Abstract
Cellulosomes are efficient cellulose-degradation systems produced by selected anaerobic bacteria. This multi-enzyme complex is assembled from a group of cellulases attached to a protein scaffold termed scaffoldin, mediated by a high-affinity protein–protein interaction between the enzyme-borne dockerin module and the cohesin module of the scaffoldin. The enzymatic complex is attached as a whole to the cellulosic substrate via a cellulose-binding module (CBM) on the scaffoldin subunit. In previous works, we have employed a synthetic biology approach to convert several of the free cellulases of the aerobic bacterium, Thermobifida fusca, into the cellulosomal mode by replacing each of the enzymes’ CBM with a dockerin. Here we show that although family six enzymes are not a part of any known cellulosomal system, the two family six enzymes of the T. fusca system (endoglucanase Cel6A and exoglucanase Cel6B) can be converted to work as cellulosomal enzymes. Indeed, the chimaeric dockerin-containing family six endoglucanase worked well as a cellulosomal enzyme, and proved to be more efficient than the parent enzyme when present in designer cellulosomes. In stark contrast, the chimaeric family six exoglucanase was markedly less efficient than the wild-type enzyme when mixed with other T. fusca cellulases, thus indicating its incompatibility with the cellulosomal mode of action.
Keywords: Cellulases, Designer cellulosome, Degradation of crystalline cellulose, Enzyme synergy, Substrate targeting and enzyme proximity, Bioenergy
Introduction
Cellulosomes are multi-enzymatic complexes produced by anaerobic bacteria that efficiently degrade plant matter to simple sugars (Bayer et al. 2004; Doi and Kosugi 2004; Demain et al. 2005). The different cellulose-degrading enzymes (cellulases) are assembled on a long backbone scaffold protein (scaffoldin) via the affinity of their integral docking module (dockerin) to complementary modules (cohesins) on the scaffoldin. In addition, the scaffoldin bears a carbohydrate-binding module (CBM), which binds the entire enzymatic complex to the cellulosic substrate. The scaffoldin is also usually attached to the cell wall by an anchoring protein, and thus the whole cell is attached to its source of carbon and energy. The efficient synergistic degradation of plant matter is thus considered to be a combination of both the targeting of the enzymatic complex to the substrate (targeting effect) and the spatial proximity of the different cellulases to each other (proximity effect).
Based on the modular nature of the cellulosomal system and its potential to increase synergism among cellulose-degrading enzymes, the concept of designer cellulosomes was proposed (Bayer et al. 1994). Designer cellulosomes are composed of dockerin-bearing cellulases (either wild type or chimaeric), which are assembled onto an artificial scaffoldin, composed of a CBM and cohesin modules from different cellulosomal species, complementary to those of the dockerins. The order and content of such designer cellulosomes can be predetermined, owing to the high species specificity of dockerin modules for their target cohesins (Pagès et al. 1997; Mechaly et al. 2000, 2001; Jindou et al. 2004), and the hydrolytic efficiency produced by the multienzyme assembly is similar to that of the parallel mixture of free enzymes.
Fierobe et al. (2001), (2002), (2005) constructed binary and ternary cellulase-containing designer cellulosomes, based on cellulases of the bacterium Clostridium cellulolyticum. In these studies, the designer cellulosomes were shown to be more efficient than the free-mode enzymes, and both targeting and a proximity effects were demonstrated.
In previous studies, we have shown the successful conversion of four out of the six cellulases, comprising the free-cellulase system of the aerobic bacterium, Thermobifida fusca, to the cellulosomal mode (Caspi et al. 2006, 2008). Each of the catalytic modules (exoglucanases Cel6B and Cel48A, and endoglucanases Cel6A and Cel5A) was synthetically fused to a dockerin module derived from a native cellulosomal enzyme, and each chimaera bound specifically to its target cohesin module and retained a certain level of cellulose-degrading ability. In addition, we have shown an efficient assembly of a two-chimaera designer cellulosome, which degraded crystalline cellulose substrates more efficiently than the parallel combination of the original free wild-type enzymes (Caspi et al. 2009).
In this communication we present a series of novel chimaeric binary and ternary designer cellulosomes, based on the converted free enzymes of T. fusca. All of the chimaeras displayed the basic substrate-targeting effect. Three of the four converted cellulases were shown to be efficiently adapted to the cellulosomal mode of activity, in which synergistic degradation of crystalline cellulosic substrates could be demonstrated. In contrast, the family six exoglucanase (Cel6B) was shown to function better as a free enzyme as opposed to the cellulosomal mode.
Materials and methods
Cloning of wild-type T. fusca enzymes
Wild-type Cel6A, Cel6B, Cel5A and Cel48A, each containing a CBM2, were cloned as described previously (Ghangas and Wilson 1988; Zhang et al. 1995; Irwin et al. 2000).
Cloning of the chimaeric proteins
The 6A-c chimaera (Cel6A catalytic module with a C-terminal C. cellulolyticum Cel5A dockerin) and the t-6B chimaera (Cel6B catalytic module with an N-terminal Clostridium thermocellum xylanase Xyn10Z dockerin) were constructed as previously described (Caspi et al. 2006). The t-5A chimaera (Cel5A catalytic module with an N-terminal C. thermocellum xylanase Xyn10Z dockerin), the f-5A chimaera (Cel5A catalytic module with an N-terminal Ruminococcus flavefaciens ScaB dockerin) and the b-48A chimaera (Cel48A catalytic module with an N-terminal Bacteroides cellulosolvens ScaA dockerin) were constructed as previously described (Caspi et al. 2008). DNA encoding the C. thermocellum dockerin Xyn10Z was amplified from C. thermocellum genomic DNA, using the primers 5- TTAAGGTACCTGAAAGCAGTTCCACAGG -3 (KpnI site in boldface) and 5- TATACTCGAGTCCGGGGAACTCTGTAATAATGC -3 (XhoI site in boldface). The amplified C. thermocellum dockerin was ligated to a KpnI-XhoI linearized pET28a that previously contained p-6A-c to form p-6A-t.
Scaf·CT, Scaf·CTF and Scaf·BT were cloned as described previously (Yaron et al. 1995; Fierobe et al. 2005; Caspi et al. 2009). Scaf·TF was produced based on the Scaf·CTF plasmid, using the primers 5- AATTCCATGGCGACAAACACACCGACAAACACACC -3 (NcoI site in boldface) and 5- GTGGCTCGAGTTAAACAATGATAGCGCC -3 (XhoI site in boldface). Primers: 5- TTAAGGATCCAGCAGTGTTTCTCCAACAACAAGTGTGC -3 (BamHI site in boldface) and 5-GGCCGGATCCTTAACTAGTAATTGGCTTATTAGTTACAGTAATGC -3 (BamHI site in boldface) were used to amplify the third cohesin of B. cellulosolvens ScaB. This PCR product was ligated into Scaf·CT containing BamHI-linearized pET9d resulting in the Scaf·CTB construct.
Protein expression and purification
Cel6A, Cel6B, Cel5A and Cel48A were prepared as previously described (Ghangas and Wilson 1988; Zhang et al. 1995; Irwin et al. 2000). The family 6, 5 and 48 derived chimaeras (6A-c, 6A-t, t-6B, f-5A and b-48A) were expressed in E. coli BL21 (λDE3) pLysS cells, and purified on a Ni column, as previously described (Caspi et al. 2006, 2008).
Purification of single cohesin scaffoldins (Scaf·C, Scaf·T, Scaf·F and Scaf·B) and double and triple cohesin scaffoldins (Scaf·BT, Scaf·CT, Scaf·TF, Scaf·CTF and Scaf·CTB) was carried out on phosphoric acid swollen cellulose (PASC) as previously reported (Caspi et al. 2006).
All purified proteins were stored in 50% (vol/vol) glycerol at −20°C. Purity of all proteins was tested by SDS–PAGE on 12% acrylamide gels. The concentration of each protein was determined by measuring its absorbance (280 nm) in 6 M guanidine hydrochloride, according to the estimated values calculated using the ProtParam tool (www.expasy.org/tools/protparam.html).
Preparation of designer cellulosomes
Molar equivalents of the desired dockerin-bearing enzymes and chimaeric scaffoldin were combined in TBS, containing 5 mM CaCl2 and 0.025% Tween20, and allowed to incubate at 37°C for 1 h to ensure proper complex formation.
Affinity-based ELISA
The procedure of Barak et al. (2005), Caspi et al. (2006), was followed. Rabbit anti-Cel6A (diluted 1:20,000 in blocking buffer) was employed as primary antibody for detection of the interaction of the 6A-t dockerin with its cohesin counterpart.
Enzyme activity
Enzyme activity was assayed on bacterial microcrystalline cellulose (BMCC) and Avicel, using a reducing sugar assay, according to the previously reported procedure (Caspi et al. 2008). Final enzyme concentrations of 1 μM of each component (combinations of wild-type or chimaeras, designer cellulosomes or CBM-complexed chimaeras) were used, and assays were performed in triplicate. Specific activity was defined as mol reducing sugar per mol enzyme per minute.
Results
Construction and expression of recombinant proteins
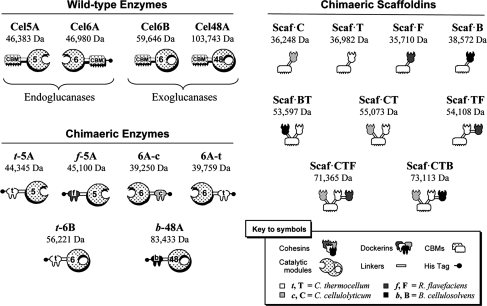
Nineteen different recombinant proteins were designed, expressed and purified in order to perform this work (see Fig. 1). Several of these were described in previous studies by our group, and others were designed specifically for the current study. The four wild-type T. fusca enzymes, Cel6A, Cel6B, Cel5A and Cel48A, were cloned and purified as described previously (Ghangas and Wilson 1988; Zhang et al. 1995; Irwin et al. 2000). Chimaeric derivatives of the two exoglucanases and two endoglucanases were designed to contain divergent dockerins in order to construct the desired designer cellulosomes required for this study. In all cases, the dockerins replaced the CBM2 at its normal position in the native protein. Preparation of dockerin-containing chimaeras of the T. fusca endoglucanase Cel5A was described earlier (Caspi et al. 2006). t-5A and f-5A consisted of the catalytic module of Cel5A appended with the respective dockerins from C. thermocellum xylanase Xyn10Z and from R. flavefaciens scaffoldin ScaB. The activities of the latter two chimaeras on crystalline cellulosic substrates were found to be analogous as reported earlier (Caspi et al. 2009), and either chimaeric enzymes could thus be used according to the dockerin specificity requirements of the chimaric scaffoldin. Chimaera 6A-c comprised the catalytic module of T. fusca endoglucanase Cel6A appended with a dockerin from the C. cellulolyticum Cel5A, as described earlier (Caspi et al. 2006). In addition, 6A-t was designed to provide another option for a 6A-based chimaera, with an alternative dockerin derived from the C. thermocellum xylanase Xyn10Z. Chimaera t-6B contained the catalytic module of T. fusca exoglucanase Cel6B with the same C. thermocellum dockerin attached to its N-terminus (Caspi et al. 2006). Chimaera b-48A comprised the catalytic module of T. fusca exoglucanase Cel48A, appended with a dockerin from the B. cellulosolvens ScaA scaffoldin (Ding et al. 2000), as described earlier (Caspi et al. 2006).
Fig. 1.
Schematic representation of the recombinant proteins used in this study. The CBM2s of the respective wild-type T. fusca enzymes (Cel5A, Cel6A, Cel6B or Cel48A) were replaced by a dockerin. The shading of each symbol denotes the source of the module: light gray (C. cellulolyticum), white (C. thermocellum), dark gray (R. flavefaciens), black (B. cellulosolvens), stippled (T. fusca). The corresponding bacterial source—C. cellulolyticum, C. thermocellum, R. flavefaciens or B. cellulosolvens—is additionally indicated by the following notations: c,t, f, or b for the dockerins and C, T, F or B for the cohesins, respectively. Chimaeras t-5A, f-5A, 6A-c, 6A-t, t-6B and b-48A included a His tag, attached distally to the respective dockerin module of the enzyme. The divergent species of cohesin-bearing scaffoldin (Scaf) included a C. thermocellum CBM3a, shown symbolically. The molecular mass is shown for each of the expressed proteins
Four different species of single cohesin-containing constructs were also prepared for this study according to previous reports (Barak et al. 2005; Haimovitz et al. 2008). The cohesins were each located C-terminally to a C. thermocellum CBM3a. Scaf·C contained the first cohesin from C. cellulolyticum CipC, Scaf·T the third cohesin from C. thermocellum CipA, Scaf·F the first cohesin from R. flavefaciens ScaB, and Scaf·B the third cohesin from B. cellulosolvens ScaB. In order to prepare higher-order designer cellulosomes, five additional artificial scaffoldins, each bearing two or three divergent cohesins, were prepared for this study: Scaf·BT, Scaf·CT, Scaf·TF, Scaf·CTF and Scaf·CTB, each containing the C. thermocellum CBM3a for substrate targeting together with the designated cohesins. The CBM3a served as a substrate-targeting agent, an affinity tag for isolation on a cellulose matrix (Morag et al. 1995) and may also improve the expression, solubility and stability characteristics of the different fusion proteins (Berdichevsky et al. 1999).
All of the purified recombinant proteins showed a single major band on SDS–PAGE (not shown), and in each case the mobility was consistent with their respective molecular mass. The data for f-5A, t-5A, 6A-c, t-6B, b-48A, scaffoldins C, T, F, B and BT were reported earlier (Caspi et al. 2006, 2008, 2009). Similar levels of purity were found for the other enzymes and scaffoldins prepared in the present study.
Specificity of the chimaeric enzyme-borne dockerins
The specificity of all chimaeric dockerin-containing enzymes to their target cohesins was demonstrated in previous works (Pagès et al. 1997; Barak et al. 2005; Caspi et al. 2006, 2008, 2009; Haimovitz et al. 2008), except for the 6A-t chimaera. To test its specificity, the same procedure using affinity-based ELISA was conducted, and the chimaera was shown to bind selectively to its Scaf·T target, and not to any of the non-matching cohesins (data not shown).
Hydrolytic activity of the dockerin-containing enzymes
Combinations of the different chimaeric enzymes were examined for their activities on two crystalline forms of cellulose, BMCC (Fig. 2) and Avicel (Fig. 3). Each combination of enzymes was tested in the following configurations: Free, whereby the chimaeric dockerin-bearing enzymes were applied in the absence of scaffoldin(s); Scaf, whereby the same enzymes were incorporated into a designer cellulosome (in which the chimaeras are all complexed on a single chimaeric scaffoldin); and CBM, in which each chimaera is attached individually to its matching cohesin-CBM fusion protein. These samples were compared to the parallel mixture of wild-type enzymes (WT).
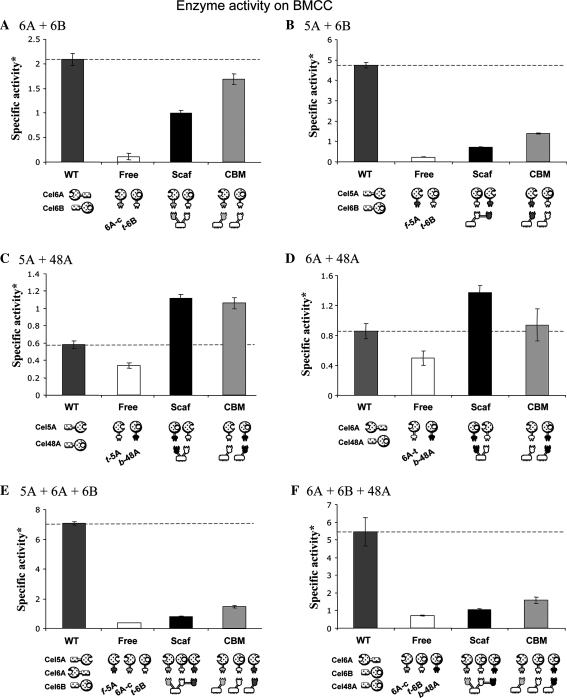
Fig. 2.
Enzymatic activity of designer cellulosomes of different compositions on BMCC. Each diagram below the bargraph represents different combinations of the designated enzymes, designated by symbols represented in Fig. 1. In all diagrams, WT indicates the mixture of the naturally secreted CBM-bearing enzymes, Free refers to the mixture of the dockerin-containing cellulase chimaeras alone, Scaf represents the designer cellulosome complexes (with the indicated chimaeric scaffoldin), and CBM denotes a mixture of single dockerin-containing enzymes complexed with matching CBM-Coh fusion proteins to restore the substrate-targeting function while retaining the free enzyme mode. Specific activity* is defined as mol reducing sugar per mol enzyme per minute. Errorbars denote standard deviations
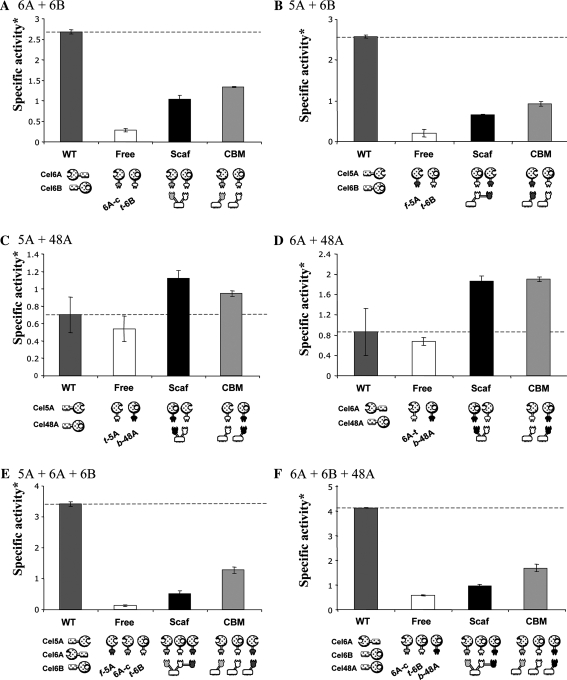
Fig. 3.
Enzymatic activity of designer cellulosomes of different compositions on Avicel. For details, see legend to Fig. 2
The combinations of chimaeric enzymes were composed of at least one endoglucanase and one exoglucanase. All possible binary combinations were tested, in addition to two combinations of ternary chimaera mixtures: 5A + 6A + 6B (two endoglucanases and an exoglucanase) and 6A + 6B + 48A (an endoglucanase and two exoglucanases).
Of the four tested enzymes, the family six exoglucanase was demonstrated to be the most crucial for synergistic cellulose degradation. Wild type mixtures containing the Cel6B cellulase (Figs. 2, 3, panels A, B, E and F) were far more active on both BMCC and Avicel compared to any combination that lacked Cel6B (either wild type or converted to the cellulosomal mode).
Interestingly, all combinations that included the chimaeric Cel6B exoglucanase (t-6B) were significantly less efficient in the degradation of both microcrystalline cellulose substrates, compared to the wild-type cellulase mixture (Figs. 2 and 3, panels A, B, E, F). In contrast, pairs of chimaeric enzymes that did not include the chimaeric family six exoglucanase (5A + 48A and 6A + 48A) hydrolyzed crystalline cellulose in a more efficient manner than the parallel combination of wild-type enzymes (Figs. 2, 3, panels C, D). A significant targeting effect was demonstrated by all tested combinations. The relatively poor cellulose-degrading activity of the free dockerin-containing chimaeras was significantly improved by adding the matching cohesin-CBM fusion protein. The latter served to restore the targeting function to the chimaeric enzymes, which was lost upon replacement of the CBM2 of the wild-type enzyme with a dockerin module. All designer cellulosomes that contained the t-6B exoglucanase exhibited an “anti-proximity effect”, i.e., restoration of the targeting function in the individual free chimaeras (CBM samples) proved more active than the analogous designer cellulosomes (Scaf samples). In contrast, designer cellulosomes lacking the family six exoglucanase appeared, in some cases (Figs. 2d, 3c), to exhibit a defined proximity effect on crystalline cellulose substrates.
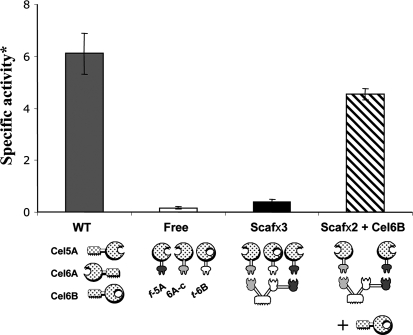
To further explore the preference of the family six exoglucanse for the free mode, a designer cellulosome sample, composed of three chimaeric enzymes—f-5A, 6A-c and t-6B, assembled on Scaf·CTF—was compared with wild-type (free) Cel6B added to a binary designer cellulosome—6A-c and f-5A, assembled on the same scaffoldin (Fig. 4). The results revealed that the mixture of wild-type Cel6B with the binary system was significantly more efficient in degrading BMCC than the ternary designer cellulosome, further supporting our contention that the family 6 exoglucanase prefers the free enzyme mode.
Fig. 4.
Incompatibility of Cel6B exoglucanase with the cellulosomal mode of action. The enzymatic activity of complexed versus free forms of Cel6A, Cel5A and Cel6B on BMCC was determined. WT indicates the mixture of the naturally secreted CBM-bearing enzymes, Free refers to the mixture of the dockerin-containing cellulase chimaeras alone, Scafx3 represents the designer cellulosome complex containing the three chimaeric enzymes, and Scafx2 + Cel6B denotes the mixture of the wild-type Cel6B cellulase in free form together with a designer cellulosome containing chimaeras of the other two enzymes. Specific activity* is expressed as mol reducing sugar per mol enzyme per minute. Errorbars denote standard deviations
Discussion
In nature, cellulosome-producing bacteria incorporate enzymes into multi-component complexes in a highly regulated process (Dror et al. 2003a, b, 2005; Bayer et al. 2004; Newcomb et al. 2007). Since the specificities of the scaffoldin-borne cohesins and enzyme-borne dockerins are virtually identical (Yaron et al. 1995; Pagès et al. 1997), selective incorporation of a given enzyme in situ is not determined at the biochemical level of the cohesin-dockerin interaction itself, but by a different undetermined cellular mechanism. Consequently, in order to control the incorporation in vitro of a given enzyme into an artificial cellulosome, we have to employ cohesins and dockerins of divergent specificities. For this purpose, the concept of designer cellulosomes was originally proposed (Bayer et al. 1994). Designer cellulosomes have since been employed as a tool to investigate cellulosome structure and function (Fierobe et al. 2001, 2002, 2005; Mingardon et al. 2007b; Caspi et al. 2009) and may eventually provide a platform for industrial application, such as for conversion of lignocellulosic materials to soluble sugars en route to biofuels (Bayer et al. 2007, 2008a, b; Wilson 2009).
Technically, the production of discrete designer cellulosomes involves the development of a set of appropriate high-affinity cohesin-dockerin pairs that exhibit different specificities, usually from different cellulosome-producing bacteria. The cohesin modules are joined together into a recombinant chimaeric scaffoldin, usually with characteristic intermodular linker segments and a CBM for introduction of the substrate-binding function. The complementary dockerin module is attached to a catalytic module from an appropriate enzyme, and the various recombinant chimaeric enzymes can then be incorporated into desired designer cellulosome complexes.
Rigorous analyses of the various modular components and the chimaeric enzymes, both alone and in complex, are necessary in order to validate the characteristics of the resultant designer cellulosomes. The enzymes may originally be cellulosomal or non-cellulosomal in nature. In the case of cellulosomal enzymes, the native dockerin module must be replaced by a divergent dockerin species; in the case of the free, non-cellulosomal enzymes, a dockerin must be added—in most instances by replacing a native CBM.
In previous work (Caspi et al. 2006, 2008, 2009), we have selected the free enzyme system of the aerobic bacterium, T. fusca (Wilson 2004), in order to provide a defined platform for conversion of distinctive cellulases into the cellulosome mode. The employment of an alien (non-cellulosomal) system for this purpose, allows us to address whether the cellulosomal mode of action is reserved for certain types or families of cellulases, or whether inclusion of enzymes into cellulosomes can be of a more general nature. In the case of the restricted number (six) of the native T. fusca cellulases, four are in families of common occurrence in cellulosomes, i.e., Cel5A, Cel9A, Cel9B and Cel48. In contrast, two additional enzymes are from family six, and to date no family six cellulase is known to be a part of any natural cellulosome; they appear only in free cellulase systems of both fungi and bacteria. The fact that T. fusca produces two family six enzymes—one an endoglucanase (Cel6A) and the other an exoglucanase (Cel6B)—enabled us to examine whether either or both of these non-cellulosomal enzyme types can function in the cellulosome mode. Moreover, we can determine how efficiently they perform on different cellulosic substrates together with other enzymes in designer cellulosomes.
In earlier work (Caspi et al. 2006, 2008), we converted both of the T. fusca family six enzymes to the cellulosome mode by removing the resident CBM and replacing it with a dockerin. The activities of the resultant chimaeric enzymes were examined individually on various cellulosic substrates. The data indicated that the chimaeric dockerin-containing endoglucanase (6A-c) would be appropriate for inclusion into designer cellulosomes, but implied that the chimaeric exoglucanase (t-6B) appeared unsuitable for the cellulosome mode. The latter dockerin-containing enzyme exhibited significantly reduced activity on crystalline cellulosic substrates, compared to the native form.
In contrast to the family six exoglucanase, the chimaeric form of the other T. fusca exoglucanase, the family 48 enzyme, displayed heightened activity on such substrates, indicating its compatibility with the cellulosome mode (Caspi et al. 2008). Indeed, a subsequent study demonstrated synergistic cellulose hydrolysis of the chimeric dockerin-containing family 48 exoglucanase when incorporated into a designer cellulosome together with the T. fusca family five endoglucanase (Caspi et al. 2009).
In the present communication, we have employed the designer cellulosome system to investigate how the various T. fusca enzymes act together in the cellulosome mode. The results demonstrate that the chimaeric T. fusca endoglucanase, 6A-c, acts in concert with the family 48 enzyme in designer cellulosomes, while displaying enhanced synergism with a defined proximity effect. On the other hand, the chimaeric exoglucanase, t-6B appears to require a CBM in the free mode for optimal activity. In a recent work, Fierobe et al. (Mingardon et al. 2007a) also studied the combination of a non-cellulosomal family six endoglucanase from the fungus, Neocallimastix patriciarum, with bacterial enzymes of cellulosomal origin. Consistent with our current study, this report showed that converted family six endoglucanases can provide efficient, synergistic hydrolysis of cellulosic substrates as components of designer cellulosomes. In view of the apparent compatibility of the family six endoglucanases with the cellulosomal mode, it is somewhat peculiar that cellulosome-producing bacteria have not acquired these particular enzymes as part of their enzyme arsenal, as they have acquired so many other enzymatic components from other families of glycoside hydrolases.
It is interesting to speculate why the family six exoglucanase was incompatible with the cellulosome mode as opposed to the endoglucanase from the same family. The answer may lie in their different active-site topographies. The endoglucanase exhibits a cleft-like active site that can accommodate any segment along the cellulose chain, whereas that of the exoglucanase is tunnel-shaped, accessible only to the non-reducing chain end of the substrate. Moreover, the processive nature of the exoglucanase may be dependent on the intimate and correct orientation of the catalytic module with the CBM. Thus, the scaffoldin-borne CBM may not suffice to facilitate the threading of the substrate into the tunnel.
The difference in compatibility of the two exoglucanases with the cellulosome mode corresponds nicely with the natural state, wherein family 48 exoglucanases are common components of all known cellulosome systems, whereas family six enzymes have not yet been found in cellulosome-producing bacteria. The differences between the two families of exoglucanases may reflect putative differences in their mechanism of action on cellulose (Teeri 1997).
Finally, the results of this study demonstrate that every prospective enzyme, destined for incorporation into designer cellulosomes, must be subjected to systematic analysis to ensure its compatibility with the cellulosome mode of action. Only following thorough examination of its characteristics can an enzyme be verified for use as a component of designer cellulosomes.
Acknowledgments
The authors are grateful for the expert assistance of Sarah Ouanounou Moraïs throughout the stages of preparation of this manuscript. This research was supported by the Brazilian friends of the Weizmann institute of science Alternative Energy Research Initiative (research grants from Mr. Charles Rothschild, Mr. Mario Fleck, and Roberto, and Renata Ruhman), and by grants from the United States-Israel Binational Science Foundation (BSF), Jerusalem, Israel and by the Israel Science Foundation (Grant nos 966/09 and 159/07). E.A.B. holds The Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry.
References
- Barak Y, Handelsman T, Nakar D, Mechaly A, Lamed R, Shoham Y, Bayer EA. Matching fusion-protein systems for affinity analysis of two interacting families of proteins: the cohesin–dockerin interaction. J Mol Recogit. 2005;18:491–501. doi: 10.1002/jmr.749. [DOI] [PubMed] [Google Scholar]
- Bayer EA, Morag E, Lamed R. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 1994;12:378–386. doi: 10.1016/0167-7799(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Bayer EA, Belaich J-P, Shoham Y, Lamed R. The cellulosomes: multi-enzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- Bayer EA, Lamed R, Himmel ME. The potential of cellulases and cellulosomes for cellulosic waste management. Curr Opin Biotechnol. 2007;18:237–245. doi: 10.1016/j.copbio.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Bayer EA, Henrissat B, Lamed R. The cellulosome: a natural bacterial strategy to combat biomass recalcitrance. In: Himmel ME, editor. Biomass recalcitrance. London: Blackwell; 2008. pp. 407–426. [Google Scholar]
- Bayer EA, Shoham Y, Lamed R. Cellulosome-enhanced conversion of biomass: on the road to bioethanol. In: Wall J, Harwood C, Demain AL, editors. Bioengergy. Washington, DC: ASM Press; 2008. pp. 75–96. [Google Scholar]
- Berdichevsky Y, Lamed R, Frenkel D, Gophna U, Bayer EA, Yaron S, Shoham Y, Benhar I. Matrix-assisted refolding of single-chain Fv-cellulose binding domain fusion proteins. Protein Expr Purif. 1999;17:249–259. doi: 10.1006/prep.1999.1125. [DOI] [PubMed] [Google Scholar]
- Caspi J, Irwin D, Lamed R, Shoham Y, Fierobe H-P, Wilson DB, Bayer EA. Thermobifida fusca family-6 cellulases as potential designer cellulosome components. Biocat Biotransform. 2006;24:3–12. doi: 10.1080/10242420600598046. [DOI] [Google Scholar]
- Caspi J, Irwin D, Lamed R, Fierobe H-P, Wilson DB, Bayer EA. Conversion of noncellulosomal Thermobifida fusca free exoglucanases into cellulosomal components: comparative impact on cellulose-degrading activity. J Biotechnol. 2008;135:351–357. doi: 10.1016/j.jbiotec.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Caspi J, Barak Y, Haimovitz R, Irwin D, Lamed R, Wilson DB, Bayer EA. Conversion of a Thermobifida fusca endoglucanase into the cellulosomal mode: effect of linker length and dockerin position. Appl Environ Microbiol. 2009;75:7335–7342. doi: 10.1128/AEM.01241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demain AL, Newcomb M, Wu JH. Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev. 2005;69:124–154. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S-Y, Bayer EA, Steiner D, Shoham Y, Lamed R. A scaffoldin of the Bacteroides cellulosolvens cellulosome that contains 11 type II cohesins. J Bacteriol. 2000;182:4915–4925. doi: 10.1128/JB.182.17.4915-4925.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi RH, Kosugi A. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat Rev Microbiol. 2004;2:541–551. doi: 10.1038/nrmicro925. [DOI] [PubMed] [Google Scholar]
- Dror TW, Morag E, Rolider A, Bayer EA, Lamed R, Shoham Y. Regulation of the cellulosomal celS (cel48A) gene of Clostridium thermocellum is growth-rate dependent. J Bacteriol. 2003;185:3042–3048. doi: 10.1128/JB.185.10.3042-3048.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror TW, Rolider A, Bayer EA, Lamed R, Shoham Y. Regulation of expression of scaffoldin-related genes in Clostridium thermocellum. J Bacteriol. 2003;185:5109–5116. doi: 10.1128/JB.185.17.5109-5116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror TW, Rolider A, Bayer EA, Lamed R, Shoham Y. Regulation of major cellulosomal endoglucanases of Clostridium thermocellum differs from that of a prominent cellulosomal xylanase. J Bacteriol. 2005;187:2261–2266. doi: 10.1128/JB.187.7.2261-2266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierobe H-P, Mechaly A, Tardif C, Belaich A, Lamed R, Shoham Y, Belaich J-P, Bayer EA. Design and production of active cellulosome chimeras: selective incorporation of dockerin-containing enzymes into defined functional complexes. J Biol Chem. 2001;276:21257–21261. doi: 10.1074/jbc.M102082200. [DOI] [PubMed] [Google Scholar]
- Fierobe H-P, Bayer EA, Tardif C, Czjzek M, Mechaly A, Belaich A, Lamed R, Shoham Y, Belaich J-P. Degradation of cellulose substrates by cellulosome chimeras: substrate targeting versus proximity of enzyme components. J Biol Chem. 2002;277:49621–49630. doi: 10.1074/jbc.M207672200. [DOI] [PubMed] [Google Scholar]
- Fierobe H-P, Mingardon F, Mechaly A, Belaich A, Rincon MT, Lamed R, Tardif C, Belaich J-P, Bayer EA. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined tri-functional scaffoldin. J Biol Chem. 2005;280:16325–16334. doi: 10.1074/jbc.M414449200. [DOI] [PubMed] [Google Scholar]
- Ghangas GS, Wilson DB. Cloning of the Thermomonospora fusca endoglucanase E2 gene in Streptomyces lividans: affinity purification and functional domains of the cloned gene product. Appl Environ Microbiol. 1988;54:2521–2526. doi: 10.1128/aem.54.10.2521-2526.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovitz R, Barak Y, Morag E, Voronov-Goldman M, Lamed R, Bayer EA. Cohesin-dockerin microarray: diverse specificities between two complementary families of interacting protein modules. Proteomics. 2008;8:968–979. doi: 10.1002/pmic.200700486. [DOI] [PubMed] [Google Scholar]
- Irwin DC, Zhang S, Wilson DB. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur J Biochem. 2000;267:4988–4997. doi: 10.1046/j.1432-1327.2000.01546.x. [DOI] [PubMed] [Google Scholar]
- Jindou S, Kajino T, Inagaki M, Karita S, Beguin P, Kimura T, Sakka K, Ohmiya K. Interaction between a type-II dockerin domain and a type-II cohesin domain from Clostridium thermocellum cellulosome. Biosci Biotechnol Biochem. 2004;68:924–926. doi: 10.1271/bbb.68.924. [DOI] [PubMed] [Google Scholar]
- Mechaly A, Yaron S, Lamed R, Fierobe H-P, Belaich A, Belaich J-P, Shoham Y, Bayer EA. Cohesin-dockerin recognition in cellulosome assembly: experiment versus hypothesis. Proteins. 2000;39:170–177. doi: 10.1002/(SICI)1097-0134(20000501)39:2<170::AID-PROT7>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Mechaly A, Fierobe H-P, Belaich A, Belaich J-P, Lamed R, Shoham Y, Bayer EA. Cohesin-dockerin interaction in cellulosome assembly: a single hydroxyl group of a dockerin domain distinguishes between non-recognition and high-affinity recognition. J Biol Chem. 2001;276:9883–9888. doi: 10.1074/jbc.M009237200. [DOI] [PubMed] [Google Scholar]
- Mingardon F, Chanal A, López-Contreras AM, Dray C, Bayer EA, Fierobe H-P. Incorporation of fungal cellulases in bacterial minicellulosomes yields viable, synergistically acting cellulolytic complexes. Appl Environ Microbiol. 2007;73:3822–3832. doi: 10.1128/AEM.00398-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingardon F, Chanal A, Tardif C, Bayer EA, Fierobe H-P. Exploration of new geometries in cellulosome-like chimeras. Appl Environ Microbiol. 2007;73:7138–7149. doi: 10.1128/AEM.01306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morag E, Lapidot A, Govorko D, Lamed R, Wilchek M, Bayer EA, Shoham Y. Expression, purification and characterization of the cellulose-binding domain of the scaffoldin subunit from the cellulosome of Clostridium thermocellum. Appl Environ Microbiol. 1995;61:1980–1986. doi: 10.1128/aem.61.5.1980-1986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb M, Chen CY, Wu JH. Induction of the celC operon of Clostridium thermocellum by laminaribiose. Proc Natl Acad Sci USA. 2007;104:3747–3752. doi: 10.1073/pnas.0700087104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès S, Belaich A, Belaich J-P, Morag E, Lamed R, Shoham Y, Bayer EA. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridiumcellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins. 1997;29:517–527. doi: 10.1002/(SICI)1097-0134(199712)29:4<517::AID-PROT11>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Teeri TT. Crystalline cellulose degradation: new insight into the function of cellobiohydrolases. Trends Biotechnol. 1997;15:160–167. doi: 10.1016/S0167-7799(97)01032-9. [DOI] [Google Scholar]
- Wilson DB. Studies of Thermobifida fusca plant cell wall degrading enzymes. Chem Rec. 2004;4:72–82. doi: 10.1002/tcr.20002. [DOI] [PubMed] [Google Scholar]
- Wilson DB. Cellulases and biofuels. Curr Opin Biotechnol. 2009;20:295–299. doi: 10.1016/j.copbio.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Yaron S, Morag E, Bayer EA, Lamed R, Shoham Y. Expression, purification and subunit-binding properties of cohesins 2 and 3 of the Clostridium thermocellum cellulosome. FEBS Lett. 1995;360:121–124. doi: 10.1016/0014-5793(95)00074-J. [DOI] [PubMed] [Google Scholar]
- Zhang S, Lao G, Wilson DB. Characterization of a Thermomonospora fusca exocellulase. Biochemistry. 1995;34:3386–3395. doi: 10.1021/bi00010a030. [DOI] [PubMed] [Google Scholar]