Abstract
The toxic equivalency factor (TEF) approach recommended by the World Health Organization is used to quantify dioxin-like exposure concentrations for mixtures of polychlorinated dibenzo-dioxins, -furans, and polychlorinated biphenyls (PCBs), including 2,3,7,8-tetrachlorodibenzofuran (TCDF) and 3,3′,4,4′,5-pentachlorobiphenyl (PCB126) relative to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Whole-genome microarrays were used to evaluate the hepatic gene expression potency of TCDF and PCB126 relative to TCDD with complementary histopathology, tissue level analysis, and ethoxyresorufin-O-deethylase (EROD) assay results. Immature ovariectomized C57BL/6 mice were gavaged with 0.001, 0.01, 0.03, 0.1, 0.3, 1, 3, 10, 30, 100, and 300 μg/kg TCDD and TEF-adjusted doses (TEF for TCDF and PCB126 is 0.1) of TCDF or PCB126 (1, 3, 10, 30, 100, 300, 1000, and 3000 μg/kg of TCDF or PCB126) or sesame oil vehicle and sacrificed 24 h post dose. In general, TCDD, TCDF, and PCB126 tissue levels, as well as histopathological effects, were comparable when comparing TEF-adjusted doses. Automated dose-response modeling (ToxResponse Modeler) of the microarray data identified 210 TCDF and 40 PCB126 genes that exhibited sigmoidal dose-response curves with comparable slopes when compared with TCDD. These similar responses were used to calculate a median TCDF gene expression relative potency (REP) of 0.06 and a median PCB126 gene expression REP of 0.02. REPs of 0.02 were also calculated for EROD induction for both compounds. Collectively, these data suggest that differences in the ability of the liganded aryl hydrocarbon receptor:AhR nuclear translocator complex to elicit differential hepatic gene expression, in addition to pharmacokinetic differences between ligands, influence their potency in immature ovariectomized C57BL/6 mice.
Keywords: TCDD, TCDF, PCB126, TEF, mouse, dose-response
Polychlorinated aromatic hydrocarbons (PCAHs), including 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and related chemicals, such as 2,3,7,8-tetrachlorodibenzofuran (TCDF) and 3,3′,4,4′,5-pentachlorobiphenyl (PCB126), are ubiquitous environmental contaminants that elicit a broad spectrum of species-specific biochemical and toxic effects. Polychlorinated dibenzo-p-dioxins (PCDDs) and dibenzofurans (PCDFs) are by-products of waste combustion, herbicide production, and other industrial processes (Commoner et al., 1987; Mason and Safe, 1986; Poland and Glover, 1973; Safe et al., 1982). In contrast, polychlorinated biphenyls (PCBs) were manufactured between 1930 and 1977 as mixtures that included PCB126 as well as other congeners (Mayes et al., 1998). PCBs were used as coolants, lubricants, and dielectric insulating fluids for transformers and capacitors (Mullin et al., 1984; Safe, 1990; Safe et al., 1982). Even though their production has ceased, they continue to be released into the environment through the mishandling of PCB-containing products (National Toxicology Program [NTP], 2006b).
Although there are 75, 135, and 209 possible PCDD, PCDF, and PCB congeners, respectively, only 7, 10, and 12 are considered dioxin-like based on their ability to bind and activate the aryl hydrocarbon receptor (AhR) (Van den Berg et al., 2006). The AhR is a cytosolic ligand–activated basic helix-loop-helix Per-Arnt-Sim domain containing transcription factor (Denison and Heath-Pagliuso, 1998; Denison et al., 2002; Poland and Knutson, 1982; Safe, 2001). Following ligand binding, chaperone proteins that maintain the AhR in an inactive state dissociate from the AhR allowing translocation to the nucleus and heterodimerization with AhR nuclear translocator (ARNT). The AhR:ARNT heterodimer complex then interacts with dioxin response elements in the regulatory regions of target genes followed by recruitment of coregulatory proteins, leading to changes in gene expression (Denison and Heath-Pagliuso, 1998; Nebert et al., 2000).
Environmental exposure to PCDDs, PCDFs, and PCBs typically occurs as a complex mixture. In order to estimate the risk associated with a mixture, the concentration and potency of each toxic PCDD, PCDF, and PCB congeners are taken into account based on its toxic equivalency factor (TEF) relative to TCDD, the most toxic congener (Ahlborg, 1994; Barnes, 1991; Birnbaum and DeVito, 1995; Haws et al., 2006; Van den Berg et al., 1998, 2006). TEFs are single point potency estimates that were developed from relative potency (REP) values calculated by comparing the effective dose (ED50) value of a single response elicited by TCDD and dividing it by the ED50 for the same response elicited by the congener of interest (Van den Berg et al., 1998, 2006). Consequently, the uncertainty of a TEF point estimate can extend over orders of magnitude, reflecting the range of REP values available for a particular congener in various model systems (Harris et al., 1993; Haws et al., 2006; Poland and Knutson, 1982; Safe, 1997, 1990; Starr et al., 1999; Van den Berg et al., 2006). TEFs are independent of dose, time point, and tissue and largely reflect biochemical effects such as enzyme induction. Although results from cancer bioassays or developmental-reproductive studies have been considered and are the critical human health risk assessment endpoints for PCDDs and PCDFs, no TEF value is based exclusively on these endpoints. Furthermore, it is assumed that at submaximal doses, the contribution of each congener is additive. There are also pharmacokinetic and distributional differences between congeners that may affect their REPs (Budinsky et al., 2006; DeVito et al., 1997, 1998; Diliberto et al., 2001; Safe, 1995).
The TEFs for TCDF and PCB126 are currently set at 0.1, indicating that they are 10 times less potent than TCDD. However, our recent studies suggest that the TEF of 0.1 does not accurately reflect the hepatic potency of TCDF and PCB126 relative to TCDD in C57BL/6 mice (Burgoon et al., 2009; Kopec et al., 2008; N'Jai et al., 2008). Although pharmacokinetic differences are an important factor, temporal and dose-dependent microarray data suggest that TCDF- and PCB126-elicited gene expression responses are also not equivalent, in terms of potency and efficacy, relative to TCDD at TEF-adjusted equivalent doses. To further investigate this hypothesis, parallel TCDD, TCDF, and PCB126 dose-response studies were conducted at 24 h to minimize the pharmacokinetic and distributional differences between these compounds. In addition to using the same species, comparable study designs and analysis methods were also used as previously reported (Burgoon et al., 2009; Kopec et al., 2008; N'Jai et al., 2008). Moreover, complementary histopathology, tissue level analysis, and ethoxyresorufin-O-deethylase (EROD) activities were assessed. Results from this study not only expand the available REP data in C57BL/6 mice but also suggest that TCDF- and PCB126-elicited differential gene expression responses are not comparable to TCDD at TEF-adjusted equivalent doses at 24 h when pharmacokinetic and distributional differences are minimized.
MATERIALS AND METHODS
Animal husbandry.
Female C57BL/6 mice, ovariectomized by the supplier on postnatal day (PND) 20, with body weights (BW) within 10% of the average, were obtained from Charles Rivers Laboratories (Kingston, NY) on PND 25. Animals were housed in polycarbonate cages containing cellulose fiber chips (Aspen Chip Laboratory Bedding, Northeastern Products, Warrensberg, NY) with 30–40% humidity and a 12-h light/dark cycle (0700 h–1900 h). Mice had free access to deionized water and Harlan Teklad 22/5 Rodent Diet 8640 (Madison, WI). Animals were dosed on PND 28 following acclimatization for 3 days. Immature mice were used because they are more responsive to AhR ligands and to facilitate comparisons with other data sets obtained using the same model, study design, and analysis methods (Boverhof et al., 2005; Kopec et al., 2008; N'Jai et al., 2008). Animals were ovariectomized to negate potential interactions with estrogens produced by the developing ovaries because some animals are approaching reproductive maturity. All procedures were carried out with the approval of the Michigan State University Institutional Animal Care and Use Committee.
Dose-response study.
A stock solution of PCB126 (99.7% purity; AccuStandard, New Haven, CT) was dissolved in acetone (J.T. Baker, Phillipsburg NJ), followed by dilution in sesame oil (Sigma, St. Louis, MO), and evaporation of the acetone using a mild stream of nitrogen gas. The PCB126 stock was further diluted in sesame oil to achieve the desired doses. TCDD and TCDF were gifts from The Dow Chemical Company (Midland, MI). Animals were orally gavaged using 1.5-inch feeding needle with a 2.25-mm ball end (Cadence Science, Lake Success, NY). Mice received 0.1 ml of a single dose of 0.001, 0.01, 0.03, 0.1, 0.3, 1, 3, 10, 30, 100, and 300 μg/kg of TCDD or 1, 3, 10, 30, 100, 300, 1000, and 3000 μg/kg of PCB126 or TCDF or 0.1 ml sesame oil vehicle and sacrificed 24 h post dose. PCB126 and TCDF doses were TEF adjusted to be equivalent to the TCDD doses (Van den Berg et al., 2006) (Table 1). Mice were sacrificed by cervical dislocation and tissue samples were removed, weighed, flash frozen in liquid nitrogen, and stored at −80°C. The right lobe of the liver was fixed in 10% neutral buffered formalin (Sigma) for histological analysis.
TABLE 1.
The 24 h Dose-Response Study Design
| Number of animals |
||||
| Dose (μg/kg) | TCDD | PCB126 | TCDF | Vehicle (sesame oil) |
| 0 | — | — | — | 7 |
| 0.001 | 4 | — | — | — |
| 0.01 | 4 | — | — | — |
| 0.03 | 4 | — | — | — |
| 0.1 | 4 | — | — | — |
| 0.3 | 4 | — | — | — |
| 1 | 4 | 4 | 4 | — |
| 3 | 4 | 4 | 4 | — |
| 10 | 4 | 4 | 4 | — |
| 30 | 4 | 4 | 4 | — |
| 100 | 4 | 4 | 4 | — |
| 300 | 4 | 4 | 4 | — |
| 1000 | — | 4 | 4 | — |
| 3000 | — | 4 | 4 | — |
Histological analysis.
Fixed liver tissues were sectioned and processed in ethanol, xylene, and paraffin using a Thermo Electron Excelsior tissue processor (Waltham, MA). Tissues were embedded in paraffin with Miles Tissue Tek II embedding center after which paraffin blocks were sectioned at 5 μm with a rotary microtome. Liver sections were placed on glass microscope slides, washed twice in xylene for 5 min, followed by four quick washes in ethanol, and rinsed in water. Slides were placed in Gill 2 hematoxylin (Thermo Fisher Scientific, Waltham, MA) for 1.5 min followed by two to three quick dips in 1% glacial acetic acid water and rinsed with running water for 2–3 min. Slides were then rinsed in ethanol and counterstained with 1% eosin Y-phloxine B solution (Sigma) followed by multiple rinses in ethanol and xylene. Coverslips were attached using aqueous mounting media. All the histological processing was performed at Michigan State University Investigative HistoPathology Laboratory, Division of Human Pathology, using a modified version of previously published procedures (Sheehan and Hrapchak, 1980).
Quantification of hepatic TCDD, PCB126, and TCDF levels.
Liver samples were processed in parallel with laboratory blanks and a reference or background sample at Wellington Laboratories Inc. (Guelph, ON, Canada). The samples (100–500 mg) were transferred to a tared screw cap culture tube and weights were recorded. Samples were then spiked with a mixture of 13C12-2,3,7,8-TCDD, 13C12-PCB126, and 13C12-2,3,7,8-TCDF surrogates and digested with HCl. Each digest was split between two screw cap tubes and 3–4 ml hexane was added to each tube followed by vigorous mixing. The tubes were centrifuged, and the organic layer was removed. The hexane extraction was performed three times per tube with the six hexane fractions combined. The hexane fraction was then split and one fraction was archived. The other fraction was processed further using a small multilayer (acid/base/neutral) silica gel column eluted with 20–25 ml of hexane. The eluate was concentrated and transferred to a conical microvial with pentane and dichloromethane rinses and allowed to dry. Immediately prior to injection on the high-resolution gas chromatograph/high-resolution mass spectrometer (HRGC/HRMS) system, a mixture of 13C12-1,2,3,4-TCDD, 13C12-PCB111, and 13C12-1,2,3,4-TCDF injection standards were added to the conical microvial. TCDD, PCB126, and TCDF analyses were performed using an Agilent (Agilent Technologies, Inc., Santa Clara, CA) 6890 series HRGC with direct capillary interface to a Waters (Milford, MA) Autospec Ultima HRMS. Chromatographic separations were carried out on a 60-m DB5 (0.25 mm internal diameter and 0.25 μm film thickness) column in constant flow mode (Helium, 1 ml/min). All injections were 1 μl using splitless injection. The mass spectrometer was operated in positive electron ionization selective ion recording mode at a mass resolving power of 10,000 or greater.
EROD assay.
Microsomes were extracted from ∼100 mg samples by differential centrifugation (Moore et al., 2009). Tissue was minced and homogenized using Tri-R Stir-R homogenizer (Tri-R Instruments, Inc., Rockville Centre, NY) in 0.05M Tris (Invitrogen, Carlsbad, CA), 1.15% KCl (J.T. Baker), pH 7.5, and then centrifuged at 4°C, 10,000 × g for 10 min. The supernatant was centrifuged at 4°C, 100,000 × g for 30 min. The microsomal pellets were resuspended in 0.01M EDTA (Invitrogen), and 1.15% KCl, pH 7.4, and recentrifuged at 100,000 × g for 60 min. Final pellets were resuspended in a stabilizing buffer (20% glycerol [J.T. Baker], 0.1M KH2PO4 [J.T. Baker], and 1mM EDTA and 1mM dithiothreitol [Roche Applied Science, Indianapolis IN], pH 7.25) and stored at −80°C. Extracted hepatic microsomes from vehicle, TCDD-, PCB126-, and TCDF-treated mice were assayed for EROD activity by monitoring the production of resorufin measured at 590 nm in a 96-well plate (Costar, Corning, NY) using a Fluoroskan Ascent fluorometer (Thermo Fisher Scientific) and a corresponding software (version 2.6). The assay was performed in 0.05M HEPES (Sigma), pH 7.8, and 3.35mM 7-ethoxyresorufin (Molecular Probes, Eugene OR). Catalytic activity was initiated by addition of 0.3mM NADPH (Sigma), and fluorescence was measured every 2 min. After 30 min, the assay was terminated by addition of 36 μg fluorescamine (Sigma) in acetonitrile. Protein concentrations were measured at 460 nm using bovine serum albumin (Roche Applied Science) as a protein standard. Linear portions of each kinetic profile analysis were used to determine picomoles of resorufin produced per minute and standardized to the total protein (mg).
RNA isolation.
Frozen liver samples (left lobe, ∼100 mg stored at −80°C) were immediately transferred to 1 ml TRIzol (Invitrogen) and homogenized using a Mixer Mill 300 tissue homogenizer (Retsch, Germany). Total RNA was isolated according to the manufacturer’s protocol with an additional acid phenol:chloroform extraction. Isolated RNA was resuspended in RNA storage solution (Ambion Inc., Austin, TX) and quantified (A260). RNA quality was assessed by determining the A260:A280 ratio and visual inspection of 2 μg on a denaturing gel.
Microarray experimental design.
Treated and vehicle RNA samples were individually hybridized to 4 × 44 K Agilent oligonucleotide microarrays (Agilent Technologies, Inc.). Three biological replicates were performed using one-color labeling (Cy3) for each dose according to the manufacturer’s protocol (Agilent Manual: G4140-90040 v. 5.7). Microarray slides were scanned at 532 nm (Cy3) on a GenePix 4000B scanner (Molecular Devices, Union City, CA). Images were analyzed for feature and background intensities using GenePix Pro 6.0 (Molecular Devices). All data were archived in TIMS dbZach data management system (Burgoon and Zacharewski, 2007).
Microarray analysis.
All microarray data in this study passed our laboratory quality assurance protocol (Burgoon et al., 2005). TCDD, PCB126, and TCDF data sets were independently normalized because of the overall size of the files using a semiparametric approach (Eckel et al., 2005). Posterior probabilities were then calculated using an empirical Bayes method on a per-gene and dose basis using model-based t values (Eckel et al., 2004). Gene expression data were then ranked and prioritized (P1(t) values > 0.99 or > 0.90 and |fold change| > 1.5) to identify differentially expressed genes. Complete dose-response microarray data sets are available as Supplementary tables 3–5.
Quantitative real-time PCR.
Quantitative real-time PCR (QRT-PCR) was performed as previously described (Boverhof et al., 2005). Briefly, 1 μg of total RNA was reverse transcribed by SuperScript II (Invitrogen) using an anchored oligo-dT primer. The complementary DNA (cDNA) (1 μl) was used as a template in a 30-μl reaction containing 0.1μM of forward and reverse gene-specific primers, 3mM MgCl2, 1mM dNTPs, 0.025 IU AmpliTaq Gold, and 1X SYBR Green PCR buffer (Applied Biosystems, Foster City, CA). Supplementary table 2 provides the names, abbreviations, accession numbers, forward and reverse primer sequences, and amplicon sizes. Amplification was conducted on an Applied Biosystems PRISM 7500 Sequence Detection System. The cDNAs were quantified using a standard curve approach, and the copy number of each sample was standardized to three housekeeping genes (Actb, Gapdh, and Hprt) (Vandesompele et al., 2002). For graphing purposes (GraphPad Prism 5.0), the relative expression levels were scaled such that the expression level of the time-matched control group was equal to one.
Dose-response modeling.
Dose-response modeling was performed using the ToxResponse Modeler (Burgoon and Zacharewski, 2008). ToxResponse Modeler performs automated dose-response modeling by identifying the best-fit model among five different mathematical models (linear, exponential, Gaussian, sigmoidal, and quadratic). The algorithm then chooses the best-fit model of the five best in-class models. The overall best-fit model is then used to calculate the ED50 values. Microarray dose-response data were first filtered using a P1(t) > 0.90 cutoff and filtered to identify genes exhibiting a sigmoidal dose-response profile. Supplementary table 6 contains the list of sigmoidal dose-responsive genes together with corresponding ED50 and REP values. REP values were calculated on a per-feature/per-gene basis using model-based ED50 values:
Statistical analysis.
All statistical analyses were performed with SAS 9.1 (SAS Institute, Cary, NC). All data (with the exception of microarray data) were analyzed by ANOVA followed by Dunnett’s or Tukey’s post hoc tests. Differences between treatment groups were considered significant when p < 0.05.
RESULTS
Liver and BWs
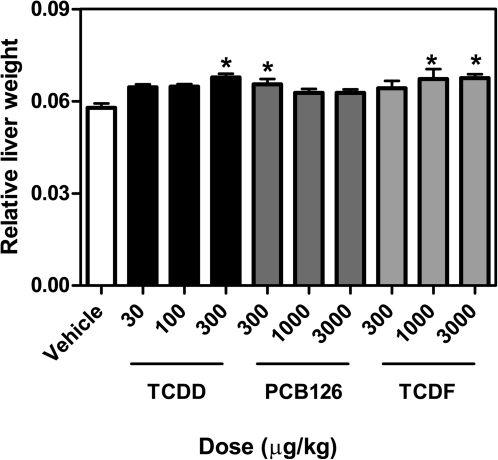
Rodents exposed to PCAHs including dioxins, PCBs, and furans exhibit temporal and dose-dependent hepatic changes characterized by increases in liver weight (Denison and Heath-Pagliuso, 1998; NTP, 2006a,b). PCB126 and TCDF have TEFs of 0.1 (Van den Berg et al., 2006), indicating that 10 times more chemical is required to elicit similar effects compared with an equivalent dose of TCDD. In our study, PCB126 and TCDF doses were TEF adjusted to theoretically match the potency of TCDD. Significant (p < 0.05) increases in relative liver weight (RLW) were observed with 300 μg/kg TCDD, 300 μg/kg PCB126, and 1000 and 3000 μg/kg TCDF in agreement with previous reports using the same compounds, animal models, and study designs (Fig. 1) (Boverhof et al., 2005, 2006; Kopec et al., 2008; N'Jai et al., 2008). No changes in BW or BW gain relative to vehicle controls were observed within 24 h, indicating no systemic toxicity or wasting syndrome response at the doses used.
FIG. 1.
Dose-dependent changes in the RLWs 24 h post dose for TCDD, PCB126, and TCDF. Results are displayed as mean ± SE of at least four independent replicates. Data were analyzed by ANOVA followed by Dunnett’s post hoc test: *p < 0.05 for vehicle versus treated samples. No additional significant treatment-related alterations in liver or organ weights were noted in the dose-response study.
Hepatic TCDD, PCB126, and TCDF Levels
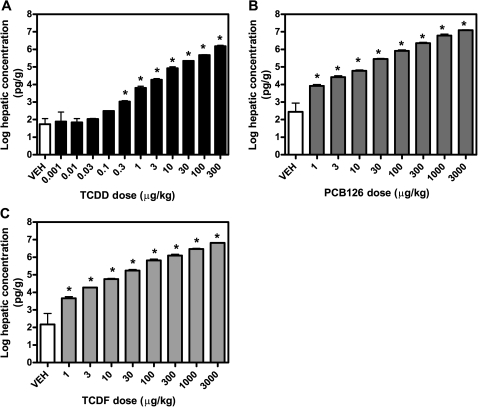
TCDD, PCB126, and TCDF levels per liver wet weight (in pg/g) were measured in three individual samples per dose using HRGC/HRMS. A dose-dependent increase in the hepatic concentration of all three compounds was observed (Figs. 2A–C and Table 2). The tissue levels of TCDD, PCB126, and TCDF were significantly (p < 0.05) higher compared to vehicle controls, except for the lowest doses of TCDD (0.001–0.1 μg/kg), because of the presence of low background levels of TCDD. At TEF-equivalent doses, the levels of all three compounds were comparable except for statistical differences at 0.1, 0.3, 100, and 300 μg/kg (Supplementary fig. 1). The use of TEF values for determining tissue concentrations increases the uncertainty and reliability of potency estimates (Van den Berg et al., 2006). Despite this limitation, the use of TEF-adjusted tissue level data suggests that the potencies of TCDD, TCDF, and PCB126 should be comparable because there were minimal differences in hepatic absorption, distribution, and metabolism between compounds at 24 h (Supplementary fig. 1). This is significant because pharmacokinetic differences between these ligands have been reported to contribute to substantial differences in potencies at later time points (Budinsky et al., 2008; DeVito and Birnbaum, 1995; Kopec et al., 2008; N'Jai et al., 2008).
FIG. 2.
Hepatic (A) TCDD, (B) PCB126, and (C) TCDF levels per g liver wet weight measured 24 h post dose using HRGC/HRMS. The data are displayed on a log scale to visualize tissue concentrations at all doses. The results are displayed as mean ± SE of at least three independent samples. Data were analyzed by ANOVA followed by Dunnett’s post hoc test: *p< 0.05 for vehicle versus treated samples.
TABLE 2.
Absolute Hepatic Tissue Levels (in pg/g or ppt per wet weight) for TCDD-, PCB126-, and TCDF-Treated Samples and Vehicle Controls Measured by HRGC/HRMS
| Absolute hepatic tissue levels (in pg/g or ppt per wet weight) |
|||
| Dose (μg/kg) | TCDD | PCB126 | TCDF |
| Vehicle (sesame oil) | 1.00 × 102 ± 9.84 × 101 | 1.22 × 103 ± 1.98 × 103 | 1.05 × 103 ± 1.68 × 103 |
| 0.001 | 2.07 × 102 ± 2.39 × 102 | — | — |
| 0.01 | 8.83 × 101 ± 7.12 × 101 | — | — |
| 0.03 | 1.09 × 102 ± 1.29 × 101 | — | — |
| 0.1 | 3.10 × 102 ± 2.06 × 101 | — | — |
| 0.3 | 1.08 × 103 ± 2.21 × 102 | — | — |
| 1 | 6.73 × 103 ± 2.21 × 103 | 8.66 × 103 ± 2.58 × 103 | 4.79 × 103 ± 1.56 × 103 |
| 3 | 1.90 × 104 ± 4.85 × 103 | 2.72 × 104 ± 9.24 × 103 | 1.88 × 104 ± 1.70 × 103 |
| 10 | 8.70 × 104 ± 2.78 × 104 | 6.08 × 104 ± 1.23 × 104 | 5.81 × 104 ± 6.36 × 103 |
| 30 | 2.25 × 105 ± 1.06 × 104 | 2.89 × 105 ± 2.72 × 104 | 1.80 × 105 ± 3.81 × 104 |
| 100 | 4.75 × 105 ± 5.08 × 104 | 8.39 × 105 ± 2.06 × 105 | 6.81 × 105 ± 2.24 × 105 |
| 300 | 1.52 × 106 ± 3.07 × 105 | 2.29 × 106 ± 4.92 × 105 | 1.29 × 106 ± 3.52 × 105 |
| 1000 | — | 6.41 × 106 ± 1.95 × 106 | 2.96 × 106 ± 4.66 × 105 |
| 3000 | — | 1.25 × 107 ± 1.01 × 106 | 6.63 × 106 ± 4.05 × 105 |
Note. The values represent mean ± SD of at least three independent replicates.
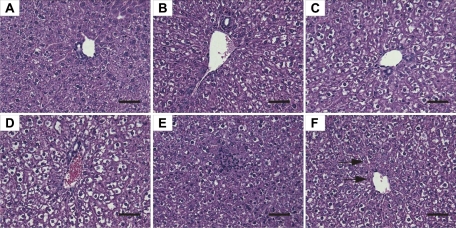
Histopathology
Hematoxylin and eosin staining was examined on the three highest TEF-equivalent doses (30, 100, and 300 μg/kg TCDD; 300, 1000, and 3000 μg/kg PCB126 and TCDF). TCDD, PCB126, and TCDF elicited dose-dependent increases in periportal hepatocellular vacuolization and multifocal mixed inflammatory infiltration (Table 3 and Fig. 3). TCDD and the equivalent TEF-matched TCDF doses elicited comparable hepatic vacuolization, whereas TEF-equivalent PCB126 doses exhibited slightly less vacuolization (Table 3). Hepatocellular single cell necrosis was present only in the highest dose groups for all three compounds with the most pronounced necrosis occurring with 3000 μg/kg TCDF (Table 3 and Fig. 3). Comparable levels of dose-dependent mixed inflammatory cell foci (lymphocytes, monocytes, and neutrophils) were observed. Qualitatively, these results suggest that TEF-matched doses of TCDD, PCB126, and TCDF elicit comparable histopathological effects at 24 h. Supplementary table 1 contains a detailed report on the incidence and severity of histopathological responses.
TABLE 3.
Dose-Dependent Incidence and Severity of Hepatic Histopathological Responses in the Vehicle, TCDD-, PCB126-, and TCDF-Treated Mice at 24 h
| Treatment and dose (μg/kg) |
||||||||||
| Vehicle |
TCDD |
PCB126 |
TCDF |
|||||||
| 0 | 30 | 100 | 300 | 300 | 1000 | 3000 | 300 | 1000 | 3000 | |
| Hepatocellular vacuolization | ||||||||||
| Average severity | 0 | 1 | 1.75 | 2.5 | 1 | 1.5 | 2 | 1.25 | 1.75 | 2.5 |
| Hepatocellular single cell necrosis | ||||||||||
| Average severity | 0 | 0 | 0 | 0.5 | 0 | 0 | 0.5 | 0 | 0 | 1.25 |
| Mixed cell infiltration | ||||||||||
| Average severity | 0 | 0.25 | 0.25 | 0.5 | 0 | 0.5 | 0.5 | 0 | 0.25 | 0.75 |
Note. The average severity scores are reported as a weighted average based on the following scoring scheme: minimal (grade of 1), slight (grade of 2), moderate (grade of 3), and/or marked (grade of 4) responses divided by the total number of examined animals. A comprehensive histopathological report with detailed incidence and severity is available in Supplementary table 1.
FIG. 3.
Representative hematoxylin and eosin–stained liver sections 224 h postexposure to PCB126, TCDF, or TCDD. Selected doses resulted in vacuolization, single cell necrosis, and/or immune cell infiltration. (A) Vehicle treatment resulted in no visible hepatic alterations. (B) 300 μg/kg PCB126 elicited minimal vacuolization compared with (C) 1000 μg/kg TCDF and (D) 300 μg/kg TCDD, which exhibited more pronounced vacuolization. (E) 3000 μg/kg TCDF resulted in immune cell infiltration as well as (F) instances of necrosis (arrow). Bars = 50 μm.
EROD Activity
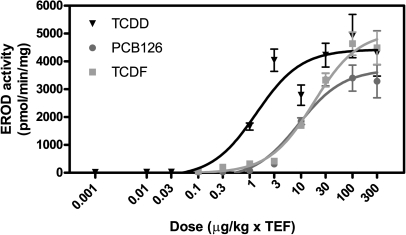
Hepatic EROD activity was assessed to anchor our results to data within the World Health Organization (WHO) REP database (Haws et al., 2006). TCDD induced a dose-dependent sigmoidal increase in EROD activity with an ED50 of 1.1 μg/kg, calculated using the ToxResponse Modeler (Burgoon and Zacharewski, 2008) (Fig. 4). TEF-matched doses of PCB126 and TCDF elicited comparable EROD induction (Fig. 4) but were less potent than TCDD (PCB126 ED50 = 53.6 μg/kg and TCDF ED50 = 54.2 μg/kg). These EROD activity REPs (0.02 for PCB126 and TCDF) are consistent with values reported in the WHO REP database (Haws et al., 2006). Moreover, in our published study examining TEF-equivalent doses of TCDD and TCDF at 72 h, the ToxResponse Modeler identified a comparable EROD REP value (TCDF EROD REP72h of 0.04) (Burgoon et al., 2009).
FIG. 4.
Dose-dependent induction of EROD activity following exposure to TEF-equivalent doses of TCDD, PCB126, and TCDF. The ToxResponse Modeler calculated ED50 values of 1.1, 53.6, and 54.2 μg/kg for TCDD, PCB126, and TCDF, respectively, yielding PCB126 and TCDF EROD REPs of 0.02. Symbols represent the mean ± SE of four independent samples. Sigmoidal dose-response curve fitting was done using GraphPad Prism 5.0.
Hepatic TCDF and PCB126 Gene Expression REPs
TCDF and PCB126 microarray data were analyzed to determine both per-gene and aggregate gene expression REP values using set theory and our ToxResponse Modeler (Figs. 5A–C). The analysis abides by assumptions underlying the TEF concept (e.g., same mechanism of action, parallel dose-response curves). TCDD, TCDF, and PCB126 microarray data sets were individually normalized (Eckel et al., 2005) and analyzed using an empirical Bayes approach to identify differentially expressed features. Compared with previously published microarray studies (Boverhof et al., 2005; Kopec et al., 2008; N'Jai et al., 2008), a more relaxed P1(t) cutoff was used (P1(t) > 0.90) to be more inclusive, yielding higher statistical power at the risk of including more false positives at this early stage in the dose-response analysis.
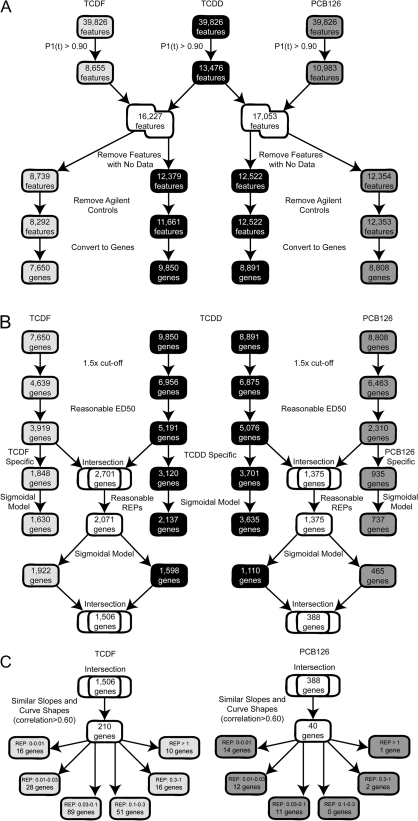
FIG. 5.
(A) Comprehensive TCDD, TCDF, and PCB126 dose-response analysis—Part 1/3. Differentially regulated features were identified using a relaxed P1(t) cutoff of 0.90 to maximize the number of differentially expressed genes under consideration. TCDD elicited ∼1.6 times more differentially expressed features compared with TCDF (13,476 vs. 8655) and ∼1.3 times more differentially expressed features compared with PCB126 (13,476 vs. 10,983). Unions were taken to identify all differentially regulated features. Features missing data at any dose, as well as Agilent controls, were removed and not considered further. Features were mapped to specific genes for further analysis. (B) Comprehensive TCDD, TCDF, and PCB126 dose-response analysis—Part 2/3. Differentially expressed genes were analyzed further if the change in expression was greater than 1.5-fold for at least one dose. Genes exhibiting a sigmoidal dose-response were identified and intersected to identify genes responsive to both TCDD and TCDF and to TCDD and PCB126 at 24 h. Genes were examined further if the model-based ED50 value, calculated by ToxResponse Modeler, was within the experimental dose range. Genes exhibiting a sigmoidal dose-response curve for both TCDD and TCDF were intersected to identify 1506 genes that exhibited an expression change greater than 1.5-fold for at least one dose, a P1(t) > 0.90, a sigmoidal profile, and an ED50 value within the experimental range. Identical analysis yielded only 388 sigmoidal genes for TCDD and PCB126. PCB126 lost a majority of differentially expressed dose-responsive genes from further consideration because the ED50s were not within the experimental dose range. (C) Comprehensive TCDD, TCDF, and PCB126 dose-response analysis—Part 3/3. In the final analysis, assumptions regarding similarities in the slopes and shapes of corresponding TCDD and TCDF as well as TCDD and PCB126 sigmoidal dose-response curves were assessed by calculating the correlation coefficients of the elicited dose-response curves. The correlation analysis identified 210 genes from TCDD versus TCDF comparisons, and only 40 genes from TCDD and PCB126 comparisons with correlation coefficients greater than 0.60. The distribution of individual gene expression REPs is provided. The median REP for hepatic gene expression in the immature ovariectomized C57BL/6 mouse at 24 h was calculated to be 0.06 and 0.02 for TCDF and PCB126, respectively.
TCDD elicited ∼1.6 times more differentially expressed features when compared to TCDF (13,476 vs. 8655) (Fig. 5A) at comparable hepatic levels (Supplementary fig. 1). This trend is similar to the 72 h dose-response analysis, where TCDD elicited the differential expression of almost twice as many features compared with TCDF, but at lower hepatic TCDF levels because of ligand-specific pharmacokinetics (Budinsky et al., 2008; Burgoon et al., 2009; DeVito and Birnbaum, 1995; Diliberto et al., 1995, 2001; Hamm et al., 2003). Likewise, TCDD differentially expressed ∼1.3 times more features compared to PCB126 (13,476 vs. 10,983) (Fig. 5A). It should be noted that TCDD consistently elicited the greatest number of differentially expressed features (and genes in subsequent analysis) compared to either TCDF or PCB126.
Unions of the TCDD plus TCDF (16,227 features) and TCDD plus PCB126 (17,053 features) data sets were taken to include all differentially regulated features for further dose-response and REP analysis. Features missing data at one or more doses as well as the Agilent included controls were removed and not considered further. Features were then mapped back to genes to investigate dose-dependent changes in expression (Fig. 5A).
Genes with a P1(t) > 0.90 were also filtered for a >1.5-fold change in expression for at least one dose relative to time-matched controls. This resulted in 5191 TCDD and 3919 TCDF genes that exhibited an expression change greater than 1.5-fold for at least one dose with a corresponding P1(t) > 0.90 and an ED50 value within the experimental dose range (Fig. 5B). This means that of the 5191 genes differentially expressed by TCDD for at least one dose, 3120 genes were not represented in the TCDF data set and of the 3919 TCDF genes, 1848 were not represented in the TCDD data set (Fig. 5B). Compared with the 72 h dose-response analysis (Burgoon et al., 2009), approximately five times more TCDD and TCDF genes were identified at 24 h (5191 TCDD and 3919 TCDF genes at 24 h compared with 1027 TCDD and 837 TCDF genes at 72 h).
The ToxResponse Modeler then identified the best linear, exponential, Gaussian, sigmoidal, and quadratic models for each dose-responsive differentially expressed gene (e.g., best in class). The model that best fit the data among the different classes (e.g., best in show) was then used to calculate an ED50 for each dose-responsive gene. Only those genes with an ED50 within the experimental dose-response range were retained for subsequent REP analysis. The intersection of 5191 TCDD and 3919 TCDF genes identified 2071 genes that were used to calculate gene-specific REPs. Furthermore, 3120 and 1848 genes were TCDD- and TCDF specific, respectively. Of those, 2137 TCDD and 1630 TCDF genes exhibited sigmoidal dose-response curves. The intersection of 2071 genes in common between TCDD and TCDF with reasonable ED50 values identified 1506 genes with sigmoidal dose-response curves (Fig. 5B).
In order to comply with WHO TEF assumptions regarding similar slopes and curve shapes, only those genes that exhibited correlation coefficient greater than 60% were considered further for gene-specific REP analysis. In this study, the coefficient was lowered compared with the 72 h TCDF dose-response analysis (correlation coefficient > 80%) (Burgoon et al., 2009) to include more genes and to account for the smaller overlap between TCDD and PCB126 data sets, thus facilitating comparisons between PCB126 and TCDF at 24 h. Correlation analysis identified 210 genes with similar slopes and curve shapes elicited by TCDD and TCDF (Fig. 5C). Sixteen of these genes had gene-specific REP values between 0 and 0.01, 28 between 0.01 and 0.03, 89 between 0.03 and 0.1, 51 between 0.1 and 0.3, 16 between 0.3 and 1, and 10 genes had REPs greater than 10. The median REP for all 210 genes was 0.06 lower than the 72 h median REP of 0.1 (0.096) calculated using 83 genes with similar slopes and curves for TCDD and TCDF (Burgoon et al., 2009). Relaxing the correlation coefficient used for the 72 h dose-response study (Burgoon et al., 2009) to 60% resulted in an almost identical median gene expression REP of 0.1 (0.112).
An identical approach was used to determine PCB126 gene-specific REPs and a median REP (Fig. 5C). In summary, only 40 genes exhibited similar slope and sigmoidal curves, of which 14 genes had REPs between 0 and 0.01, 12 between 0.01 and 0.03, 11 between 0.03 and 0.1, 2 between 0.3 and 1, and only 1 gene had a REP greater than 1. The median PCB126 gene expression REP for the 40 sigmoidal genes was 0.02. The PCB126 data set lost a majority of genes from further consideration because the ED50s were not within the experimental dose range. The low number of dose-responsive genes considered for the PCB126 REP analysis can be attributed to the lower number of differentially expressed genes with sufficient potency when compared with TCDF and TCDD.
REPs based on tissue levels were also calculated for a subset of responsive genes used to determine REPs based on administered dose (Supplementary table 6). However, a vast majority of tissue level–based REPs could not be calculated because of differences in curve shapes and slopes. Overall, the median tissue level gene expression REPs for TCDF and PCB126 were comparable with administered dose–based REPs for the limited number of genes considered (TCDF REPadministered dose = 0.06, TCDF REPtissue level = 0.04, and PCB126 REPsadministered dose and tissue level = 0.02). Note that approximately 60% of the tissue level–based REPs were within an order of magnitude of the administered dose–based REPs.
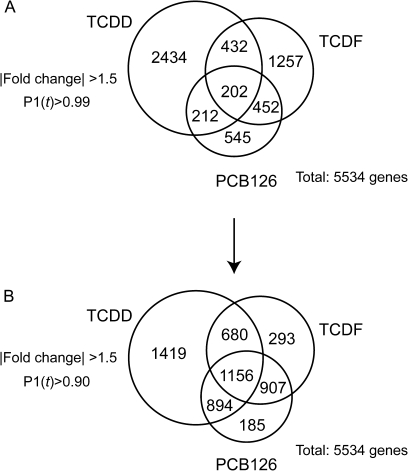
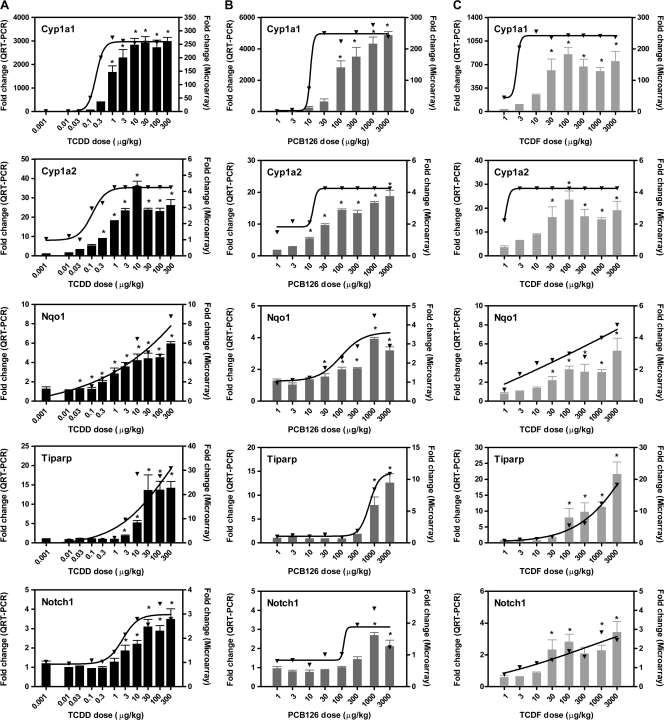
Microarray analysis suggests that there were a number of TCDF- and PCB126-specific responses. Three-way Venn analysis of the 3280, 2343, and 1411 gene expression changes elicited by TCDD, TCDF, and PCB126 (P1(t) > 0.99 and |fold change| > 1.5), respectively, identified only 202 genes differentially expressed by all three compounds (Fig. 6A), However, after relaxing the statistical cutoff (P1(t) > 0.90 and |fold change| > 1.5) (Fig. 6B), almost all PCB126- and TCDF-elicited gene expression changes overlapped, consistent with the conserved AhR-mediated mechanism of action. Overlaps between TCDD versus TCDF and TCDD versus PCB126 also significantly increased. The high number of TCDD-specific genes could be attributed to a wider range of doses used in the study (11 doses of TCDD vs. 8 doses of PCB126 and TCDF), and the greater potency and efficacy of TCDD even at TEF-adjusted doses of TCDF and PCB126. These results are in agreement with reports suggesting that TCDF- and PCB126-elicited responses are a subset of TCDD-regulated genes and consistent with a common AhR-mediated mode of action (Kopec et al., 2008; N'Jai et al., 2008). At later time points, TCDD elicits more pronounced histopathological responses and changes in serum clinical chemistry compared with TCDF and PCB126. A more thorough discussion of the association between differential gene expression and pathology has been previously reported (Boverhof et al., 2005, 2006; Kopec et al., 2008; N'Jai et al., 2008). QRT-PCR confirmed the dose-dependent changes in expression for a subset of AhR-regulated genes (Fig. 7). The classic members of the “AhR gene battery” involved in phase I and phase II metabolism including Cyp1a1, Cyp1a2, and Nqo1 (Nebert et al., 1990) were induced. In addition, Tiparp previously reported to exhibit robust AhR-dependent expression (Ma et al., 2001; Tijet et al., 2006) and Notch1 implicated in cell differentiation and suggested to have a role in normal AhR signaling during liver development (Boverhof et al., 2005; Harper et al., 2003) were also significantly expressed. The results from QRT-PCR analysis were in good agreement with the microarray data.
FIG. 6.
Three-way Venn analysis of TCDD, PCB126, and TCDF elicited differentially expressed genes at (A) stringent (|fold change| > 1.5, P1(t) > 0.99) and (B) relaxed statistical criteria (|fold change| > 1.5, P1(t) > 0.90).
FIG. 7.
QRT-PCR verification of selected AhR-regulated genes: Cyp1a1, Cyp1a2, Nqo1, Tiparp, and Notch1 for (A) TCDD, (B) PCB126, and (C) TCDF at 24 h. The same RNA samples used in the dose-response microarray studies were also used for QRT-PCR analysis. All fold changes were calculated relative to vehicle controls. Bars (left y-axis) and lines (right y-axis) represent QRT-PCR and microarray data, respectively. The genes are represented by their official gene symbols. Bars represent the mean ± SE of four independent QRT-PCR samples. QRT-PCR data were analyzed by ANOVA followed by Dunnett’s post hoc test: *p < 0.05 for vehicle versus treated samples. ED50 values for microarray and QRT-PCR mRNA levels are provided in Supplementary table 7.
DISCUSSION
In this study, the dose-dependent hepatic gene expression effects of TEF-adjusted equipotent doses of TCDF and PCB126 (WHO TEF = 0.1) relative to TCDD (WHO TEF = 1) were examined at 24 h. In addition, complementary histopathology and EROD activity were assessed in order to associate dose-dependent changes in gene expression to higher order effects at comparable TEF-adjusted TCDD, TCDF, and PCB126 tissue levels. Unlike comparable studies at later time points, the TEF-adjusted tissue levels for TCDD, TCDF, and PCB126 were comparable at 24 h except for slight statistical differences at 0.1, 0.3, 100, and 300 μg/kg (dose × TEF). Therefore, differences in pharmacokinetics cannot be used to explain the weaker responses elicited by equipotent TEF-adjusted doses of TCDF and PCB126. By minimizing the differences in TEF-adjusted levels of TCDD, TCDF, and PCB126 in the liver, we further investigated qualitative and quantitative gene expression differences that may contribute to the weaker responses elicited by TCDF and PCB126.
Overall, the results of this 24 h study are consistent with our previous 72 h studies using a similar model, the same oligonucleotide microarrays, and the same analysis methods (Burgoon et al., 2009; Kopec et al., 2008; N'Jai et al., 2008). At the highest doses, 3000 μg/kg TCDF, 3000 μg/kg PCB126, and 300 μg/kg TCDD elicited comparable increases in RLW accompanied by similar levels of hepatocellular vacuolization and minimal evidence of necrosis and immune cell infiltration at 24 h. Consequently, TEF-adjusted equipotent doses of TCDF and PCB126 elicited comparable phenotypic level effects when compared with TCDD. However, at later time points (72 and 168 h), with more apparent differences in tissue levels, TCDD, TCDF, and PCB126 elicited different levels of vacuolization, necrosis, and immune cell infiltration (Boverhof et al., 2005; Burgoon et al., 2009; Kopec et al., 2008; N'Jai et al., 2008), consistent with reported ligand-specific pharmacokinetics (DeVito et al., 1998).
Despite comparable levels of vacuolization, necrosis, and immune cell infiltration at 24 h, there were significant differences in EROD activity induced by TCDF, PCB126, and TCDD. TCDF and PCB126 induced comparable EROD activity levels, resulting in ED50 values of 53.6 and 54.2 μg/kg, respectively. The EROD ED50 for TCDD was 1.1 μg/kg comparable with the ED50 of 2.5 μg/kg in B6 mice at 24 h (Ma et al., 1992) and 1.6 μg/kg in C57BL/6 mice at 7 days (Lin et al., 1991). This is also in agreement with the TCDF EROD ED50 at 72 h in terms of maximum EROD induction as well as the ED50 values and REPs (Burgoon et al., 2009). In addition, ED50 values for TCDD, TCDF, and PCB126 induction of Cyp1a1 messenger RNA (mRNA) levels (0.09, 2.5, and 3.9 μg/kg, respectively) were consistently lower compared with the corresponding EROD values (1.1, 53.6, and 54.2 μg/kg, respectively). The 0.09 μg/kg TCDD Cyp1a1 mRNA ED50 from this study is almost identical to that reported by Abel et al. (1996) in C57BL/6 mice at 24 h (0.08 μg/kg). Others have also reported lower ED50 values for Cyp1a1 mRNA compared with EROD activity in B6C3F1 at the same time point (mRNA ED50 of 1.6 μg/kg and EROD ED50 of 5.3 μg/kg) (Narasimhan et al., 1994). Therefore, dose-dependent changes in gene expression may provide a more sensitive indicator of exposure, and possibly toxicity, provided an association can be established between gene function and the etiology of an adverse effect, assuming that a change in gene expression correlates with protein expression and activity.
TEF-adjusted equipotent doses of TCDD, TCDF, and PCB126 did not elicit comparable gene expression changes. TCDD elicited the greatest number of differentially expressed genes in comparison with TCDF and PCB126 at TEF-adjusted equipotent doses. Furthermore, TCDF- and PCB126-elicited changes in gene expression were a subset of the genes differentially expressed by TCDD. Suggestions of TCDF- and PCB126-specific gene expression responses have been previously shown to be a statistical artifact. When hard cutoff values (P1(t) > 0.99 and |fold change| > 1.5) are relaxed (P1(t) > 0.90 and |fold change| > 1.5), almost all PCB126- and TCDF-elicited genes become a subset of genes differentially expressed by TCDD (Kopec et al., 2008; N'Jai et al., 2008) (Fig. 6). However, the quantitatively lower number of TCDF- and PCB126-elicited differentially expressed genes cannot be fully attributed to differences in ligand pharmacokinetics. HRGC/HRMS analysis indicates that the hepatic levels of TCDD, TCDF, and PCB126 at 24 h were essentially equipotent when TEF adjusted. Consequently, the ability of the liganded AhR:ARNT complex to efficiently recruit the same complement of coactivators may also be an important factor for overall congener potency. For example, gene transactivation is reported to be congener structure-, coactivator-, and cell context dependent (Zhang et al., 2008).
In order to calculate gene-specific REP values, TCDD-, TCDF-, and PCB126-elicited gene expression changes with a P1(t) > 0.90 and |fold change| > 1.5 were analyzed using our automated dose-response modeler (Burgoon and Zacharewski, 2008). REPs for only 210 TCDF versus TCDD genes and 40 PCB126 versus TCDD genes were calculated in order to comply with WHO guidelines requiring sigmoidal dose-response curves with similar slopes and shapes (Haws et al., 2006; Van den Berg et al., 2006). Overall, the TCDF median REP values at 24 h (0.06) and 72 h (0.1) are similar. This difference may be attributed, in part, to the use of a wider dose range and higher doses of TCDF and TCDD in the 24 h study and to the use of intact mice in the 72 h study. In contrast, the PCB126 REP of 0.02 reflects its weaker potency even at TEF-adjusted doses. PCB126 treatment also resulted in a lower number of differentially regulated genes exhibiting a sigmoidal dose-response when compared with TCDD. Many PCB126-elicited gene expression changes were excluded because of poor induction (< 1.5-fold) or because they did not exhibit a sigmoidal dose-response curve. Alternatively, REPs based on points of departure (Burgoon and Zacharewski, 2008), lowest observable adverse effect level and/or no observable adverse effect level could be also considered (Van den Berg et al., 2006).
In summary, this study expands the available hepatic REP data for TCDF and PCB126 using the immature ovariectomized C57BL/6 mice. It further suggests that at TEF-adjusted doses and at equipotent TEQ hepatic tissue levels, TCDF and PCB126 elicit weaker responses compared to TCDD. Therefore, in addition to pharmacokinetic differences, congener structure also plays an important role in gene expression potency and efficacy. Further associations of specific dose-responsive genes to toxic events are compromised by limited gene annotation and an incomplete understanding of the mechanisms involved in the etiology of the toxic responses elicited by TCDD and related compounds.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Environmental Health Sciences Superfund Basic Research Program (P42ES04911); The Dow Chemical Company (219109).
Supplementary Material
Acknowledgments
The authors would like to thank Agnes Forgacs and Ed Dere for critical reading of this manuscript and Suntae Kim for the technical support.
References
- Abel J, et al. Dose-response relationship of cytochrome P4501b1 mRNA induction by 2,3,7,8-tetrachlorodibenzo-p-dioxin in livers of C57BL/6J and DBA/2J mice. Arch. Toxicol. 1996;70:510–513. doi: 10.1007/s002040050307. [DOI] [PubMed] [Google Scholar]
- Ahlborg UG. Human health risk assessment and risk perception related to the Baltic Sea. Arch. Toxicol. Suppl. 1994;16:53–59. doi: 10.1007/978-3-642-78640-2_6. [DOI] [PubMed] [Google Scholar]
- Barnes DG. Toxicity equivalents and EPA's risk assessment of 2,3,7,8-TCDD. Sci. Total Environ. 1991;104:73–86. doi: 10.1016/0048-9697(91)90008-3. [DOI] [PubMed] [Google Scholar]
- Birnbaum LS, DeVito MJ. Use of toxic equivalency factors for risk assessment for dioxins and related compounds. Toxicology. 1995;105:391–401. doi: 10.1016/0300-483x(95)03237-a. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, et al. Temporal and dose-dependent hepatic gene expression patterns in mice provide new insights into TCDD-Mediated hepatotoxicity. Toxicol. Sci. 2005;85:1048–1063. doi: 10.1093/toxsci/kfi162. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, et al. Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 mice. Toxicol. Sci. 2006;94:398–416. doi: 10.1093/toxsci/kfl100. [DOI] [PubMed] [Google Scholar]
- Budinsky RA, et al. Recommended relative potency factors for 2,3,4,7,8-pentachlorodibenzofuran: the impact of different dose metrics. Toxicol. Sci. 2006;91:275–285. doi: 10.1093/toxsci/kfj125. [DOI] [PubMed] [Google Scholar]
- Budinsky RA, et al. A pilot study of oral bioavailability of dioxins and furans from contaminated soils: impact of differential hepatic enzyme activity and species differences. Chemosphere. 2008;70:1774–1786. doi: 10.1016/j.chemosphere.2007.08.035. [DOI] [PubMed] [Google Scholar]
- Burgoon LD, et al. Protocols for the assurance of microarray data quality and process control. Nucleic Acids Res. 2005;33:e172. doi: 10.1093/nar/gni167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoon LD, et al. Automated dose-response analysis of the relative hepatic gene expression potency of TCDF in C57BL/6 mice. Toxicol. Sci. 2009;112:221–228. doi: 10.1093/toxsci/kfp180. [DOI] [PubMed] [Google Scholar]
- Burgoon LD, Zacharewski TR. dbZach toxicogenomic information management system. Pharmacogenomics. 2007;8:287–291. doi: 10.2217/14622416.8.3.287. [DOI] [PubMed] [Google Scholar]
- Burgoon LD, Zacharewski TR. Automated quantitative dose-response modeling and point of departure determination for large toxicogenomic and high-throughput screening data sets. Toxicol. Sci. 2008;104:412–418. doi: 10.1093/toxsci/kfn083. [DOI] [PubMed] [Google Scholar]
- Commoner B, et al. The origin and health risks of PCDD and PCDF. Waste Manag. Res. 1987;5:327–346. [Google Scholar]
- Denison MS, Heath-Pagliuso S. The Ah receptor: a regulator of the biochemical and toxicological actions of structurally diverse chemicals. Bull. Environ. Contam. Toxicol. 1998;61:557–568. doi: 10.1007/pl00002973. [DOI] [PubMed] [Google Scholar]
- Denison MS, et al. Ligand binding and activation of the Ah receptor. Chem. Biol. Interact. 2002;141:3–24. doi: 10.1016/s0009-2797(02)00063-7. [DOI] [PubMed] [Google Scholar]
- DeVito MJ, Birnbaum LS. The importance of pharmacokinetics in determining the relative potency of 2,3,7,8-tetrachlorodibenzo-p-dioxin and 2,3,7,8-tetrachlorodibenzofuran. Fundam. Appl. Toxicol. 1995;24:145–148. doi: 10.1006/faat.1995.1016. [DOI] [PubMed] [Google Scholar]
- DeVito MJ, et al. Dose-response relationships for polyhalogenated dioxins and dibenzofurans following subchronic treatment in mice. I. CYP1A1 and CYP1A2 enzyme activity in liver, lung, and skin. Toxicol. Appl. Pharmacol. 1997;147:267–280. doi: 10.1006/taap.1997.8261. [DOI] [PubMed] [Google Scholar]
- DeVito MJ, et al. Dose-response relationships for disposition and hepatic sequestration of polyhalogenated dibenzo-p-dioxins, dibenzofurans, and biphenyls following subchronic treatment in mice. Toxicol. Sci. 1998;46:223–234. doi: 10.1006/toxs.1998.2530. [DOI] [PubMed] [Google Scholar]
- Diliberto JJ, et al. Dose-response relationships of tissue distribution and induction of CYP1A1 and CYP1A2 enzymatic activities following acute exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in mice. Toxicol. Appl. Pharmacol. 1995;130:197–208. doi: 10.1006/taap.1995.1025. [DOI] [PubMed] [Google Scholar]
- Diliberto JJ, et al. Subchronic exposure of [3H]-2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in female B6C3F1 mice: relationship of steady-state levels to disposition and metabolism. Toxicol. Sci. 2001;61:241–255. doi: 10.1093/toxsci/61.2.241. [DOI] [PubMed] [Google Scholar]
- Eckel JE, et al. Empirical Bayes gene screening tool for time-course or dose-response microarray data. J. Biopharm. Stat. 2004;14:647–670. doi: 10.1081/BIP-200025656. [DOI] [PubMed] [Google Scholar]
- Eckel JE, et al. Normalization of two-channel microarray experiments: a semiparametric approach. Bioinformatics. 2005;21:1078–1083. doi: 10.1093/bioinformatics/bti105. [DOI] [PubMed] [Google Scholar]
- Hamm JT, et al. A mixture of dioxins, furans, and non-ortho PCBs based upon consensus toxic equivalency factors produces dioxin-like reproductive effects. Toxicol. Sci. 2003;74:182–191. doi: 10.1093/toxsci/kfg107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JA, et al. Notch signaling in development and disease. Clin. Genet. 2003;64:461–472. doi: 10.1046/j.1399-0004.2003.00194.x. [DOI] [PubMed] [Google Scholar]
- Harris M, et al. Comparative potencies of Aroclors 1232, 1242, 1248, 1254, and 1260 in male Wistar rats–assessment of the toxic equivalency factor (TEF) approach for polychlorinated biphenyls (PCBs) Fundam. Appl. Toxicol. 1993;20:456–463. doi: 10.1006/faat.1993.1056. [DOI] [PubMed] [Google Scholar]
- Haws LC, et al. Development of a refined database of mammalian relative potency estimates for dioxin-like compounds. Toxicol. Sci. 2006;89:4–30. doi: 10.1093/toxsci/kfi294. [DOI] [PubMed] [Google Scholar]
- Kopec AK, et al. Comparative toxicogenomic examination of the hepatic effects of PCB126 and TCDD in immature, ovariectomized C57BL/6 mice. Toxicol. Sci. 2008;102:61–75. doi: 10.1093/toxsci/kfm289. [DOI] [PubMed] [Google Scholar]
- Lin FH, et al. The effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on the hepatic estrogen and glucocorticoid receptors in congenic strains of Ah responsive and Ah nonresponsive C57BL/6J mice. Toxicol. Appl. Pharmacol. 1991;108:129–139. doi: 10.1016/0041-008x(91)90276-k. [DOI] [PubMed] [Google Scholar]
- Ma Q, et al. TCDD-inducible poly(ADP-ribose) polymerase: a novel response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem. Biophys. Res. Commun. 2001;289:499–506. doi: 10.1006/bbrc.2001.5987. [DOI] [PubMed] [Google Scholar]
- Ma X, et al. Protein tyrosine phosphorylation as an indicator of 2,3,7,8-tetrachloro-p-dioxin exposure in vivo and in vitro. Biochem. Biophys. Res. Commun. 1992;189:59–65. doi: 10.1016/0006-291x(92)91525-u. [DOI] [PubMed] [Google Scholar]
- Mason G, Safe S. Synthesis, biologic and toxic effects of the major 2,3,7,8-tetrachlorodibenzo-p-dioxin metabolites in the rat. Toxicology. 1986;41:153–159. doi: 10.1016/0300-483x(86)90196-4. [DOI] [PubMed] [Google Scholar]
- Mayes BA, McConnell EE, Neal BH, Brunner MJ, Hamilton SB, Sullivan TM, Peters AC, Ryan MJ, Toft JD, Singer AW, et al. Comparative carcinogenicity in Sprague-Dawley rats of the polychlorinated biphenyl mixtures Aroclors 1016, 1242, 1254, and 1260. Toxicol. Sci. 1998;41:62–76. doi: 10.1093/toxsci/41.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JN, Newsted JL, Hecker M, Zwiernik MJ, Fitzgerald SD, Kay DP, Zhang X, Higley EB, Aylward LL, Beckett KJ, et al. Hepatic P450 enzyme activity, tissue morphology and histology of mink (Mustela vison) exposed to polychlorinated dibenzofurans. Arch. Environ. Contam. Toxicol. 2009;57:416–425. doi: 10.1007/s00244-008-9241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin MD, et al. High-resolution PCB analysis: synthesis and chromatographic properties of all 209 PCB congeners. Environ. Sci. Technol. 1984;18:468–476. doi: 10.1021/es00124a014. [DOI] [PubMed] [Google Scholar]
- N'Jai A, et al. Comparative temporal toxicogenomic analysis of TCDD- and TCDF-mediated hepatic effects in immature female C57BL/6 mice. Toxicol. Sci. 2008;103:285–297. doi: 10.1093/toxsci/kfn053. [DOI] [PubMed] [Google Scholar]
- Narasimhan TR, et al. Relative sensitivities of 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced Cyp1a-1 and Cyp1a-2 gene expression and immunotoxicity in female B6C3F1 mice. Fundam. Appl. Toxicol. 1994;23:598–607. doi: 10.1006/faat.1994.1146. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program (NTP) NTP technical report on the toxicology and carcinogenesis studies of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (CAS No. 1746-01-6) in female Harlan Sprague-Dawley rats (Gavage Studies) Natl. Toxicol. Program Tech. Rep. Ser. 2006a;521 4–232. [PubMed] [Google Scholar]
- National Toxicology Program (NTP) NTP toxicology and carcinogenesis studies of 3,3′,4,4′,5-pentachlorobiphenyl (PCB 126) (CAS No. 57465-28-8) in female Harlan Sprague-Dawley rats (Gavage Studies) Natl. Toxicol. Program Tech. Rep. Ser. 2006b;520 4–246. [PubMed] [Google Scholar]
- Nebert DW, et al. Cellular responses to oxidative stress: the [Ah] gene battery as a paradigm. Environ. Health Perspect. 1990;88:13–25. doi: 10.1289/ehp.908813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW, et al. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem. Pharmacol. 2000;59:65–85. doi: 10.1016/s0006-2952(99)00310-x. [DOI] [PubMed] [Google Scholar]
- Poland A, Glover E. Studies on the mechanism of toxicity of the chlorinated dibenzo-p-dioxins. Environ. Health Perspect. 1973;5:245–251. doi: 10.1289/ehp.7305245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland A, Knutson JC. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Safe S. Polychlorinated biphenyls (PCBs), dibenzo-p-dioxins (PCDDs), dibenzofurans (PCDFs), and related compounds: environmental and mechanistic considerations which support the development of toxic equivalency factors (TEFs) Crit. Rev. Toxicol. 1990;21:51–88. doi: 10.3109/10408449009089873. [DOI] [PubMed] [Google Scholar]
- Safe S. Limitations of the toxic equivalency factor approach for risk assessment of TCDD and related compounds. Teratog. Carcinog. Mutagen. 1997;17:285–304. [PubMed] [Google Scholar]
- Safe S. Molecular biology of the Ah receptor and its role in carcinogenesis. Toxicol. Lett. 2001;120:1–7. doi: 10.1016/s0378-4274(01)00301-0. [DOI] [PubMed] [Google Scholar]
- Safe S, Robertson LW, Safe L, Parkinson A, Bandiera S, Sawyer T, Campbell MA. Halogenated biphenyls: molecular toxicology. Can. J. Physiol. Pharmacol. 1982;60:1057–1064. doi: 10.1139/y82-151. [DOI] [PubMed] [Google Scholar]
- Safe SH. Modulation of gene expression and endocrine response pathways by 2,3,7,8-tetrachlorodibenzo-p-dioxin and related compounds. Pharmacol. Ther. 1995;67:247–281. doi: 10.1016/0163-7258(95)00017-b. [DOI] [PubMed] [Google Scholar]
- Sheehan DC, Hrapchak BB. Theory and Practice of Histotechnology. 2nd ed. St. Louis, MO: Mosby; 1980. [Google Scholar]
- Starr TB, et al. The trouble with TEFs. Environ. Health Perspect. 1999;107:A492–A493. doi: 10.1289/ehp.107-1566591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijet N, et al. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol. Pharmacol. 2006;69:140–153. doi: 10.1124/mol.105.018705. [DOI] [PubMed] [Google Scholar]
- Van den Berg M, Birnbaum L, Bosveld AT, Brunstrom B, Cook P, Feeley M, Giesy JP, Hanberg A, Hasegawa R, Kennedy SW, et al. Toxic equivalency factors (TEFs) for PCBs, PCDDs, PCDFs for humans and wildlife. Environ. Health Perspect. 1998;106:775–792. doi: 10.1289/ehp.98106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg M, Denison M, De Vito M, Farland W, Feeley M, Fiedler H, Hakansson H, Hanberg A, Haws L, Rose M, Safe S, et al. The 2005 World Health Organization reevaluation of human and mammalian toxic equivalency factors for dioxins and dioxin-like compounds. Toxicol. Sci. 2006;93:223–241. doi: 10.1093/toxsci/kfl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, et al. Ligand-dependent interactions of the Ah receptor with coactivators in a mammalian two-hybrid assay. Toxicol. Appl. Pharmacol. 2008;227:196–206. doi: 10.1016/j.taap.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.