Abstract
Human fingertip microflora is transferred to touched objects and may provide forensically relevant information on individual hosts, such as on geographic origins, if endogenous microbial skin species/strains would be retrievable from physical fingerprints and would carry geographically restricted DNA diversity. We tested the suitability of physical fingerprints for revealing human host information, with geographic inference as example, via microbial DNA fingerprinting. We showed that the transient exogenous fingertip microflora is frequently different from the resident endogenous bacteria of the same individuals. In only 54% of the experiments, the DNA analysis of the transient fingertip microflora allowed the detection of defined, but often not the major, elements of the resident microflora. Although we found microbial persistency in certain individuals, time-wise variation of transient and resident microflora within individuals was also observed when resampling fingerprints after 3 weeks. While microbial species differed considerably in their frequency spectrum between fingerprint samples from volunteers in Europe and southern Asia, there was no clear geographic distinction between Staphylococcus strains in a cluster analysis, although bacterial genotypes did not overlap between both continental regions. Our results, though limited in quantity, clearly demonstrate that the dynamic fingerprint microflora challenges human host inferences for forensic purposes including geographic ones. Overall, our results suggest that human fingerprint microflora is too dynamic to allow for forensic marker developments for retrieving human information.
Electronic supplementary material
The online version of this article (doi:10.1007/s00414-009-0352-9) contains supplementary material, which is available to authorized users.
Keywords: Microbial forensics, Fingertip microflora, Microbial DNA analysis, Microbial fingerprint, Forensic, PFGE, Staphylococcus
Introduction
Microbes can negatively interfere with the postmortem assessment of alcohol abuse and in this way pose problems for forensic investigators [1]. However, microbial forensics is often chiefly associated with the detection of highly pathogenic microbes to which humans are deliberately exposed in cases of biological terrorism [2, 3]. However, human fingertip microflora left behind on touched objects at crime scenes may potentially contain forensically relevant information that may be useful for human host inferences accessible via microbial DNA fingerprinting of physical fingerprints. For example, if endogenous microbial skin species/strains with a geographically restricted distribution could be retrieved from touched objects via microbial DNA analysis, the geographic origin of the human host individual could be determined indirectly. Information about the geographic region of origin can be relevant in suspect-less forensic cases where the evidence DNA sample does not match either a suspect’s DNA profile or any in a criminal DNA database. In such cases, geographic information derived from crime scene samples is expected to reduce the potential pool of suspects by allowing police investigations to concentrate on specific groups of people, i.e., those from a restricted geographic region. Numerous human genetic markers have been suggested for inferring human genetic ancestry mostly to the continental level [4–7], and a recent study indicated that inferring the subregion of origin of an unknown European may be feasible from autosomal genetic data [8]. However, direct ancestry inference based on human genetic markers is currently far from perfect, initiating the question whether microbial DNA may be used to supplement human DNA markers in reliable ancestry reconstruction of unknown persons. Recently, it has been shown that the gastric pathogenic bacteria Helicobacter pylori has intimately coevolved with its human host [9, 10]. Although this example may be of limited direct relevance for forensics, because samples containing H. pylori are usually not found at crime scenes (with the exception of bodies in cases of missing persons), it shows that in principle human geographic signatures are inferable from microbial genomes. The human skin is a complex microbial ecosystem consisting of multiple niches, which can differ drastically from each other [11]. Interactions between skin microbes and the human host, as well as between the microbial occupants, are still poorly understood. The current knowledge on skin microbiota primarily derives from cultivation-based studies [12, 13], although molecular fingerprinting techniques have been employed more recently [14, 15]. If a comparable relationship exists between humans and their skin microbiota, as has been observed for H. pylori, new methods for human geographic origin determination could be developed based on DNA analysis of fingertip microflora, with interesting new applications to molecular analyses of physical fingerprints left at crime scenes.
Material and methods
Volunteers and microbiological procedures
Five Dutch European subjects from Rotterdam and three volunteers from Dhaka, Bangladesh, placed their right index finger on a Colombia III blood agar plate (bioMerieux, Marcy-l’Etoile, France). Fingerprint sampling for Dutch volunteers was performed in Rotterdam whereas that of Bangladeshi volunteers was performed in Dhaka. After washing this finger with hospital soap and air drying, another fingerprint, from the same finger, was placed next to the first. Whereas the first fingerprint was expected to provide a mixture of transient and resident microbiota, the second one was assumed to provide resident microbiota only as soap removes the upper dead-skin layers including their transient microflora. After overnight incubation at 37°C, colonies with different morphologies were isolated, with 20 per fingerprint (two each) from the five Dutch individuals (n = 200 strains). Three weeks later, this procedure was repeated for assessment of bacterial persistence in the Dutch volunteers. Agar plates from Bangladeshi volunteers were shipped by courier to Rotterdam after overnight incubation at 37°C for further analyses. Given the diversity in genotypes encountered in the time-wise resampling experiments using Dutch volunteers (see below), this procedure was not repeated for the volunteers from Bangladesh. Gram staining and the determination of catalase activity were applied for the identification of staphylococci using commercially available test kits (bioMérieux, Marcy-l’Etoile, France). Staphylococcus epidermidis sensu stricto was identified among the isolates by a species-specific polymerase chain reaction assay as described elsewhere [16]. A coagulase test with Slidex™ Staph Plus (bioMérieux, Marcy-l’Etoile, France) was performed to detect Staphylococcus aureus isolates. Staphylococcus spp. were classified with the ID 32 Staph API system (bioMérieux, Marcy-l’Etoile, France). Strains were suspended in 15% glycerol in water and stored at −80°C.
Genotyping bacterial isolates
In order to generate DNA fingerprints for the most relevant bacterial isolates, all staphylococcal isolates were subjected to pulsed-field gel electrophoresis (PFGE). PFGE can be used to electrophoretically separate DNA restriction fragments ranging in length between 50 and 1,000 kb. Bacteria were embedded in low melting agarose. After lysostaphin and proteinase treatment and SmaI macrorestriction [17], PFGE was performed in a 1% InCert agarose (FMC) gel in 0.5× TBE at 14°C, using a constant electric field of 6 V cm−1 with pulse ramping from 0.1 to 30 s at a 60°/−60° angle for 18 h. PFGE patterns were analyzed for levels of similarity with BioNumerics 4.5 (Applied-Maths, Sint Martens Latem, Belgium).
Results and discussion
Developing forensic DNA markers for human host inferences from physical fingerprints relies on the examination of the transient exogenous microflora left behind together with the physical fingerprint. However, any human host inference from fingertip microflora using physical fingerprints would require that the transient microflora is representative of the resident endogenous microflora of the same individual and is not simply reflecting microbial species/strains picked up from the environment. To test this prerequisite, we investigated and compared by means of DNA fingerprinting techniques the transient superficial skin microbiota and the resident endogenous microflora of human fingertips within several individuals. In addition, we collected data at two time points to investigate time-wise microflora persistency within individuals. To this end, we used samples from five Dutch volunteers and gram staining as well as catalase activity testing revealed that all bacterial skin isolates that were selected were Gram-positive cocci. Most isolates were Staphylococcus spp., a small number of colonies represented Micrococci (~5% of total isolates), and the vast majority (97%) of staphylococci were coagulase negative. Healthy skin is known to harbor gram-positive pleomorphic bacilli, including Corynebacterium, Brevibacteria, and Propionibacteria sp. [18], but these were not detected here. After the species identification, all staphylococci were genetically fingerprinted using PFGE. PFGE is the most discriminatory typing method that is currently available for typing of staphylococci [19]. Transient microflora (TM) was collected from physical fingerprints before washing with soap and endogenous microflora (RM) after soap washing (see “Materials and methods” section for details). The TM showed higher numbers of PFGE types than those seen in the RM in three of the five Dutch subjects (Table 1). This can be explained by nonendogenous species picked up from the environment and being transferred. Significant variability between the subjects was seen for both the TM and RM, as has been previously reported for other skin areas [10, 13]. Notably, the RM types encountered after washing were very similar intraindividually but very different interindividually. This suggests that certain types of bacteria are able to efficiently and persistently colonize the skin of a given individual. Within the RM, all Dutch subjects had one or two dominant types, mostly Staphylococcus warneri species. These types were also present within the TM of three of the five subjects, while S. epidermidis was present at various time points in all of the subjects TM. Only one subject (subject 1) showed the major RM type as the major TM type. In several subjects, the dominant RM type did not appear at all in the TM, although there was considerable overlap in nondominant species between the TM and RM in many subjects. In general, PFGE types within the RM and within the TM for the same subject appeared consistent over time. Although the degree of variability changed in the 3-week period for every subject, in some subjects, it changed more drastically than in others. At time point 1, overlapping PFGE types were seen in four of the five subjects in the TM and RM. However, by time point 2, only one subject (subject 4) showed any overlap. In one subject (subject 4), the major RM type did not occur in the TM of the same sampling time point 2 but was present in considerable frequency in the TM of the earlier time point 1. Thus, although we see some resemblance between RM and TM as well as some time-wise consistency, the pattern detected is far from perfect. By viewing our results only qualitatively (ignoring type frequencies), RM signals were detectable from the TM analyses in only 50% of the Dutch experiments (combining individuals and time points), which appears discouraging as basis for developing molecular markers for forensic applications to infer individual host information.
Table 1.
Staphylococcus species and PFGE types from Dutch fingerprints before washing with soap representing elements of the transient (TM) microflora and after washing representing resident fingertip microflora (RM)
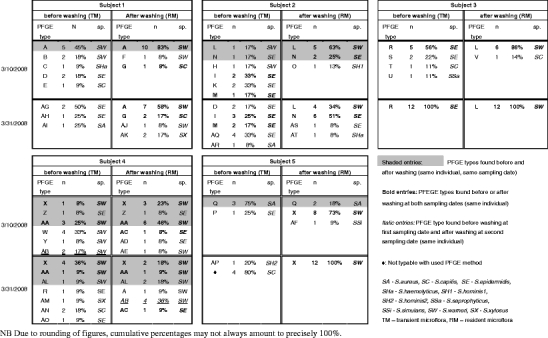
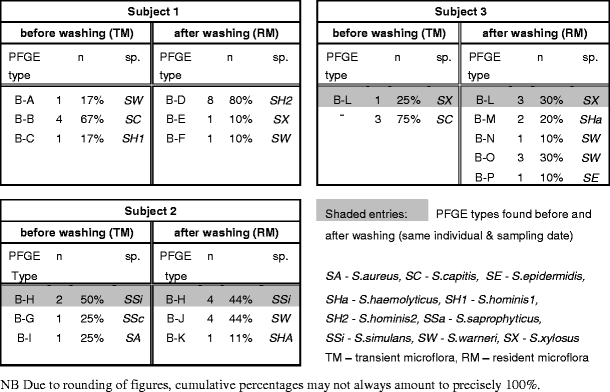
We chose geographic inference as a test example for the type of human host information that would be interesting to retrieve from physical fingerprints by means of microbial DNA analysis for forensic applications. A prerequisite for developing microbial skin markers for inferring human geographic origins would be that the endogenous fingertip microflora of humans from different parts of the world shows differences in their genetic diversity and that retrievable microbial genotypes cluster according to geographic origin of the host individuals. To test this, we obtained fingerprint microflora from native inhabitants of Bangladesh collected in Dhaka, Bangladesh, and compared their microbiological profiles with those of the Dutch volunteers. Fingertip microflora of three Bangladeshi volunteers were isolated and processed in the same manner as for the Dutch volunteers, although only at one time point (three individuals, two fingerprints each and 20 bacterial isolates per fingerprint, hence 3 × 2 × 20 = 120 strains; Table 2). Correspondence between RM and TM was observed in two of the three subjects for one PFGE type, although the persistent PFGE type found differed between the subjects. Many PFGE types were different between RM and TM. Notably, subject 3 from Bangladesh showed an unexpectedly higher variety of staphylococci in the RM compared to TM. Comparing PFGE types between Dutch and Bangladeshi subjects revealed some differences, e.g., Bangladeshi skin microbiota contained less S. warneri and much less S. epidermidis types compared with the Dutch ones. Also, more Micrococcus spp. were found among the TM in Bangladeshi (45%) than among the Dutch (7%). Furthermore, none of the PFGE types from the Bangladesh fingerprints were seen in the Dutch collection, indicating extensive geographic heterogeneity within the Staphylococcus species. However, when each PFGE type obtained during this study was subjected to Dice-based cluster analysis for visualizing geographic structure in the skin staphylococci (see Supplementary material Fig. 1), the Bangladeshi PFGE patterns efficiently mixed with the Dutch ones. Hence, information about geographic origin of the Dutch and Bangladeshi donors was not obvious among genotype data of the staphylococci inhabiting the human fingertip skin. Although in general a limited number of five and three persons tested would not allow for cluster analysis, the full absence of clusters as identified in the present study already shows that individual geographic inferences for forensics purposes is not possible using the approach employed here.
Table 2.
Staphylococcus species and PFGE types from Bangladeshi fingerprints before washing with soap representing elements of the transient (TM) microflora and after washing representing resident fingertip microflora (RM)
To conclude, we see only limited geographic differentiation between microbial DNA fingerprints from Dutch Europeans and Bangladeshi south Asians, indicating that geographic inferences of human hosts from fingertip microbial DNA analysis is not feasible. Furthermore, our results from DNA profiling of transient and resident fingertip microflora show that human fingertip microflora is too dynamic and thus does not fulfill the criteria required for forensic marker developments to infer any human host information from physical fingerprints.
Electronic supplementary material
(DOC 662 kb)
Acknowledgements
We are grateful to all volunteers for participation and thank Kaye Ballantyne for useful comments on the manuscript. The study was supported by the Erasmus University Medical Center Rotterdam and additionally by funds from the Netherlands Forensic Institute.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Baranowski S, Serr A, Thierauf A, Weinmann W, Grosse Perdekamp M, Wurst FM, Halter CC. In vitro study of bacterial degradation of ethyl glucuronide and ethyl sulphate. Int J Legal Med. 2008;122:389–393. doi: 10.1007/s00414-008-0229-3. [DOI] [PubMed] [Google Scholar]
- 2.Budowle B, Murch R, Chakraborty R. Microbial forensics: the next forensic challenge. Int J Legal Med. 2005;119:317–330. doi: 10.1007/s00414-005-0535-y. [DOI] [PubMed] [Google Scholar]
- 3.Enserink M, Ferber D. Microbial forensics. Report spells out how to fight biocrimes. Science. 2003;299:1164–1165. doi: 10.1126/science.299.5610.1164b. [DOI] [PubMed] [Google Scholar]
- 4.Lao O, van Duijn K, Kersbergen P, de Knijff P, Kayser M. Proportioning whole-genome single-nucleotide-polymorphism diversity for the identification of geographic population structure and genetic ancestry. Am J Hum Genet. 2006;78:680–690. doi: 10.1086/501531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips C, Salas A, Sanchez JJ, et al. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci Int Genet. 2007;1:273–280. doi: 10.1016/j.fsigen.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Kosoy R, Nassir R, Tian C, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2008;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Hum Mutat. 2008;29:648–658. doi: 10.1002/humu.20695. [DOI] [PubMed] [Google Scholar]
- 8.Lao O, Lu TT, Nothnagel M, et al. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 9.Linz B, Balloux F, Moodley Y, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moodley Y, Linz B, Yamaoka Y, et al. The peopling of the Pacific from a bacterial perspective. Science. 2009;323:527–530. doi: 10.1126/science.1166083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredricks DN. Microbial ecology of human skin in health and disease. J Investig Dermatol Symp Proc. 2001;6:167–169. doi: 10.1046/j.0022-202x.2001.00039.x. [DOI] [PubMed] [Google Scholar]
- 12.Leyden JJ, McGinley KJ, Nordstrom KM, Webster GF. Skin microflora. J Invest Dermatol. 1987;88:65s–72s. doi: 10.1111/1523-1747.ep12468965. [DOI] [PubMed] [Google Scholar]
- 13.Roth RR, James WD. Microbial ecology of the skin. Annu Rev Microbiol. 1988;42:441–464. doi: 10.1146/annurev.mi.42.100188.002301. [DOI] [PubMed] [Google Scholar]
- 14.Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci U S A. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grice EA, Kong HH, Renaud G, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martineau F, Picard FJ, Roy PH, Ouellette M, Bergeron MG. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J Clin Microbiol. 1996;34:2888–2893. doi: 10.1128/jcm.34.12.2888-2893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linhardt F, Ziebuhr W, Meyer P, Witte W, Hacker J. Pulsed-field gel electrophoresis of genomic restriction fragments as a tool for the epidemiological analysis of Staphylococcus aureus and coagulase-negative staphylococci. FEMS Microbiol Lett. 1992;74:181–185. doi: 10.1111/j.1574-6968.1992.tb05363.x. [DOI] [PubMed] [Google Scholar]
- 18.Chiller K, Selkin BA, Murakawa GJ. Skin microflora and bacterial infections of the skin. J Investig Dermatol Symp Proc. 2001;6:170–174. doi: 10.1046/j.0022-202x.2001.00043.x. [DOI] [PubMed] [Google Scholar]
- 19.Schouls LM, Spalburg EC, van Luit M, Huijsdens XW, Pluister GN, van Santen-Verheuvel MG, van der Heide HG, Grundmann H, Heck ME, de Neeling AJ. Multiple-locus variable number of tandem repeat analysis of Staphylococcus aureus: comparison with pulsed field gel electrophoresis and spa typing. PLoS ONE. 2009;4:e5082. doi: 10.1371/journal.pone.0005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 662 kb)


