon usage using the Gene Designer Software (DNA2.0, Menlo Park, CA, USA). For secretion the signal sequence of the yeast mating factor α, also codon optimized, was added by overlap extension PCR (polymerase chain reaction). The obtained construct was ligated into the multiple cloning site (MCS, EcoRI/NotI) of our E. coli/P.pastoris shuttle vector pPpT2 (Fig. 1). The AOX1 promoter and terminator sequences, which were synthesized by GenScript (Piscataway, NJ, USA) based on the sequence information from GenBank (accession no. U96967), were used for the construction of the plasmid pPpT2. To allow selection against the antibiotic zeocin we used the Ble gene from S. hindusdanus (GenBank accession no. A31898), which was optimized for expression in E. coli and P. pastoris (Leto Software, Entelechon Corp.). In the referred plasmid pPpT2 the zeocin resistance gene was under the control of a 3′ 34 bp truncated 552 bp long fragment of the ILV5 promoter and the AOD terminator (466 bp), both derived from P. pastoris, strain CBS7435. For bacterial replication the pUC origin (pBR322) was used. For expression of the resistance marker in E. coli a synthetic prokaryotic consensus promoter (P EM72) was embedded between the 3′ truncated eukaryotic promoter and the start of the resistance gene.
Fig. 1.
Pichia pastoris shuttle vector pPpT2 (3,546 bp), P AOX1, AOX1 promoter; AOX1TT, AOX1 terminator; MCS (multiple cloning site), EcoRI, SpeI, AscI, NotI; P ILV5, promoter of the P. pastorisILV5 gene; P EM72, synthetic E. coli promoter; Zeocin Syn, synthetic Zeocin gene; AODTT, terminator of the P. pastoris AOD gene; pUC ori, origin of replication
Chemicals and media
Oligonucleotide primers were obtained from Eurogentec (Seraing, Belgium). For plasmid isolation the GeneJET™ Plasmid Miniprep Kit of Fermentas (Burlington, Ontario, Canada) was used. PCR fragment purification was done with the Wizard® SV Gel and PCR Clean-Up System of Promega (Madison, WI, USA). Chemicals were purchased if not stated otherwise from Becton, Dickinson and Company (Franklin Lakes, NJ, USA), Fresenius Kabi Austria (Graz, Austria), Carl Roth (Karlsruhe, Germany), and Sigma–Aldrich (St Louis, MO, USA).
E. coli complex media was composed of 10 g l−1 trypton, 5 g l−1 yeast extract and 5 g l−1 NaCl. For selection in E. coli a concentration of 25 μg ml−1 zeocin was used. Complex Pichia pastoris media (YPD) contained 10 g l−1 yeast extract, 20 g l−1 peptone and 20 g l−1 glucose. For antibiotic selection 100 μg ml−1 zeocin were used. 15 g l−1 agar was added for plate media. Buffered minimal media BMD (1%), BMM2 and BMM10 consisted per liter of 200 ml 1 M potassium phosphate buffer (pH 6), 13.4 g yeast nitrogen base without amino acids, 0.0004 g l−1 biotin and 11 g l−1 glucose or 1 or 5% (v/v) methanol, respectively. BMGY media used for inocolum cultures for fedbatch cultivations contained as carbon source 10 g l−1 glycerol (Invitrogen, Carlsbad, CA, USA). Batch mineral medium contained per litre 30 g glucose, 0.17 g CaSO4_2H2O, 2.86 g K2SO4, 0.64 g KOH, 2.32 g MgSO4_7H2O, 0.22 g NaCl, 0.6 g EDTA disodium salt and 4.25 ml H3PO4 (85%). Additionally 0.00087 g biotin and 4.35 ml PTM1 mineral salt solution were added to the media after sterilisation. PTM1 mineral salt solution (Invitrogen, Carlsbad, CA, USA) contained per litre 5 ml H2SO4 (69%), 3.84 g CuSO4, 0.08 g NaI, 3.0 g MnSO4_H2O, 0.2 g Na2MoO4_2H2O, 0.02 g H3BO3, 105 0.92 g CoCl2_6H2O, 20.0 g ZnCl2 and 65.0 g FeSO4_7H2O. 12 ml PTM1 salt solution and 0.0024 g biotin were added to the feed solutions. To avoid foam antifoam A (10%) was added in a concentration of 100 μl l−1. The glucose substrate feed contained 632 g kg−1 α-d(+)-glucose monohydrate, the methanol substrate feed 99% methanol.
AOX1 promoter variants
The knowledge about possible regulatory sites on the AOX1 promoter provided the starting point for this work (Hartner et al. 2008). Representative deletion variants and multiplications of cis-acting elements from this previous study as well as new promoter variants were compared in this study with respect to technological aspects for the expression of porcine trypsinogen.
The promoter variant d1 as well as the improved combination variant d1+ increased expression in screening studies, using green fluorescent protein (GFP) as reporter, to 133 and 166%, if compared to the wild type (WT) AOX1 promoter on single copy level after 72 h of methanol induction (Hartner et al. 2008). Similar positive effects were found employing these constructs for other intracellular and secreted model enzymes. In contrast, under the same conditions the promoter variants d6 and d6* resulted in lower expression (44 and 55%, respectively relative to the wild type promoter), but significant expression started already before induction. Chosen short promoter variants were named AOX176-Mat1-Mc and AOX176-201-214. Single copy transformants of these variants showed very low methanol induced GFP expression (5–10%), but the ability to express protein already before induction. From these results it can be speculated that the short synthetic promoters might also be regulated via a glucose depletion-derepression mechanism as it was described for the variants d6 and d6*.
For further studies about the apparently important region d1 a new PAOX1 double deletion variant, namely dHap2345-1z1, was generated based on construct dHap2345-1 (Hartner et al. 2008). In addition, a new cis-acting element core promoter fusion called AOX176-Rap1 was synthesized. Although no significant effect on promoter activity was found by Hartner et al. (2008) by doubling the putative Rap1p binding region, the previous deletion studies indicated an importance of the Rap1 region showing only 34% residual activity upon deletion. For this reason the putative Rap1p binding region was also included in further promoter design as described below.
Design of the AOX1 promoter variants
Sequence numeration was done in consensus to the natural AOX1 promoter in upstream direction starting at the start codon of the AOX1 open reading frame (CG(−1)A(+1)TG). Variant d1 consisted of a deletion of 66 bp between position −711 and −776. The variant d1+ was based on d1 and contained an additional multiplication of 14 bases (201–214, located at position −189). Variant d6 was made by a deletion of 30 bases at position −223. Construct d6* contained an additional deletion of 2 base pairs, located 3′ of d6 (∆TA/−208/−209). Construct dHap2345-1z1 was a double deletion variant with deleted regions between −746/−750 and −753/−755. The short synthetic promoters consisted of the putative strong positive acting elements Rap1 (−599/−615), Mat1-Mc (−253/−269) and 201–214 (−189/−202) fused to the core (basal) promoter element AOX176 (168 bp). Figure 2 shows these variants in comparison to the wild type AOX1 promoter.
Fig. 2.
Sequence alignment of the AOX1 wild type (WT) promoter and AOX1 promoter variants, a wild type PAOX1 (WT), deletion variants (d6, d1), double deletion variants (d6*, dHap2345-1z1), b synthetic variants AOX176-Rap1 (Rap1), AOX176-Mat1-Mc (Mat1) and AOX176-201-214 (214); underlined: EcoRI, BglII, BspTI restriction sites, (−1): 3′ promoter end, rectangle: cis-acting elements (Rap1, Mat1-Mc, 201–214)
Generation of the AOX1 promoter variants
The employed AOX1 promoter variants d1, d1+, d6 and d6* obtained from Hartner and Glieder (2005) were amplified by PCR using following primers (restriction sites underlined): PAOX1Bglfw 5′-TTCAGATCTAACATCCAAAGACGAAAGG-3′, PAOX1Ecorv 5′-TTGAATTC TTTCAATAATTAGTTGTTTTTTG-3′). For the PCR amplifications Phusion™ High-Fidelity DNA Polymerase distributed by New England Biolabs (Ipswich, MA, USA) was used. Promoter variant dHap2345-1z1 was generated by site directed mutagenesis (SDM) according to Wang and Malcolm (2002). Also, small synthetic promoter variants were amplified by PCR, but using PfuUltra™ polymerase (Stratagene Inc.). To generate the small promoter variants the positive acting elements Mat1-Mc, Rap1, 201–214 were attached to the core promoter fragment AOX176 (168 bp, pPpT2) by long high quality PCR primers. According to Hartner et al. (2008) a BspTI restriction site was introduced as linker between the positive acting element and the core promoter fragment. Following primers were used for amplification: Mat1-McAOX176fw 5′-AAAGATCTTTATGCATTGTCTCCACCTTAAGGACAGCAATATATAAACAGAAG-3′, Rap1AOX176fw 5′-AAAGATCTTTACACCCGAACATCACCTTAAGGACAGCAATATATAAACAGAAG-3′, 201-214AOX176fw 5′-AAAGATCTCATGATCAAAATTTCATGATCAAAATTTCTTAAGGACAGCAATATATAAACAGAAG-3′ (reverse primer PAOX1Ecorv, restriction sites underlined, positive acting elements bold, core promoter binding site italic). Amplified promoters were digested with BglII/EcoRI and ligated into pPpT2. Fermentas enzymes (Burlington, Ontario, Canada) were used for all cloning steps.
All experiments were performed using the Pichia pastoris strain CBS7435, obtained from CBS fungal biodiversity centre. Subcloning was done using the E. coli strain DH5a-T1R (Invitrogen, Carlsbad, CA, USA).
Transformation, screening and rescreening
The condensed protocol of Lin-Cereghino et al. (2005) was used for Pichia pastoris transformations. Therefore 1 μg of linearized plasmid DNA was prepared for each construct and electroporation was done using following parameters: 1.5 kV, 25 μF and 200 Ω. For regeneration 1 ml of cold sorbitol was added. Transformed cells were regenerated for 2 h at 28°C and plated on selective media.
Pichia pastoris was transformed with 9 different trypsinogen expression constructs, including 8 different promoter variants (d1, d1+, d6, d6*, dHap2345-1z1, AOX176-Rap1, AOX176-Mat-1MC, AOX176-201-214) and PAOX1 wild type. 88 transformants of each construct were picked for screening and expression analysis. Screening was done in 96 deep well plates according to Weis et al. (2004). Clones were grown 60 h in 300 μl BMD1% media. Induction was performed by addition of 250 μl BMM2 media followed by methanol pulses of 50 μl BMM10 after 12 and 24 h of methanol induction (in rescreening also at 48, 72 and 96 h). Samples (50 μl) were taken before induction and subsequently every 24 h. From samples cells were harvested by centrifugation at 4,000 rpm and 4°C for 10 min in an Eppendorf (Hamburg, Deutschland) 5810R centrifuge. Obtained supernatant was used for activity measurements.
Seven clones of each construct representing the whole activity range in the initial primary screen were chosen for rescreening. In the rescreening process, also performed in 96 deep well plates, selected clones were inoculated 4 times each and the standard deviation of the activity values was calculated.
Fed batch cultivations in bioreactors
Fedbatch cultivations were performed using the 1.5 l fedbatch-pro® bioreactor system (DASGIP AG, Juelich, Germany). Cultivation parameters were as follows: 28°C, pH 6. Total batch volume was 650 ml, inoculum volume 50 ml. In the batch phase cells were grown on glucose with a calculated amount of 7.8 gC. The batch phase (phase A) was followed by a fedbatch phase (phase B) in which glucose was fed exponentially with a rate of 1.4*e(0.2*t) gC h−1 for 12.25 h. Fedbatch studies using methanol in the production phase (phase C) were characterized by a constant methanol feed of 1.7 gC h−1. Fedbatch studies substituting methanol with glucose in phase C were characterized by a constant glucose feed rate of 2.68 gC h−1.
Final cultivation experiments were performed using the Infors Multifors system (Infors AG, Bottmingen-Basel, Germany). Using this cultivation system the batch and fedbatch feeds were adjusted according to the pO2 signal of the culture. The total batch volume was 500 ml, inoculum volume 50 ml. To delay product degradation the production temperature was reduced to 20°C. In cultivation studies using methanol for induction the glycerol based batch and fed batch phases were followed by a methanol feed of 0.3 gC h−1. Said methanol feed was applied for 20 h. Subsequently a constant methanol feed of 1.6 gC h−1 was set. In fedbatch cultivations using glycerol and glucose in the production phase feed parameters were as follows: 1.6 gC h−1 (glycerol) and 1.8 gC h−1 (glucose). Note, for cultivations using glucose in the production phase, glucose was also used as substrate during growth phase.
Trypsin activity assay
Trypsin activity was measured photometrically in microplates on a Spectramax Plus 384 plate reader (Molecular Devices, Sunnyvale, CA, USA) using N-a-benzoyl-L-arginine p-nitroanilide (BApNA) as a substrate (Hayakawa et al. 1979). Shortly, 5 μl of supernatant were activated by enterokinase for 3 h at 37°C (0.5 μl enterokinase 0.5 U ml−1, 11 μl TEA buffer). 100 μl of BApNA (0.5 mmol l−1) were added to start the reaction and kinetics were recorded for 5 min at 405 nm. TEA (triethanolamine) buffer for trypsinogen activity assay was used in a concentration of 0.15 mol l−1 and a pH of 8.2. For screening and rescreening trypsinogen activity was measured with and without activation by enterokinase. Absolute trypsinogen activity values were calculated by subtracting the activated values from the auto-activated ones.
Specific productivity
Specific productivity (qp) (i.e. given in ‘units’ per gram of biomass and hour) was used to compare the best clones comprising different promoter constructs. It was computed from activity (dcp) and biomass (x) data for the time of increasing product concentration (dt) according to Eq. (1).
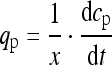 |
1 |
Protein analysis
Total protein concentration was estimated using the Coomassie (Bradford) Protein Assay Kit of Pierce (Thermo Fisher Scientific Inc., Rockford, IL USA). Electrophoretic separation of protein samples was done using the Agilent (Santa Clara CA, USA) Bioanalyzer 2100. 4 μl of supernatant were denatured for 5 min at 95°C with Agilent sample buffer (2 μl, 3.5% 1 M DTT). Samples were centrifuged and 84 μl of deionized H20 were added. Chip preparation was done according to the Protein 80 Kit manual. Additionally, the Caliper Life Science (Hopkinton, MA 01748 USA) LabChip ® GX II was used to determine protein concentration, following the HT Protein Express LabChip® Assay user instructions.
Real time PCR
For copy number estimation Real Time (RT)-PCR was performed using the ABI PRISM 7300 Real Time PCR System and Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), following a procedure described by Abad et al. (2010). The Pichia pastorisARG4 gene was used as a reference for quantification. Following primers were used for amplification of the reference gene and inserted construct: ARG4-RTfw (TCCTCCGGTGGCAGTTCTT), ARG4-RTrv (TCCATT GACTCCCGTTTGAG), AOX1-RTfw (GAAGCTGCCCTGTCTTAAACCTT), AOX1-RTrv (CAAAAGCTTGTCAATTGGAACCA). The PCR program was performed according to Hartner et al. (2008). DNA preparation was adapted from Hoffman and Winston (1987).
Results
Rescreening
Trypsinogen activities of the best clones obtained from rescreening after 0, 24, 48, 72 and 96 h of methanol induction, were normalized by the best obtained wild type PAOX1 transformant which was induced for 96 h (Table 1). 100% wild type activity was equivalent to 16 U l−1 OD−1. Best results in rescreening were obtained using variants d1+ and d6*, showing 218% und 202% relative activity after 72 h of methanol induction. Interestingly, both constructs showed a decrease in activity after 96 h of induction, similar to the WT construct. Variant d1 showed in contrast highest activity after 96 h with 185% relative activity. Following the expression landscape of this construct, maximum values may not have been reached. With regard to expression profiles, variant d1+ showed the highest increase in activity over a short period of time. For this construct, the relative trypsinogen activity increased from 7 to 172% within the first 24 h of induction. Compared to the WT strain, expression was increased more than twofold at this time point. The analyses for the synthetic construct AOX176-Rap1 showed the possibility to produce trypsinogen up to the end of the glucose batch, with the best clone reaching 77% relative activity. Interestingly, this clone was also methanol inducible, reaching 164% activity after 72 h of methanol induction.
Table 1.
Relative promoter comparison, activities of the best clones from rescreening are given in relation to the value of the WT-strain at 96 h (i.e. 100%) and the time of methanol induction—resp. time 0 h at the end of glucose batch; standard deviations were calculated using data from four different wells
| Promoter | 0 h | 24 h | 48 h | 72 h | 96 h |
|---|---|---|---|---|---|
| WT | 8 ± 8 | 86 ± 14 | 127 ± 14 | 136 ± 15 | 100 ± 21 |
| d1 | 4 ± 9 | 83 ± 11 | 86 ± 10 | 152 ± 15 | 185 ± 45 |
| d1+ | 7 ± 4 | 172 ± 55 | 189 ± 61 | 218 ± 23 | 167 ± 40 |
| d6* | −6 ± 10 | 124 ± 30 | 141 ± 39 | 202 ± 44 | 123 ± 31 |
| Rap1_G3 | 77 ± 16 | 96 ± 22 | 100 ± 29 | 164 ± 24 | 80 ± 14 |
| Rap1_A7 | 52 ± 10 | 46 ± 35 | 44 ± 17 | −5 ± 51 | 49 ± 6 |
| Rap1_C8 | 39 ± 8 | 36 ± 8 | 49 ± 15 | 172 ± 64 | 49 ± 5 |
| Rap1_B3 | 46 ± 8 | 68 ± 15 | 96 ± 17 | 80 ± 45 | 61 ± 5 |
Due to the strong derepression effect of construct AOX176-Rap1, indicating an altered regulation pattern, four AOX176-Rap1 clones (G3, A7, C8, B3) of different expression strengths were selected for further studies. Excluded from further analyses were the following constructs: d1, d6, dHap2345-1z1, AOX176-Mat1-Mc, AOX176-201-214. Promoter construct d1 was excluded from further analysis due to its lower specific productivity compared to construct d1+. Variant d6 was not considered because of its similarity in expression profile to variant d6*. Variant dHap2345-1z1 was excluded due to low activity under both conditions, methanol induction as well as glucose derepression. The short synthetic constructs AOX176-Mat1-Mc and AOX176-201-214 were not selected as both constructs were outranked in expression by the promoter AOX176-Rap1.
Copy number estimation
To analyze a possible correlation between expression level and the number of integrated expression cassettes (copy number), the copy numbers were determined by RT-PCR for the best clones of each construct. Additionally the clones A7, C8 and B3 of construct AOX176-Rap1 were analyzed. Interestingly, it was shown that all long mutation and deletion PAOX1 variants selected through screening and rescreening contained only one copy of the integration cassette. In contrast, construct AOX176-Rap1 seemed to perform best with more integrated copies, with clone G3 having over 4 copies of the expression cassette integrated into the P. pastoris genome. The other AOX176-Rap1 strains A7, C8 and B3 contained 3–4, 2 and again 4 copies, respectively. Strains and copy numbers are shown in Table 2.
Table 2.
P. pastoris trypsinogen expression strains with corresponding copy numbers
| Construct | Copy number |
|---|---|
| WT | 1 |
| d1 | 1 |
| d1+ | 1 |
| d6* | 1 |
| Rap1-G3 | >4 |
| Rap1-A7 | 3–4 |
| Rap1-C8 | 2 |
| Rap1-B3 | 4 |
Fedbatch cultivations in bioreactors
To confirm the data of the 96 deep well screening and rescreening, parallel fedbatch cultivations were performed in bioreactors. Specific productivity, activity increase, production time and biomass growth were calculated for all strains or promoter constructs. However, it has to be considered that specific productivity decreases with longer production times and hence do not give a full picture of the abilities of a certain promoter.
Fedbatch with methanol
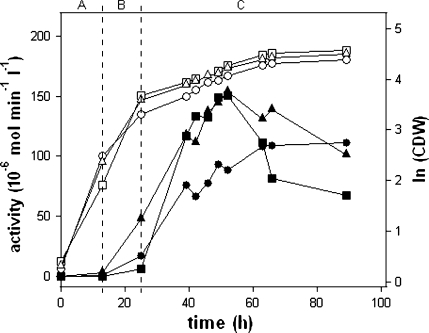
Trypsinogen expression of the best transformants of the promoter variants d1+, d6* and AOX176-Rap1 was analyzed employing methanol induced fedbatch cultivations. In the cultivations a maximal activity of 150 U l−1 was reached for the constructs d1+ and AOX176-Rap1, 55 h after process start (Fig. 3). For both constructs a maximal protein titer of ~700 mg l−1 protein was determined by protein chip analysis. Subsequently the expected degradation of trypsinogen started, which led to reduced absolute trypsinogen activity values after ~60 h process time. After 100 h of cultivation there was almost 100% activity without prior activation by enterokinase.
Fig. 3.
Expression of porcine trypsinogen using AOX1 promoter variants, a growth in batch with glucose, b growth in fedbatch with glucose, c production phase in fedbatch with methanol, open symbols logarithm of cell dry weight, closed symbols trypsinogen activity, promoters: d1+ (squares), d6* (circles), AOX176-Rap1 (triangles)
Variant d6*, in contrast to the tightly repressed promoter d1+, showed a derepression effect and produced 15 U l−1 active enzyme until the end of growth in fedbatch with glucose. Compared to variant d1+ expression was improved ~ threefold at this point of time. Regulated over glucose depletion, variant d6* was still methanol inducible, though weaker. This resulted in a calculated specific productivity of 0.03 U g−1 h−1. The effect of induction via derepression was enhanced several fold by variant AOX176-Rap1 expressing 45 U l−1 of active enzyme until the end of fedbatch phase B. In addition, variant AOX176-Rap1 was also strongly methanol inducible reaching values similar to the d1+ based transformant. Variant d1+ and AOX176-Rap1 reached comparative specific productivity levels of 0.07 and 0.06 U g−1 h−1, but variant d1+ was highest inducible by methanol in a short period of time (28 h) confirming the results obtained in rescreening.
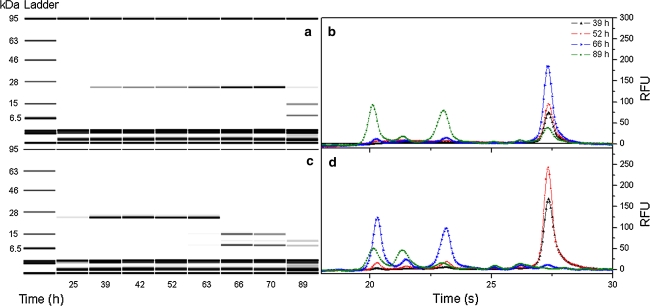
Protein chip analysis of the variants d6* and AOX176-Rap1 revealed a clearly visible trypsinogen/trypsin band at a size of ~24.5 kDa, but only up to a certain point of time (Fig. 4). In correlation with the results from activity measurements, product degradation started after a certain process time. Once trypsinogen was cleaved an autocatalytic cascade started seemingly. According to protein chip analysis we assume that produced trypsin subsequently cleaved all its zymogen to trypsin (23.6 kDa, β-trypsin), which was then degraded into its two α-trypsin chains (~13 and ~10 kDa) or even further fragmented.
Fig. 4.
Protein chip analyses of the methanol induced fedbatch cultures with the promoters d6* (a, b) and AOX176-Rap1 (c, d); a, c gel images visualizing promoter dependent trypsinogen degradation upon a certain production time, b, d corresponding fluorescence detection based chromatograms used for protein concentration estimation at different cultivation times, RFU (relative fluorescence units)
The start point of autoproteolytic degradation differed depending on the promoter used. Employment of the low producer d6*, with a corresponding maximal activity of 120 U l−1, delayed the start point of degradation significantly. In fact, the production time was extended almost twofold for this variant. Starting in fedbatch phase B, a total production time of 57 h was reached. In contrast, using the construct AOX176-Rap1 protein degradation started already after 60 h process time, with a total production time of 39 h. However, though degradation started earlier employing this construct, more active protein (700 mg l−1) was produced compared to the construct d6* (400 mg l−1).
Fedbatch with glucose
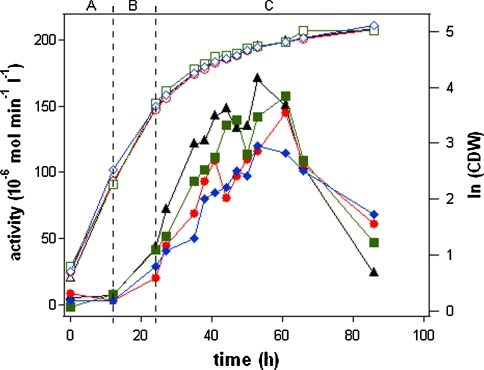
To further analyze the strong derepression effect of the synthetic construct AOX176-Rap1 a second round of fedbatch cultivations was performed employing the strains AOX176-Rap1 G3, A7, C8, B3. Here, the intention was to show the ability of such constructs to express significant amounts of protein via an inducible glucose depletion/derepression based process. Within this series strain AOX176-Rap1 G3 produced up to 170 U l−1 active enzyme at maximum level (Fig. 5). With a production time of 53 h a specific productivity of 0.03 U g−1 h−1 was calculated for this strain. For comparison AOX176-Rap1 G3 induced by methanol reached 150 U l−1 and 0.06 U g−1 h−1, respectively. Also the other AOX176-Rap1 constructs produced significant amounts of active enzyme. Strains A7, B3 and C8 showed maximal activities of 145, 160 and 120 U l−1 with corresponding productivities of 0.02, 0.03 and 0.02 U g−1 h−1.
Fig. 5.
Expression of porcine trypsinogen using synthetic promoters, a growth in batch with glucose, b growth in fedbatch with glucose, c production phase in fedbatch with glucose, open symbols logarithm of cell dry weight, closed symbols trypsinogen activity, AOX176-Rap1 variants: G3 (triangles), A7 (circles), B3 (squares), C8 (rhomboids)
Product degradation started for all strains between 50 and 70 h process time in the following order: G3, A7/B3 and C8 (data not shown). In accordance to the previous results the degradation started first for the high production strain G3 (170 U l−1) and last in case of the low producing transformant C8 (120 U l−1). Again, after about 100 h process time almost all trypsinogen was activated to trypsin.
Final fedbatch comparison
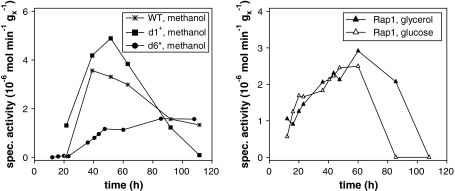
To confirm the obtained results and for further analysis of product formation and its degradation a final cultivation series using different substrates for induction was performed. In agreement with the results described above different production windows were found on account of different promoters (Fig. 6). Interestingly, construct d1+ increased activity per biomass up to 135% reaching 4.9 U g−1X. In addition, promoter d1+ produced already some trypsinogen before induction (1.3 U g−1X). This observation was also confirmed by protein chip analysis showing 25 mg l−1 protein at this time point. Surprisingly, employing the promoter variant d6* did not only result in a prolonged production phase, but also no degradation demonstrated by increasing absolute trypsinogen activity over 110 h of cultivation time (Fig. 6). Employing construct d6* a specific activity of 1.6 U g−1X was reached at maximum level. For comparison the PAOX1 wild type transformant reached a value of 3.6 U g−1X. Cultivations substituting methanol with glucose or glycerol employing promoter variant AOX176-Rap1 (Rap1_Gluc and Rap1_Glyc) showed maximum specific activities between 2.5 and 2.9 U g−1X, which matched with about 75% of the methanol induced WT value. In detail, Rap1_Gluc reached 2.5 and Rap1_Glyc 2.9 U g−1X. Expression profiles of Rap1_Gluc and Rap1_Glyc showed delayed product degradation starting between 60 and 85 h of process time. In comparison, for the constructs WT and d1+ degradation started already between 40 and 60 h of cultivation.
Fig. 6.
Specific trypsinogen activities in fedbatch cultures, left production phase with methanol (WT, d1+, d6*), right production phase with glycerol or glucose (AOX176-Rap1)
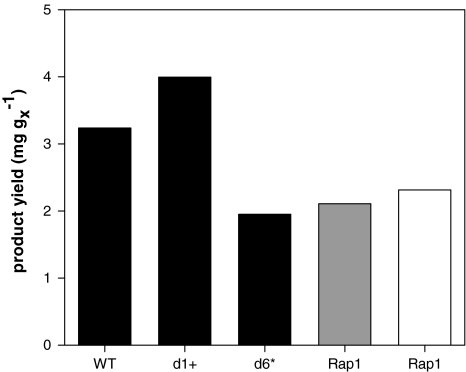
In addition protein yields were calculated (mg g−1X). In agreement to already reported results, variant d1+ performed best reaching a value of 3.99 mg g−1X, while the low producer d6* reached only 1.95 mg g−1X. Strategies employing AOX176-Rap1 on glucose or glycerol reached 2.31 and 2.11 mg g−1X, respectively. Protein yields are summarized in Fig. 7.
Fig. 7.
Product yields obtained in fedbatch cultures with different promoters (WT, d1+, d6*, AOX176-Rap1) applying different carbon sources in the production phase: methanol (black), glycerol (gray), glucose (white), protein concentration determination: Caliper capillary electrophoresis
Discussion
In this study we have shown the effect of differently regulated, laboratory generated, full length and short synthetic promoter variants on the expression of porcine trypsinogen. Briefly, protein titer could be improved up to 135% by using a stronger AOX1 promoter variant. In addition, derepressed promoter variants allowed the production of trypsinogen substituting methanol induction by a simple derepression process. Thus, the different regulatory features of construct AOX176-Rap1 now enable to avoid toxic and flammable methanol in a still tightly regulated cultivation process. Moreover, it was possible to shift the time-span of production and proteolytic product degradation.
Evaluating the screening procedure, it can be said that good correlation was found between the results obtained in rescreening and fedbatch cultivations. Product degradation was already indicated in micro scale, showing decreased activity values at 96 h of induction. Still, it is possible that the observed effect was enhanced by, or just a result of dilution through substrate addition. Interestingly, all expression strains based on full length promoter variants and giving the best results in the screening experiments contained only one single integrated copy of the expression cassette. More copies seemed to influence trypsinogen expression negatively, and were therefore not identified as hit during the screening procedure. Low copy numbers might compensate for toxicity effects of trypsinogen. Only the short synthetic construct AOX176-Rap1 showed a positive correlation between copy number and activity with the best clone having over 4 copies. These results might be explained by the fact that 1 copy of construct AOX176-Rap1 expresses a much lower level of trypsinogen compared to the other constructs, hence is far under the level of toxicity influence. This is in agreement with the results of the stronger full length promoter variants.
Rescreening results from deep well plate cultivations were confirmed by fedbatch cultivations. Generally, up-scaling resulted in an ~tenfold improvement in trypsinogen production per volume. In methanol induced fedbatch cultivations the expression and degradation profile changed depending on the promoter used. As a result of the altered regulation of construct AOX176-Rap1, finally 10 times more trypsinogen was produced until the end fedbatch phase B (see Fig. 3). The previously described d6* promoter variant showed induction by derepression with a threefold increase at this point of time. Furthermore, employing this variant allowed to delay or even abolish product degradation. This might be due to the lower specific productivity of the d6* based strain (on methanol), assuming trypsinogen degradation to be at least partly titer connected. Although variant d6* was outranked in final yield by other promoter constructs, employing this variant significantly improved product homogeneity and thereby quality in regard to production time. Surprisingly, with promoter variant d1+ some active protein was produced until the end of fedbatch phase B (see Fig. 6), if grown on glycerol. Said effect was not found in cultivations using glucose for growth. However, further experiments are needed to confirm or disprove this particular characteristic of promoter construct d1+. Meanwhile it can only be speculated that different carbon sources, here glucose and glycerol, do effect the regulation of d1+ differently. The strong derepression effect of the strains based on construct AOX176-Rap1 was verified in fedbatch studies using both glucose and glycerol as feed substrates during production phase. It was shown that about 75% of the induced WT level can be reached via a regulated glucose depletion mechanism. In addition, employing AOX176-Rap1 in glucose or glycerol driven cultivations led to delayed product degradation. Similar to variant d6* lower productivities were found, indicating decelerated specific productivity to be critical for high product quality.
Promoter variant AOX176-Mat1-Mc was not further studied in bioreactor experiments since promoter variant d1+ showed to be better in methanol induced expression and promoter variant AOX176-Rap1 under derepression conditions. Nevertheless the Mat1-Mc sequence fused to a core promoter led to a remarkable response upon methanol induction (~50% of d1+ driven expression). Also multiple Mxr1p binding sites fused to a core promoter might be useful to improve methanol induced expression by small synthetic promoters. However, though Mxr1p seems to be a key factor the full mechanism of methanol induced regulation is not yet understood. DNA footprinting studies involving such small synthetic promoters in future could facilitate the clarification of AOX1 promoter regulation which might involve several additional factors yet to be identified.
Although a variety of different synthetic promoter variants have proven their ability to successfully express recombinant trypsinogen, there is also room for improvement addressing other bottlenecks. Endogenous proteases might play an important role in the activation process. Gel images obtained from capillary electrophoresis showed that once the proteolytic cascade started, trypsin is further fragmented rapidly. Active protease might be deleterious to the cell and cause a production stop. An approach similar to Hanquier et al. (2003) with mutated protease cleavage sites could eliminate the problem of degradation and might enhance protein production. Other bottlenecks, like transport or folding, can also be limiting factors. Co-expression of helper proteins or chaperones like PDI (protein disulfide isomerase) might facilitate expression, as well.
Conclusions
Well-studied synthetic promoters that vary in their strength and regulation mechanism offer new opportunities in recombinant protein expression. As for the expression of porcine trypsinogen using the strong WT AOX1 promoter, the single copy clones performed best, space for expression optimization using the same regulatory system is limited. Said limitation can be overcome by changing the promoter itself, which was demonstrated in this study by employing novel synthetic promoters to optimize the expression of the problematic reporter trypsinogen.
Acknowledgments
We want to express our gratitude to VTU Technology for scientific and financial support. Furthermore the research was partially supported by the Swiss Academy of Engineering Sciences (SATW) in scope of the “Industrial Biotechnology” program (TK-07-19).
Ethical standards We herewith declare that the experiments comply with the current laws of the country Austria.
Conflict of interest statement This work was financed by VTU Technology. R. Weis is employed by VTU Technology. VTU Technology holds the patent WO2006089329 (Hartner and Glieder (2005) Mutant AOX1 promoters. WO2006089329). The majority of the promoters described in this study are part of this patent. Except those facts the authors for this journal article herewith declare that there is no conflict of interest and no financial relationship between the authors and the funding organization.
References
- Abad S, Kitz K, Hörmann A, Schreiner U, Hartner FS, Glieder A. Real-time PCR-based determination of gene copy numbers in Pichia pastoris. Biotechnol J. 2010;5:413–420. doi: 10.1002/biot.200900233. [DOI] [PubMed] [Google Scholar]
- Chance R, Di Marchi R, Frank B, Shields JE (1997) Process for preparing insulin analogs. US5700662
- Cregg JM, Barringer KJ, Hessler AY, Madden KR. Pichia pastoris as a host system for transformations. Mol Cell Biol. 1985;5:3376–3385. doi: 10.1128/mcb.5.12.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg JM, Vedvick TS, Raschke WC. Recent advances in the expression of foreign genes in Pichia pastoris. Biotechnology (NY) 1993;11:905–910. doi: 10.1038/nbt0893-905. [DOI] [PubMed] [Google Scholar]
- Cregg JM, Cereghino JL, Shi J, Higgins DR. Recombinant protein expression in Pichia pastoris. Mol Biotechnol. 2000;16:23–52. doi: 10.1385/MB:16:1:23. [DOI] [PubMed] [Google Scholar]
- Frank B, Prouty W, Richard H, Walden M (1995) Process for transforming a human insulin precursor to human insulin. US5457066
- Hanquier J, Sorlet Y, Desplancq D, Baroche L, Ebtinger M, Lefevre JF, Pattus F, Hershberger CL, Vertes AA. A single mutation in the activation site of bovine trypsinogen enhances its accumulation in the fermentation broth of the yeast Pichia pastoris. Appl Environ Microbiol. 2003;69:1108–1113. doi: 10.1128/AEM.69.2.1108-1113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartner FS, Glieder A (2005) Mutant AOX1 promoters. WO2006089329
- Hartner FS, Ruth R, Langenegger D, Johnson SN, Hyka P, Lin-Cereghino GP, Lin-Cereghino J, Kovar K, Cregg JM, Glieder a. Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res. 2008;36(12):e76. doi: 10.1093/nar/gkn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasslacher M, Schall M, Hayn M, Bona R, Rumbold K, Luckl J, Griengl H, Kohlwein SD, Schwab H. High-level intracellular expression of hydroxynitrile lyase from the tropical rubber tree Hevea brasiliensis in microbial hosts. Protein Expr Purif. 1997;11:61–71. doi: 10.1006/prep.1997.0765. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Kondo T, Yamazaki Y, Ito K, Iinuma Y, Okumura N, Sakakibara A, Naruse S, Toda Y, Aoki I. Diagnostic significance of serum immunoreactive trypsin in pancreatic diseases. Nippon Shokakibyo Gakkai Zasshi. 1979;76:1513–1521. [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Inan M, Meagher MM, Benson AK (2004) Alcohol oxidase 1 regulatory nucleotide sequences for heterologous gene expression in yeast. US2004137591
- Kranthi BV, Kumar R, Kumar NV, Rao DN, Rangarajan PN. Identification of key DNA elements involved in promoter recognition by Mxr1p, a master regulator of methanol utilization pathway in Pichia pastoris. Biochim Biophys Acta. 2009;1789:460–468. doi: 10.1016/j.bbagrm.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Lin-Cereghino J, Wong WW, Xiong S, Giang W, Luong LT, Vu J, Johnson SD, Lin-Cereghino GP. Condensed protocol for competent cell preparation and transformation of the methylotrophic yeast Pichia pastoris. Biotechniques. 2005;38:44–48. doi: 10.2144/05381BM04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Cereghino GP, Godfrey L, La Cruz BJ, Johnson S, Khuongsathiene S, Tolstorukov I, Yan M, Lin-Cereghino J, Veenhuis M, Subramani S, Cregg JM. Mxr1p, a key regulator of the methanol utilization pathway and peroxisomal genes in Pichia pastoris. Mol Cell Biol. 2006;26:883–897. doi: 10.1128/MCB.26.3.883-897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud RM, Kossiakoff AA, Chambers JL. Mechanisms of zymogen activation. Annu Rev Biophys Bioeng. 1977;6:177–193. doi: 10.1146/annurev.bb.06.060177.001141. [DOI] [PubMed] [Google Scholar]
- Verdier JM. Regulatory DNA-binding proteins in yeast: an overview. Yeast. 1990;6:271–297. doi: 10.1002/yea.320060402. [DOI] [PubMed] [Google Scholar]
- Wang W, Malcolm BA. Two-stage polymerase chain reaction protocol allowing introduction of multiple mutations, deletions, and insertions, using QuikChangeTM site-directed mutagenesis. Methods Mol Biol. 2002;182:37–43. doi: 10.1385/1-59259-194-9:037. [DOI] [PubMed] [Google Scholar]
- Weis R, Luiten R, Skranc W, Schwab H, Wubbolts M, Glieder A. Reliable high-throughput screening with Pichia pastoris by limiting yeast cell death phenomena. FEMS Yeast Res. 2004;5:179–189. doi: 10.1016/j.femsyr.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Werten MW, Bosch TJ, Wind RD, Mooibroek H, Wolf FA. High-yield secretion of recombinant gelatins by Pichia pastoris. Yeast. 1999;15:1087–1096. doi: 10.1002/(SICI)1097-0061(199908)15:11<1087::AID-YEA436>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Xiong AS, Yao QH, Peng RH, Zhang Z, Xu F, Liu JG, Han PL, Chen JM. High level expression of a synthetic gene encoding Peniophora lycii phytase in methylotrophic yeast Pichia pastoris. Appl Microbiol Biotechnol. 2006;72:1039–1047. doi: 10.1007/s00253-006-0384-8. [DOI] [PubMed] [Google Scholar]
- Xuan Y, Zhou X, Zhang W, Zhang X, Song Z, Zhang Y. An upstream activation sequence controls the expression of AOX1 gene in Pichia pastoris. FEMS Yeast Res. 2009;9:1271–1282. doi: 10.1111/j.1567-1364.2009.00571.x. [DOI] [PubMed] [Google Scholar]