Abstract
Association mapping using DNA-based markers is a novel tool in plant genetics for the analysis of complex traits. Potato tuber yield, starch content, starch yield and chip color are complex traits of agronomic relevance, for which carbohydrate metabolism plays an important role. At the functional level, the genes and biochemical pathways involved in carbohydrate metabolism are among the best studied in plants. Quantitative traits such as tuber starch and sugar content are therefore models for association genetics in potato based on candidate genes. In an association mapping experiment conducted with a population of 243 tetraploid potato varieties and breeding clones, we previously identified associations between individual candidate gene alleles and tuber starch content, starch yield and chip quality. In the present paper, we tested 190 DNA markers at 36 loci scored in the same association mapping population for pairwise statistical epistatic interactions. Fifty marker pairs were associated mainly with tuber starch content and/or starch yield, at a cut-off value of q ≤ 0.20 for the experiment-wide false discovery rate (FDR). Thirteen marker pairs had an FDR of q ≤ 0.10. Alleles at loci encoding ribulose-bisphosphate carboxylase/oxygenase activase (Rca), sucrose phosphate synthase (Sps) and vacuolar invertase (Pain1) were most frequently involved in statistical epistatic interactions. The largest effect on tuber starch content and starch yield was observed for the paired alleles Pain1-8c and Rca-1a, explaining 9 and 10% of the total variance, respectively. The combination of these two alleles increased the means of tuber starch content and starch yield. Biological models to explain the observed statistical epistatic interactions are discussed.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-010-1389-3) contains supplementary material, which is available to authorized users.
Introduction
Association or linkage disequilibrium mapping using DNA-based markers is a novel tool in plant genetics for the analysis of complex traits (Gupta et al. 2005; Holland 2007). In association mapping studies, phenotypes and genotypes are evaluated in populations of individuals descending from multiple crosses between different parents rather than in progeny of controlled crosses between two parental genotypes. In the latter, the genotypic and phenotypic variations present in the two parents are analyzed, whereas the former captures the genotypic and phenotypic variations of many individuals (Mackay and Powell 2007). Association genetics is particularly useful in cultivated potato (Solanum tuberosum), a tetraploid, non-inbred crop species, where linkage analysis in experimental, tetraploid populations is complicated by tetrasomic inheritance (Luo et al. 2001). Moreover, potato breeding is based on intercrossing multiple, heterozygous parents followed by multi-year selection in the segregating first filial generation. The heterozygous F1 genotypes are vegetatively propagated. Among the numerous characters that are subject to selection, tuber traits such as yield, starch content, starch yield and chip color are of primary importance. Starch is the major storage compound and constitutes 10–25% of the tuber fresh weight. Starch yield (starch weight per unit area) is a relevant character when producing potato starch for industrial uses. For the food processing industry, the color of potato chips and French fries is an important quality criterion, which depends on the tuber content of the reducing sugars glucose and fructose. During deep frying at high temperatures, reducing sugars and amino acids undergo a non-enzymatic Maillard reaction, which results in dark brown product color due to polyphenol formation (Habib and Brown 1957; Townsend and Hope 1960). Tuber storage at low temperatures prevents sprouting but leads to increased conversion of starch in sugars (‘cold sweetening’) as an adaptive response to cold stress (Burton 1969; Isherwood 1973). Tuber yield, starch and sugar content are complex traits that are controlled by multiple genetic and environmental factors. Their quantitative evaluation in tubers harvested from field trials is routine in breeding programs, provided sufficient tuber quantities are available from individual genotypes. With the exception of yield (tuber fresh weight per unit area), these quantitative traits result from the synthesis, degradation and transport of carbohydrates. The genes and biochemical pathways involved in carbohydrate metabolism are at functional level among the best studied in plants (Frommer and Sonnewald 1995). Taken together, these properties make tuber yield, starch and sugar content attractive model traits for association genetics in potato using the candidate gene approach, i.e., natural DNA polymorphisms in genes known to be functional in the trait of interest are selectively tested for association with quantitative variation of this trait in populations of individuals related by descent.
The number of associations discovered in plants between DNA polymorphisms at individual candidate loci and complex traits is steadily growing (Ducrocq et al. 2008; Eckert et al. 2009; Ehrenreich et al. 2007; Gebhardt et al. 2004; Gonzalez-Martinez et al. 2007; Li et al. 2005, 2008; Olsen et al. 2004; Pajerowska-Mukhtar et al. 2009; Remington et al. 2001; Saidou et al. 2009; Thornsberry et al. 2001). However, whereas the importance of epistasis in the genetic architecture of complex traits has been emphasized (Carlborg and Haley 2004; Phillips 2008) and a theoretical basis has been developed (Jannink 2007), there are very few experimental data so far on the contribution of epistasis to explain phenotypic variation in plant association mapping populations (Manicacci et al. 2009; Stracke et al. 2009). Epistasis is defined here as the statistical deviation from the additive combination of two loci in their effects on a phenotype (Fisher 1918; Phillips 2008).
In a previous study, we identified DNA variation in genes that function in carbohydrate metabolism or transport, which was tested for association with tuber yield, starch content, starch yield and chip color in a population of tetraploid varieties and breeding clones derived from advanced potato breeding programs. Highly significant and robust associations between individual candidate gene alleles and the traits were discovered (Li et al. 2008). In this paper, we report on the results of analyzing the genotypic and phenotypic data of this association mapping experiment for two-way epistatic interactions.
Materials and methods
Plant material
The analysis of two-way epistatic interactions was based on the phenotypic and genotypic data of 243 tetraploid individuals, which included 34 varieties as standards and 90, 96 and 23 clones from the breeding programs for chips, starch and table potatoes of the companies Böhm-Nordkartoffel Agrarproduktion (BNA), SAKA Pflanzenzucht (SAR) and NORIKA (NOR), respectively. Further details of this population referred to as population ‘ALL’ are described elsewhere (Li et al. 2008).
Phenotypic data
Tubers of the individuals of the ALL population were evaluated in 2 years as described (Li et al. 2008) for starch content (TSC, percent fresh weight), yield (TY, dt/ha = deciton per hectare, 1 dt = 100 kg), starch yield (TSY = TSC × TY, dt/ha), chip quality after harvest (CQA, score between 1 and 9, 1 = very bad chip quality; 9 = very good chip quality) and after 3–4 months storage at 4°C (CQS). Tuber starch content was determined by measuring specific gravity. Tuber yield was determined by the tuber weight. Tuber starch yield is the product of TSC × TY. Chip quality was assessed by visually scoring the chip color after deep frying of 1.2–2.0 mm tuber slices in oil at 160–180°C for 2–3 min.
Genotypic data
Genotyping of the population ALL has been described (Li et al. 2008). In brief, 36 loci on all potato chromosomes except chromosome I were evaluated for the presence or absence of 190 single strand conformation polymorphism (SSCP), simple sequence repeat (SSR), cleaved amplified polymorphic sequence (CAPS), sequence characterized amplified region (SCAR) and allele specific amplification (ASA) DNA fragments. Twenty-two loci encoded genes related to carbohydrate metabolism or transport. The remaining loci were anonymous markers such as SSRs. All DNA fragments were scored as present (1) or absent (0) in each individual, with some missing values.
Statistical analysis
As a preliminary to the association analysis, population substructure was evaluated using the software STRUCTURE (Pritchard et al. 2000) as described (Li et al. 2008). For an assumed number of groups K between 1 and 30 we found that the likelihood increased steadily with increasing K, and we found no evidence of population structure for the present set of genotypes. We also explored the genetic relationships with the first two principal components, which accounted for more than 40% of the total genetic variation, and these components represented a grouping similar to the origin of the genotypes. Therefore, we included the factor ‘origin’ in our association analyses. Origin had four levels corresponding to the genotype groups “SAR”, “BNA” and “NOR” and the group “Standards”.
Analysis of epistatic interactions was thus performed using the regression model:
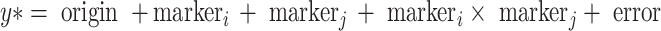 |
where y* stands for adjusted trait means as described (Li et al. 2008). Factors markeri and markerj had two levels, indicating whether a DNA fragment was present or absent. The epistatic effect between the two markers markeri and markerj was tested by a partial t test, after correcting for origin and the additive main effects of the two markers. We tested epistasis between all possible marker pairs. The proportion of genetic variation explained by an epistatic interaction was calculated as the relative increase in R 2 after the interaction effect is added to the model including origin and marker main effects.
Correction for multiple testing:
We included a correction for multiple testing aiming at the false discovery rate (FDR) (Storey and Tibshirani 2003): the proportion of false positives in the set of epistatic interactions for which the null hypothesis of no interaction is rejected. The P values resulting from testing epistasis in our regression model were converted into so-called q values that estimate FDR, according to the method of Allison et al. (2002), available in Genstat’s procedure FDRMIXTURE [GenStat (2005) GenStat 8rd edn. Release 8.1. VSN International Ltd., Oxford, UK]. In this approach, the P values are assumed to come from a mixture of two distributions, a uniform density corresponding to P values obtained from H 0 tests, and a Beta density for P values obtained from H 1 tests. The method estimates the proportion of true H 0 tests and the parameters of the Beta density, and q values are produced according to the fitted parameters. Here, we report epistatic interactions with an FDR of q ≤ 0.20.
Results
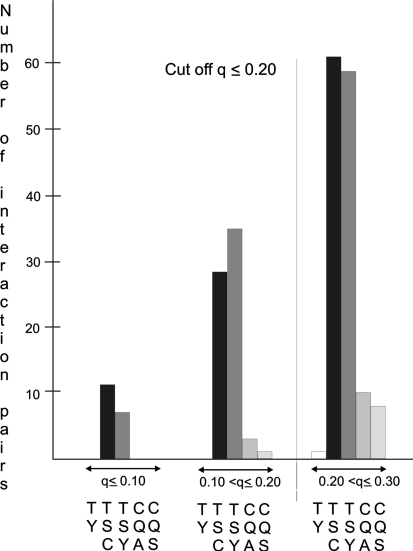
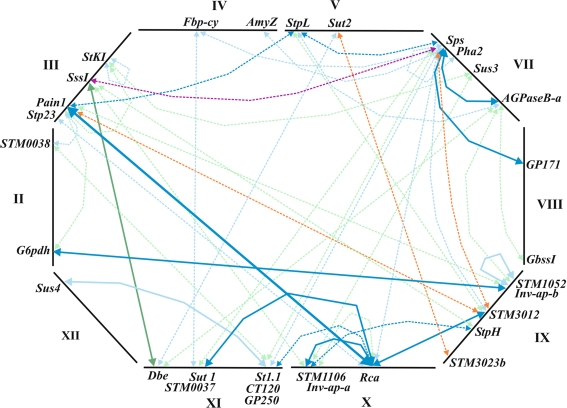
Pairwise tests for epistatic interactions between 190 polymorphic DNA fragments derived from 36 loci resulted in 17.956 tests each for tuber starch content (TSC), tuber yield (TY), tuber starch yield (TSY), chip quality after harvest (CQA) and after cold storage (CQS). Experiment-wide FDRs (q values) were calculated by applying the multiple testing correction described in “Materials and methods”. The number of interaction pairs with 0.20 < q ≤ 0.30, 0.10 < q ≤ 0.20 and q ≤ 0.10 is shown in Fig. 1. When choosing as cut-off value q < 0.20, no interaction pair remained for tuber yield and only few interaction pairs remained for chip quality. The majority of interaction pairs was found for tuber starch content and starch yield (Fig. 1). Interaction pairs for TSC and TSY partially overlapped (the same marker pair showed interaction for TSC and TSY) and some were redundant. Redundant interaction pairs resulted from different DNA fragments amplified from different regions of the same gene that were in very high or absolute linkage disequilibrium with each other and showed therefore similar interactions (Li et al. 2008). When redundant interaction pairs were removed from the data set, 50 marker pairs remained (Table 1; Supplementary Table 1), 13 of which were significant at q ≤ 0.10 for either TSC or TSY or both (Table 2). These 13 interaction pairs explained between 5 and 10% of the total variance (Table 2). Eight of the 20 different alleles forming the 13 interaction pairs had also shown main effects for at least one of the traits CQA, CQS, TSC, TSY and TY (Li et al. 2008). The most significant interaction (q < 0.05) for both TSC and TSY involved the alleles Rca-1a and Pain1-8c at two loci encoding ribulose-bisphosphate carboxylase activase and soluble acid invertase, respectively (Table 2). Mean and standard errors for TSC and TSY of the four marker genotype classes of the 13 most significant marker pairs in are shown in Table 3. The 13 most significant interaction pairs involved 15 loci (Table 2), whereas the 50 interaction pairs involved 31 of the 36 loci genotyped in the ALL population (Supplementary Table 1; Fig. 2). Alleles at the loci Sps (sucrose phosphate synthase), Rca (ribulose-bisphosphate carboxylase activase) and Pain1 (soluble acid invertase) on chromosomes VII, X and III, respectively, were most frequently participating in interactions (Fig. 2). The Sps alleles 7c, 7d and 7e were partner in 13 interactions, the Pain1 alleles 8c/9a (8c and 9a are in strong LD with each other), 5b and 5d were partner in 8 interactions and the Rca alleles 1a and 3b were partner in 11 interactions. Thirty of the 50 interaction pairs involved at least one allele of these three loci (Table 2; Supplementary Table 1).
Fig. 1.
Number of interaction pairs associated with TY (white bar), TSC (black bar), TSY (dark gray bar), CQA and CQS (light gray bars) at the experiment-wide false discovery rates q ≤ 0.10, 0.10 < q ≤ 0.20 and 0.20 < q ≤ 0.30. The cut-off value q < 0.205 was chosen as threshold for reporting significant epistatic interactions
Table 1.
Number of pairwise epistatic interactions at q ≤ 0.20
| Traits | Number of interaction pairs |
|---|---|
| TSC | 13 |
| TSY | 17 |
| TSC and TSY | 16 |
| CQA | 3 |
| TSC, TSY and CQS | 1 |
| Σ = 50 |
Table 2.
Pairwise epistatic interactions significant at q ≤ 0.10 for TSC and/or TSY
| Locus 1 | Chromosome number | Locus 2 | Chromosome number | Allele 1 | Allele 2 | TSC | TSY | ||
|---|---|---|---|---|---|---|---|---|---|
| q | R 2 (%) | q | R 2 (%) | ||||||
| Pain1 | III | Rca | X | Pain1 - 8c a | Rca - 1a a | 0.0336 | 9.13 | 0.0185 | 10.27 |
| Pain1 | III | Rca | X | Pain1-5d | Rca - 1a a | 0.0745 | 6.74 | 0.0977 | 5.81 |
| STM3012 | IX | Rca | X | STM3012-a | Rca - 1a a | 0.0608 | 7.23 | 0.1188 | 5.53 |
| STM3012 | IX | Rca | X | STM3012-b | Rca - 1a a | 0.0888 | 6.57 | 0.1065 | 5.84 |
| STM1106 | X | Rca | X | STM1106-m | Rca - 1a a | 0.0947 | 6.58 | 0.1130 | 5.72 |
| STM1106 | X | Rca | X | STM1106-f | Rca - 1a a | 0.1832 | 5.15 | 0.0472 | 7.25 |
| Sut1 | XI | Rca | X | Sut1-7b | Rca - 1a a | 0.0817 | 7.07 | 0.1855 | 4.86 |
| STM1052 | IX | Inv-ap-b | IX | STM1052-a | InvGE-6i | 0.0611 | 7.60 | >0.3 | 2.72 |
| G6pdh | II | Inv-ap-b | IX | G6pdh-4b | InvGF - 4d a | 0.1834 | 5.13 | 0.0575 | 6.81 |
| AGPaseB-a | VII | Sps | VII | AGPsb-6a | Sps - 7c a | 0.0730 | 7.55 | 0.0794 | 6.59 |
| GP171 | VIII | Sps | VII | GP171 - a a | Sps-7e a | 0.1000 | 6.01 | 0.0639 | 6.44 |
| St1.1 | XI | Sus4 | XII | St1.1 - HinfI a | Sus4-2a a | 0.0878 | 6.47 | >0.3 | 2.89 |
| SssI | III | Dbe | XI | SssI - 7a a | Dbe - 5c a | 0.2744 | 4.08 | 0.1063 | 5.66 |
aThe same marker allele showed a significant (q ≤ 0.05) main effect on at least one of the traits CQA, CQS, TSC, TSY and TY (Li et al. 2008)
Table 3.
Number of individuals, phenotypic mean and standard errors (SE) of marker genotype classes 11 (both alleles present), 00 (both alleles absent), 10 (allele 1 present, allele 2 absent) and 01 (allele 1 absent, allele 2 present) for epistatic interactions with q ≤ 0.10
| Allele 1 | Allele 2 | No. of individuals per genotype class | TSC genotype class mean (SE) (%) | TSY genotype class mean (SE) (dt/ha) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 00 | 10 | 01 | 11 | 00 | 10 | 01 | 11 | 00 | 10 | 01 | ||
| Pain1-8c | Rca-1a | 14 | 93 | 32 | 57 | 21.4 (0.8) | 17.1 (0.4) | 18.1 (0.5) | 15.4 (0.5) | 138.5 (6.0) | 104.4 (2.6) | 107.8 (3.7) | 96.6 (3.3) |
| Pain1-5d | Rca-1a | 9 | 145 | 16 | 69 | 21.1 (1.0) | 17.3 (0.3) | 16.8 (0.8) | 15.9 (0.4) | 134.9 (7.3) | 104.8 (2.0) | 99.7 (5.5) | 100.4 (3.0) |
| STM3012-a | Rca-1a | 46 | 63 | 96 | 34 | 15.3 (0.5) | 16.9 (0.4) | 17.3 (0.3) | 18.5 (0.6) | 96.4 (3.5) | 101.4 (2.8) | 105.3 (2.4) | 116.2 (4.1) |
| STM3012-b | Rca-1a | 22 | 122 | 32 | 57 | 19.3 (0.7) | 17.2 (0.3) | 16.9 (0.5) | 15.7 (0.4) | 122.6 (4.9) | 104.1 (2.2) | 100.4 (3.8) | 98.7 (3.2) |
| STM1106-m | Rca-1a | 4 | 150 | 7 | 72 | 22.4 (1.5) | 17.3 (0.3) | 15.8 (1.1) | 16.2 (0.4) | 145.3 (10.7) | 104.6 (1.9) | 94.3 (8.0) | 101.6 (2.9) |
| STM1106-f | Rca-1a | 7 | 144 | 13 | 69 | 20.7 (1.1) | 17.2 (0.3) | 16.6 (0.8) | 16.1 (0.4) | 137.8 (8.1) | 104.6 (1.9) | 95.7 (5.9) | 137.8 (2.9) |
| Sut1-7b | Rca-1a | 58 | 52 | 100 | 17 | 16.0 (0.4) | 16.7 (0.4) | 19.4 (0.7) | 17.3 (0.3) | 100.3 (3.2) | 102.7 (3.0) | 123.8 (5.4) | 104.5 (2.4) |
| STM1052-a | InvGE-6i | 118 | 64 | 17 | 31 | 16.8 (0.3) | 16.5 (0.4) | 18.9 (0.7) | 18.9 (0.6) | nsa | ns | ns | ns |
| G6pdh-4b | InvGF-4d | 23 | 93 | 73 | 51 | 15.1 (0.6) | 16.8 (0.4) | 17.4 (0.4) | 17.7 (0.5) | 89.9 (4.5) | 99.6 (2.5) | 110.6 (2.5) | 106.8 (3.2) |
| AGPsb-6a | Sps-7c | 76 | 51 | 38 | 61 | 16.7 (0.4) | 16.1 (0.4) | 18.4 (0.5) | 17.8 (0.4) | 101.4 (2.6) | 98.5 (2.9) | 108.2 (3.4) | 115.6 (2.9) |
| GP171-a | Sps-7e | 97 | 20 | 18 | 107 | 16.1 (0.3) | 15.4 (0.7) | 18.1 (0.3) | 17.6 (0.7) | 97.7 (2.4) | 92.9 (5.0) | 109.8 (2.2) | 114.0 (2.4) |
| St1.1-HinfI | Sus4-2a | 81 | 38 | 89 | 34 | 16.6 (0.3) | 15.1 (0.5) | 17.7 (0.3) | 17.7 (0.5) | ns | ns | ns | ns |
| SssI-7a | Dbe-5c | 76 | 31 | 70 | 48 | ns | ns | ns | ns | 100.0 (2.5) | 93.0 (3.8) | 116.1 (2.6) | 100.9 (3.2) |
aNot significant with the threshold q ≤ 0.20
Fig. 2.
Map of pairwise epistatic interactions associated with TSC, TSY and chip quality (CQA or CQS). Marker pairs significant at q ≤ 0.10 and 0.10 < q ≤ 0.20 are connected by solid and broken-line arrows, respectively. Dark blue arrows Associations with TSC and TSY, light blue arrows associations with TSC, blue-green arrows associations with TSY, orange arrows associations with CQA, purple arrow association with TSC, TSY and CQS. Details of the interaction pairs can be found in supplementary Table 1. The positions of the 31 interacting loci on potato chromosomes II–V, VII–XII are shown schematically, for further details see Li et al. (2008). Loci encoding candidate genes are (clock-wise starting with chromosome II): G6pdh cytoplasmic glucose-6-phosphate dehydrogenase, Stp23 plastidic L-type α-glucan phosphorylase, Pain1 vacuolar soluble acid invertase, SssI soluble starch synthase I, StKI clustered gene family of Kunitz-type enzyme inhibitors, Fbp-cy cytoplasmic fructose-1,6-bisphosphatase, AmyZ α-amylase, StpL plastidic l-type starch phosphorylase, Sut2 sucrose sensor or transporter, Sps sucrose phosphate synthase, Pha2 plasma membrane H+ -ATPase 2, Sus3 sucrose synthase 3, AGPaseB-a ADP-glucose pyrophosphorylase, B subunit, GbssI granule-bound starch synthase I, Inv-ap-b tandem duplicated apoplastic invertase genes InvGE and InvGF, StpH cytoplasmic H-type starch phosphorylase, Rca ribulose-bisphosphate carboxylase/oxygenase activase, Inv-ap-a duplicated apoplastic invertase genes, Sut1 sucrose transporter 1, Dbe debranching enzyme, Sus4 sucrose synthase 4. STM0038, STM1052, STM3012, STM3023b, STM1106 and STM0037 are SSR loci. Loci without known function in carbohydrate metabolism are GP171, GP250, CT120 and St1.1
Discussion
When searching for statistical epistatic interactions, the problem of false positives due to multiple testing is aggravated. At the same time, the power of the two-way test statistic is reduced compared with single marker association tests. A too stringent multiple testing correction might obscure true interactions, whereas no correction inflates the number of false positives. In this paper we report epistatic interactions with an experiment-wide FDR of 20% (q ≤ 0.20) (Kraakman et al. 2004). With this threshold, 50 marker pairs were associated with at least one of the five quantitative traits. Twenty percent or around ten of those pairs are expected to be false positives. With the more stringent threshold of 10% FDR (q ≤ 0.10) 13 marker pairs remained, one or two of which are expected to be false positives.
False positive associations may also arise due to population substructure and/or genetic relationships between genotypes. The analysis of the marker data with the software STRUCTURE (Li et al. 2008; Pritchard et al. 2000) did not provide clear information on the number of groups/subpopulations. Further analysis of the marker data showed that the first principal components represented a grouping coinciding with a grouping due to the origin of the genotypes. Thus, not much evidence was found for strong substructure in the association mapping population ALL. This is likely due to a rather homogeneous genetic background of the ALL population representing middle European commercial varieties and breeding clones, and the generally very high molecular diversity observed among heterozygous, tetraploid potato genotypes (Li et al. 2005). When analyzing the data with a model that corrects for kinship (as derived from marker data) about the same epistatic interactions were obtained as with the model correcting for origin, the results of which are presented in this paper. We checked a posteriori whether the 50 epistatic interactions obtained with this model could be explained either by subtle genetic relations in the population, or by other QTL main effects. First, we tested the 50 epistatic interactions with a model that included 20 principal components representing marker-based relationships between genotypes (Price et al. 2006). Eighty-nine percent of the epistatic interactions still yielded low P values (P < 0.01). In a second check we tested the epistatic interactions in a model containing multi-QTL main effects. Ninety-two percent of the epistatic interactions again yielded low P values (P < 0.01). Therefore, we cannot exclude that some epistatic effects are confounded with the other main effect QTLs. The most significant interaction between the alleles Rca-1a and Pain1-8c was detected with every test statistic applied to the dataset.
These findings demonstrate that epistasis indeed plays a role in the variation of the complex tuber traits in the analyzed population of tetraploid potato varieties and breeding clones.
Forty seven marker pairs were associated with TSC and/or TSY, three with CQA and none with TY. This differs from our previous findings when we tested all markers individually for association with the same traits (Li et al. 2008). The most significant marker alleles then were associated with chip quality (CQA and CQS), TSC and TSY, and in one case also with TY. Within the limitations of the association mapping experiment by population size and number of loci tested, chip quality was therefore not strongly influenced by epistatic interactions, in contrast to TSC and TSY. The low number of markers associated with tuber yield, both in single and pair wise tests, indicates that genomic regions other than the ones tagged by the 36 tested loci are more important for natural variation of tuber yield. The choice of candidate loci based on their function in carbohydrate metabolism and transport was more suitable to detect associations with chip quality, tuber starch content and starch yield than with tuber yield.
As a good set of functional candidate genes for yield is not available, whole genome association mapping is more appropriate to dissect tuber yield.
Three loci, Sps, Pain1 and Rca, were exceptional with respect to the number of epistatic interactions they showed with each other and with other loci. The genes encoded at these loci may be either directly causal for the observed effects, or the associations are indirect due to linkage disequilibrium with the causal genes that are physically linked but different. Linkage disequilibrium in potato can extend over few Centimorgans in the genetic material used here for association mapping (Li et al. 2008). Nevertheless, the central functional roles of Rca, Sps and Pain1 in carbon fixation and partitioning into sugars and starch suggest a possible explanation for the observed frequency of epistatic interactions. Photosynthetic CO2 fixation in chloroplasts is catalyzed by ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco). The activity of Rubisco is regulated by Rubisco activase (RCA) (Mate et al. 1993; Portis 1990; Salvucci and Ogren 1996). Fixed carbon is transitory deposited in chloroplasts as starch and after starch degradation transported as triose-phosphate into the cytoplasm of leaf mesophyll cells, where it is partitioned in sugars, other metabolites and respiratory pathways. Cytoplasmic sucrose phosphate synthase (SPS) has a key role in the synthesis of sucrose (Huber and Huber 1992; Stitt et al. 1988; Winter et al. 2000), the metabolite by which carbon is mainly exported from the leaves and transported through the vascular system to the developing tuber, where it is stored as starch in the amyloplasts of parenchyma cells (Frommer and Sonnewald 1995). Several invertase isoforms are present in source as well as sink tissues and cleave sucrose irreversibly in glucose and fructose in different cellular compartments, the apoplastic space, the cytoplasm and the vacuole. By controlling the ratio of sucrose to hexoses, invertases are crucial for the strength of metabolic sinks such as tubers, for growth and for carbon partitioning. High sucrose-to-hexose ratios favor the channeling of carbon into storage compounds whereas low sucrose-to-hexose ratios promote cell division and growth (Sturm and Tang 1999; Tymowska-Lalanne et al. 1998; Winter et al. 2000). The Pain1 locus encodes a vacuolar invertase, which together with apoplastic invertases controls the ratio of sucrose to hexose in tubers (Zrenner et al. 1996). Thus, the Sps, Pain1 and Rca loci are linked with each other and with many other enzymes through the metabolic flux from photosynthetic (source) to storage (sink) organs. Natural allelic DNA variation in the coding and/or non-coding regions of a gene can cause non-destructive functional modification of the gene product, by modulating its expression, translation, posttranslational modification, transport, degradation and/or catalytic properties. When connected with many other proteins in a metabolic network, the effect of allelic variation at one locus is likely to depend on the allelic variation at a second and more loci (Alcazar et al. 2009). Higher order interactions were not tested in the present data set.
Interestingly, the most significant epistatic interaction was observed between the alleles Rca-1a and Pain1-8c (q < 0.05). This interaction explained 9 and 10% of the total variance of TSC and TSY, respectively. When tested individually, both alleles were associated with tuber quality traits. The Rca-1a allele decreased, on average, chip quality (CQA and CQS), whereas the Pain1-8c allele increased, on average, CQA, CQS, TSC and TSY (Li et al. 2008). The combination of both alleles had no effect on chip quality any more but increased tuber starch content by 3–5% and starch yield by 20–40 dt/ha, compared with the other three allele combinations (Table 3). The following model is compatible with these observed effects: The Rca-1a allele activates Rubisco more compared to other Rca alleles, which leads to a higher rate of CO2 fixation in plants carrying this allele. On the other hand, the Pain1-8c allele encodes an invertase that is less effective than other invertase alleles in sucrose cleavage, thereby shifting the balance towards higher sucrose-to-hexose ratios in carrier plants. The combination of a more active Rca allele with a less effective invertase allele partitions more carbon into starch and increases tuber starch content and starch yield. In contrast, the combination of a more active Rca allele with ‘normal’ invertase alleles leads to lower sucrose-to-hexose ratios, higher contents of glucose and fructose, thereby decreasing chip quality, which is inversely correlated with glucose and fructose content. Vice versa, a less effective invertase allele in combination with ‘normal’ Rca alleles can still have positive effects on chip quality, tuber starch content and starch yield. The allele dosage might be an important factor in interaction models in polyploid species. However, the allele dosage could not be scored when using SSCP for detection of DNA polymorphisms. More research is needed to verify this model and to elucidate the mechanisms that determine the quality of natural alleles in the context of metabolic networks.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was funded by the German Ministry for Research and Education under the GABI (Genome analysis in the biological system plant) program (Grant no. 0313038, GABI-CHIPS) and by the Max-Planck society. Part of the work was carried out in the Department for Plant Breeding and Genetics, headed by Maarten Koornneef. The authors thank Benjamin Stich for helpful comments.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
L. Li and M.-J. Paulo equally contributed to the work and share first authorship.
References
- Alcazar R, Garcia AV, Parker JE, Reymond M. Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc Natl Acad Sci. 2009;106:334–339. doi: 10.1073/pnas.0811734106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DB, Gadbury GL, Heo M, Fernández JR, Lee C-K, Prolla TA, Weindruch R. A mixture model approach for the analysis of microarray gene expression data. Comput Stat Data Anal. 2002;39:1–20. doi: 10.1016/S0167-9473(01)00046-9. [DOI] [Google Scholar]
- Burton WG. The sugar balance in some British potato varieties during storage II: the effects of tuber age, previous storage temperature, and intermittent refrigeration upon low-temperature sweetening. Eur Potato J. 1969;12:81–95. doi: 10.1007/BF02364726. [DOI] [Google Scholar]
- Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies? Nat Rev Genet. 2004;5:618–625. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- Ducrocq S, Madur D, Veyrieras J-B, Camus-Kulandaivelu L, Kloiber-Maitz M, Presterl T, Ouzunova M, Manicacci D, Charcosset A. Key impact of Vgt1 on flowering time adaptation in maize: evidence from association mapping and ecogeographical information. Genetics. 2008;178:2433–2437. doi: 10.1534/genetics.107.084830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert A, Bower AD, Wegrzyn JL, Pande B, Jermstad KD, Krutovsky KV, St. Clair JB, Neale DB (2009) Association genetics of coastal Douglas-fir (Pseudotsuga menziesii var. menziesii, Pinaceae) I. Cold-hardiness related traits. Genetics. http://genetics.109.102350 [DOI] [PMC free article] [PubMed]
- Ehrenreich IM, Stafford PA, Purugganan MD. The genetic architecture of shoot branching in Arabidopsis thaliana: a comparative assessment of candidate gene associations vs. quantitative trait locus mapping. Genetics. 2007;176:1223–1236. doi: 10.1534/genetics.107.071928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The correlations between relatives on the supposition of Mendelian inheritance. Trans R Soc Edinb. 1918;52:399–433. [Google Scholar]
- Frommer WB, Sonnewald U. Molecular analysis of carbon partitioning in solanaceous species. J Exp Bot. 1995;46:587–607. doi: 10.1093/jxb/46.6.587. [DOI] [Google Scholar]
- Gebhardt C, Ballvora A, Walkemeier B, Oberhagemann P, Schüler K. Assessing genetic potential in germplasm collections of crop plants by marker-trait association: a case study for potatoes with quantitative variation of resistance to late blight and maturity type. Mol Breed. 2004;13:93–102. doi: 10.1023/B:MOLB.0000012878.89855.df. [DOI] [Google Scholar]
- Gonzalez-Martinez SC, Wheeler NC, Ersoz E, Nelson CD, Neale DB. Association genetics in Pinus taeda L. I. Wood property traits. Genetics. 2007;175:399–409. doi: 10.1534/genetics.106.061127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Rustgi S, Kulwal P. Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol Biol. 2005;57:461–485. doi: 10.1007/s11103-005-0257-z. [DOI] [PubMed] [Google Scholar]
- Habib A, Brown HD. The role of reducing sugars and amino acids in browning of potato chips. Food Technol. 1957;11:85–89. [Google Scholar]
- Holland JB. Genetic architecture of complex traits in plants. Curr Opin Plant Biol. 2007;10:156–161. doi: 10.1016/j.pbi.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Huber SC, Huber JL. Role of sucrose-phosphate synthase in sucrose metabolism in leaves. Plant Physiol. 1992;99:1275–1278. doi: 10.1104/pp.99.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isherwood FA. Starch-sugar interconversion in Solanum tuberosum. Phytochemistry. 1973;12:2579–2591. doi: 10.1016/0031-9422(73)85060-5. [DOI] [Google Scholar]
- Jannink J-L. Identifying quantitative trait locus by genetic background interactions in association studies. Genetics. 2007;176:553–561. doi: 10.1534/genetics.106.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman ATW, Niks RE, Van den Berg PMMM, Stam P, Van Eeuwijk FA. Linkage disequilibrium mapping of yield and yield stability in modern spring barley cultivars. Genetics. 2004;168:435–446. doi: 10.1534/genetics.104.026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Strahwald J, Hofferbert HR, Lubeck J, Tacke E, Junghans H, Wunder J, Gebhardt C. DNA variation at the invertase locus invGE/GF is associated with tuber quality traits in populations of potato breeding clones. Genetics. 2005;170:813–821. doi: 10.1534/genetics.104.040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Paulo MJ, Strahwald J, Lübeck J, Hofferbert HR, Tacke E, Junghans H, Wunder J, Draffehn A, van Eeuwijk F, Gebhardt C. Natural DNA variation at candidate loci is associated with potato chip color, tuber starch content, yield and starch yield. Theor Appl Genet. 2008;116:1167–1181. doi: 10.1007/s00122-008-0746-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZW, Hackett CA, Bradshaw JE, McNicol JW, Milbourne D. Construction of a genetic linkage map in tetraploid species using molecular markers. Genetics. 2001;157:1369–1385. doi: 10.1093/genetics/157.3.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay I, Powell W. Methods for linkage disequilibrium mapping in crops. Trends Plant Sci. 2007;12:57–63. doi: 10.1016/j.tplants.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Manicacci D, Camus-Kulandaivelu L, Fourmann M, Arar C, Barrault S, Rousselet A, Feminias N, Consoli L, Frances L, Mechin V, Murigneux A, Prioul J-L, Charcosset A, Damerval C. Epistatic interactions between Opaque2 transcriptional activator and its target gene CyPPDK1 control kernel trait variation in maize. Plant Physiol. 2009;150:506–520. doi: 10.1104/pp.108.131888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mate CJ, Hudson GS, von Caemmerer S, Evans JR, Andrews TJ. Reduction of ribulose bisphosphate carboxylase activase levels in tobacco (Nicotiana tabacum) by antisense RNA reduces ribulose bisphosphate carboxylase carbamylation and impairs photosynthesis. Plant Physiol. 1993;102:1119–1128. doi: 10.1104/pp.102.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KO, Halldorsdottir SS, Stinchcomb JR, Weinig C, Schmitt J, Purugganan MD. Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics. 2004;167:1361–1369. doi: 10.1534/genetics.103.024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar K, Stich B, Achenbach U, Ballvora A, Lübeck J, Strahwald J, Tacke E, Hofferbert H-R, Ilarionova E, Bellin D, Walkemeier B, Basekow R, Kersten B, Gebhardt C. Single nucleotide polymorphisms in the Allene Oxide Synthase 2 gene are associated with field resistance to late blight in populations of tetraploid potato cultivars. Genetics. 2009;181:1115–1127. doi: 10.1534/genetics.108.094268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC. Epistasis—the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portis ARJ. Rubisco activase. Biochim Biophys Acta. 1990;1015:15–28. doi: 10.1016/0005-2728(90)90211-L. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci U S A. 2001;98:11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidou A-A, Mariac C, Luong V, Pham J-L, Bezancon G, Vigouroux Y. Association studies identify natural variation at PHYC linked to flowering time and morphological variation in Pearl Millet. Genetics. 2009;182:899–910. doi: 10.1534/genetics.109.102756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvucci M, Ogren W. The mechanism of Rubisco activase: insights from studies of the properties and structure of the enzyme. Photosynth Res. 1996;47:1–11. doi: 10.1007/BF00017748. [DOI] [PubMed] [Google Scholar]
- Stitt M, Wilke I, Feil R, Heldt HW. Coarse control of sucrose-phosphate synthase in leaves: alterations of the kinetic properties in response to the rate of photosynthesis and the accumulation of sucrose. Planta. 1988;174:217–230. doi: 10.1007/BF00394774. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Haseneyer G, Veyrieras J-B, Geiger H, Sauer S, Graner A, Piepho H-P. Association mapping reveals gene action and interactions in the determination of flowering time in barley. Theor and Appl Genet. 2009;118:259–273. doi: 10.1007/s00122-008-0896-y. [DOI] [PubMed] [Google Scholar]
- Sturm A, Tang G-Q. The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 1999;4:401–407. doi: 10.1016/S1360-1385(99)01470-3. [DOI] [PubMed] [Google Scholar]
- Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES. Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet. 2001;28:286–289. doi: 10.1038/90135. [DOI] [PubMed] [Google Scholar]
- Townsend LR, Hope GW. Factors influencing the color of potato chips. Can J Plant Sci. 1960;40:58–64. doi: 10.4141/cjps60-007. [DOI] [Google Scholar]
- Tymowska-Lalanne Z, Kreis M, Callow JA (1998) The plant invertases: physiology, biochemistry and molecular biology. In: Advances in botanical research. Academic Press, Dublin, pp 71–117
- Winter H, Huber SC, Sharkey T. Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit Rev Biochem Mol Biol. 2000;35:253–289. doi: 10.1080/10409230008984165. [DOI] [PubMed] [Google Scholar]
- Zrenner R, Schüler K, Sonnewald U. Soluble acid invertase determines the hexose-to-sucrose ratio in cold-stored potato tubers. Planta. 1996;198:246–252. doi: 10.1007/BF00206250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.