Abstract
Homeotic class B genes GLOBOSA (GLO)/PISTILLATA (PI) and DEFICIENS (DEF)/APETALA3 (AP3) are involved in the development of petals and stamens in Arabidopsis. However, functions of these genes in the development of floral organs in torenia are less well known. Here, we demonstrate the unique floral phenotypes of transgenic torenia formed due to the modification of class B genes, TfGLO and TfDEF. TfGLO-overexpressing plants showed purple-stained sepals that accumulated anthocyanins in a manner similar to that of petals. TfGLO-suppressed plants showed serrated petals and TfDEF-suppressed plants showed partially decolorized petals. In TfGLO-overexpressing plants, cell shapes on the surfaces of sepals were altered to petal-like cell shapes. Furthermore, TfGLO- and TfDEF-suppressed plants partially had sepal-like cells on the surfaces of their petals. We isolated putative class B gene-regulated genes and examined their expression in transgenic plants. Three xyloglucan endo-1,4-beta-d-glucanase genes were up-regulated in TfGLO- and TfDEF-overexpressing plants and down-regulated in TfGLO- and TfDEF-suppressed plants. In addition, 10 anthocyanin biosynthesis-related genes, including anthocyanin synthase and chalcone isomerase, were up-regulated in TfGLO-overexpressing plants and down-regulated in TfGLO-suppressed plants. The expression patterns of these 10 genes in TfDEF transgenic plants were diverse and classified into several groups. HPLC analysis indicated that sepals of TfGLO-overexpressing plants accumulate the same type of anthocyanins and flavones as wild-type plants. The difference in phenotypes and expression patterns of the 10 anthocyanin biosynthesis-related genes between TfGLO and TfDEF transgenic plants indicated that TfGLO and TfDEF have partial functional divergence, while they basically work synergistically in torenia.
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-010-0574-z) contains supplementary material, which is available to authorized users.
Keywords: Torenia, TfGLO, TfDEF, Class B gene, Anthocyanin
Introduction
Agricultural biotechnology supports human life with numerous transgenic crops that could not be generated by classical breeding. Similarly, until now classical breeding could not generate certain varieties of ornamental flowers. For example, blue carnations (Fukui et al. 2003) and roses (Katsumoto et al. 2007), considered impossible to generate, have been produced by genetic engineering. We performed ion beam irradiation of genetically modified torenia (Sasaki et al. 2008) and screened transgenic torenia modified by various transcriptional factors of Arabidopsis thaliana (Mitsuda et al. 2008) to produce and provide unique flowers to the public. Although genetic engineering can be used to produce various floral traits, desired phenotypes are not always obtained. In ornamental flowers, information regarding genomic sequences and other factors necessary for successful molecular engineering is particularly limited. We used torenia to develop an efficient way to produce unique flowers by genetic engineering because it is a useful ornamental flowering plant characterized by simplicity, small genome size (2n = 18, 171 Mbp) (Kikuchi et al. 2006), and adaptability to tissue culture and transformation (Aida et al. 2000a). However, little information is available on floral development and flower formation, including floral organ identity genes in torenia.
During floral development in A. thaliana, identities of four different organ types are determined by combining three classes of floral organ identity genes known as MADS-box transcription factor genes. Functions of these genes have been summarized in the ABC model of floral development. A-, B-, and C-function genes control floral organ identities (Coen and Meyerowitz 1991; for review Ma 1994). Whorl 1 sepals are specified by A-function genes and whorl 2 petals by A- and B-function genes. Whorl 3 stamens are specified by B- and C-function genes and whorl 4 carpels by C-function genes. Class B genes comprise DEFICIENS (DEF)/APETALA3 (AP3) and GLOBOSA (GLO)/PISTILLATA (PI), and their functions are basically conserved among eudicots (Whipple et al. 2004; for review Soltis et al. 2007 and Becker and Theissen 2003). In A. thaliana, ap3 and pi mutations caused abnormal differentiation of whorls 2 and 3, resulting in petals and stamens being converted to sepals and carpels, respectively (Bowman et al. 1989; Bowman et al. 1991).
DEF/AP3 and GLO/PI proteins interact directly to form heterodimers (Goto and Meyerowitz 1994; Riechmann et al. 1996a) and thereafter activate target gene expression by binding to their promoters (Theissen and Saedler 2001). The sequence generally bound by MADS-box proteins is CC(A/T)6GG known as a CArG motif (for review Riechmann and Meyerowitz 1997). The heterodimeric DEF/AP3 and GLO/PI proteins also bind to this motif (Riechmann et al. 1996a). In A. thaliana and Antirrhinum majus, downstream genes regulated by class B genes have been identified by microarray analysis using ap3, pi, and def mutants (Bey et al. 2004; Zik and Irish 2003; Wellmer et al. 2004). Furthermore, in A. thaliana and A. majus, a pair of class B genes influenced the development of cells, such that they converted to petals and stamens (for review Rijpkema et al. 2007). On the other hand, duplicated class B genes have been identified in Petunia hybrida and tomato (Vandenbussche et al. 2004; de Martino et al. 2006). Furthermore, several higher plant species have duplicated class B genes in their genomes (Kramer et al. 1998; for review Soltis et al. 2007).
In higher plants, a large number of genes are duplicated and exist redundantly (for review Moore and Purugganan 2005), including transcription factors (Riechmann et al. 2000; Mitsuda and Ohme-Takagi 2009). This redundancy sometimes makes it difficult to analyze their functions by single-gene knockout mutations or RNAi because the multiplied gene(s) compensates for the function of the gene that has been targeted for knockout or knockdown (for review Shikata and Ohme-Takagi 2008). To solve this problem, a strong gene-silencing system specific to transcription factors called chimeric repressor gene-silencing technology (CRES-T) has been proven to be a useful tool. The chimeric repressor, in which transcription factors are fused to the 12-amino acid repression domain sequence called SRDX, dominantly suppresses the activity of target transcription factors to prevent expression of downstream genes, even if there are endogenous and functionally redundant transcription factors (Hiratsu et al. 2003; for review Shikata and Ohme-Takagi 2008; Mitsuda and Ohme-Takagi 2009). Therefore, the chimeric repressor produces phenotypes that are generally observed only when redundant transcription factors are mutated simultaneously. The transgenic phenotype generated by a SRDX-fused construct of a class C gene, AGAMOUS (AG), was consistent with the ag mutant phenotype (Hiratsu et al. 2003). Chimeric repressors of many transcription factors, such as Atmyb23, NAC secondary wall thickening promoting factor 1 (NST1), NST2, and TCP3, have been reported to suppress target gene expression (Matsui et al. 2004; Mitsuda et al. 2005; Koyama et al. 2007). Although the target gene repression system has been developed in A. thaliana, SRDX functions in many other plant species such as rice (Mitsuda et al. 2006), tomato (Itkin et al. 2009), and torenia (Narumi et al. 2008).
In this study, we focused on torenia class B genes and demonstrated profiles of their functions because we were interested in understanding the biotechnological controls of floral traits. To examine the functions of the genes in torenia, we generated four types of transgenic plants, TfGLO- and TfDEF-overexpressing and TfGLO- and TfDEF-suppressed plants. TfGLO-overexpressing plants showed ectopic accumulation of anthocyanin and petal-like cells on the surfaces of sepals, while TfDEF-overexpressing plants showed no phenotypic change. TfGLO-suppressed plants showed distinctive serrations on petal margins and TfDEF-suppressed plants showed partially decolorized petals. Furthermore, both TfGLO- and TfDEF-suppressed plants partially had sepal-like cells on the surfaces of petals. These results indicated that TfGLO and TfDEF play important roles in floral organ development. Based on the microarray data of A. thaliana (Zik and Irish 2003), we isolated several putative downstream genes regulated by TfGLO or TfDEF. Expression profiles of isolated downstream genes in TfGLO and TfDEF transgenic plants were similar. In addition, we isolated 10 anthocyanin biosynthesis-related genes and investigated their expression in TfGLO and TfDEF transgenic plants. We found that these two genes differentially regulate expression of anthocyanin biosynthesis-related genes. Sepals of TfGLO-overexpressing plants showed accumulation of the same type of anthocyanin as petals of wild-type plants (Aida et al. 2000b), thus suggesting that the sepals were partially converted to petals. While TfGLO and TfDEF cooperatively function in floral development, functional divergence in floral phenotypes and downstream gene regulation between the two class B genes were observed in the transgenic torenia and will be discussed.
Materials and methods
Plant materials
Torenia fournieri Lind. (Crown Violet) was grown at 25°C in an air-conditioned greenhouse. Plant materials were maintained in a plant box supplemented with 1/2 Murashige and Skoog medium containing 0.32% gellan gum. These materials were reproduced vegetatively by herbaceous cutting at 25°C under fluorescent light (16L/8D, 85 μmol m−2 s−1) following the procedure described by Aida and Shibata (2001).
Phylogenetic analysis
The sequences of class B genes were obtained from GenBank (see Supplementary Tables S1 for accession numbers). Full length of each amino acid sequence for class B genes was used for phylogenetic analysis. Protein sequences were aligned using GENETYX ver.8.0.0 (GENETYX CORPORATION, Tokyo, Japan) and refined little by hand, taking both nucleotide and amino acid sequences around MADS and K domains, PI motif, PI motif-derived sequence, and the euAP3 motif (Kramer et al. 1998; Yang et al. 2003a; Yang and Jack 2004) into consideration. After these sequences were aligned, they were used for phylogenetic analysis with neighbor-joining method in GENETYX ver.8.0.0. The statistical significance was tested by bootstrap analysis for 10,000 replicates.
Cloning of cDNA from torenia
Genes described below were isolated from T. fournieri Lind. (Crown Violet). Total RNA was prepared from torenia petals using TRIzol (Invitrogen, Tokyo, Japan). cDNA was synthesized from total RNA using a cDNA synthesis kit (Toyobo, Osaka, Japan). It was then used for gene cloning followed by RT-PCR.
We isolated the following putative downstream genes of torenia class B genes: endo-xyloglucan transferase (TfXEG1, TfXEG2, and TfXEG3), Lys-ketoglutarate reductase (TfLKR) and chlorophyll a/b-binding protein (TfCAB1, TfCAB2, and TfCAB3). Primers (Supplementary Table S2) were designed based on the sequences of AtXEG (At4g30270), AtLKR (At4g33150), and AtCAB (At1g29930). The putative downstream genes were isolated using a two-step PCR approach with primer sets for degenerate PCR and 3′RACE PCR. Primers for degenerate PCR were designed based on the highly conserved regions of each gene and those for 3′RACE PCR were based on the sequences that were isolated by degenerate PCR. We isolated the following anthocyanin biosynthesis-related genes: anthocyanin synthase (TfANS), chalcone isomerase (TfCHI), chalcone synthase (TfCHS1 and TfCHS2), dihydroflavonol 4-reductase (TfDFR), flavanone hydroxylase (TfF3H), flavonoid 3′-hydroxylase (TfF3′H1 and TfF3′H2), flavonoid 3′,5′-hydroxylase (TfF3′5′H), and UDP glucose:flavonoid 3-O-glucosyltransferase (Tf3GT). Primer sequences (Supplementary Table S2) were designed based on TfANS (AB044091), A. majus CHI (AmCHI; M68326), TfCHS (AB012923), TfDFR (AB012924), TfF3H (AB211958), TfF3′H (AB057672), TfF3′5′H (AB012925), and Perilla frutescens 3GT (Pf3GT; AB002818), which except the sequences for AmCHI and Pf3GT have been isolated from related torenia cultivars. These 10 anthocyanin biosynthesis-related genes were isolated using a two-step PCR approach, similar to that described above for putative downstream genes. Among these 10 isolated genes, the sequence of isolated TfF3′H1 was identical to that of the existing clone (AB057672). In this study, we have termed this clone (AB057672) TfF3′H1. The nucleotide sequences of the cloned cDNA, except for TfF3′H1, have been registered in the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/Welcome-j.html), and their accession numbers are described in Supplementary Table S3.
Production of recombinant proteins in Escherichia coli
Open reading frames of TfGLO (AB548150) and TfDEF (AB359951) were amplified using PCR with primer sets of Torenia GLO1 EcoIF and Torenia GLO1 His XhoR and Torenia DEF1 EcoIF and Torenia DEF1 His XhoR, respectively (Supplementary Table S4). These primers attach EcoRI or XhoI sites to the 5′- or 3′-ends, respectively. The amplified DNA fragments were cloned into the corresponding sites of the pET32a vector (Novagen, Madison, WI, USA) to obtain pET32a-TfGLO and pET32a-TfDEF plasmids, which enable production of TfGLO and TfDEF with a thioredoxin (Trx) tag at the N terminus and a His6 tag at the N and C termini.
The pET32a-TfGLO and pET32a-TfDEF plasmids were used to transform competent cells of Escherichia coli strain Origami™ B (DE3) (Novagen). Induction of recombinant proteins was performed as follows: 0.1 mM isopropyl β-d-thiogalactopyranoside at 20°C for 12 h. Purification of His6-tagged proteins using a Ni Sepharose™ 6 fast flow column (GE Healthcare Bio-Sciences, Piscataway, NJ, USA) was performed according to the manufacturer’s instructions. Purified proteins were desalted on a Zeba™ desalt spin column (Thermo Scientific, Waltham, MA, USA) equilibrated with 20 mM Tris–HCl (pH 7.5) containing 10% glycerol. Qualities of the purified proteins were confirmed by SDS-PAGE and immunoblot analysis.
Production of recombinant proteins by in vitro translation
The coding regions of TfGLO and TfDEF were amplified using primer sets of Torenia GLO1 1F and GLO Flag R and Torenia DEF1 1F and DEF Flag R, respectively (Supplementary Table S4). Amplified fragments were attached with adenine using 10× A-attachment mix (Toyobo) in TA cloning. These fragments were cloned into pT7Blue T-Vectors (Novagen) to obtain pT7Blue-TfGLO and pT7Blue-TfDEF plasmids, which permit production of TfGLO and TfDEF with FLAG tags at the C terminus.
For in vitro translation experiments, proteins were synthesized using the TNT T7 Quick for PCR DNA kit (Promega, Tokyo, Japan). TfGLO and TfDEF fragments were amplified using the primer sets of GLO TnT F and M13 new R with pT7Blue-TfGLO as a template and of DEF TnT F and M13 new R with pT7Blue-TfDEF as a template, respectively (Supplementary Table S4). Qualities of synthesized proteins were confirmed by immunoblot analysis, and 1.5-μl protein solutions were used in this experiment.
Immunoprecipitation and immunoblot analyses of recombinant class B proteins
Immunoprecipitation analyses were performed with according to the procedures described by Riechmann et al. (1996a) and Goto and Meyerowitz (1994), with some modifications. The in vitro translation solution was diluted with an equal volume of 2× buffer A. A mixture of 20-μl diluted protein solution and 2-μg Trx fusion protein was incubated at 4°C for 60 min to enable protein association. Then, 400 μl of ice-cold buffer B and 0.5 μl of Trx Tag™ monoclonal antibody (Novagen) were added, and the samples were incubated at 4°C for 60 min. nProtein A sepharose™ 4 fast flow (GE Healthcare) (100 μl equilibrated in the buffer B) was then added and incubation was continued for 60 min. Sepharose beads were collected by centrifugation (500 rpm for 5 min) and was washed five times with RIPA buffer [50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100, 0.2% SDS, 4 mg/ml BSA], once with LiCl-urea buffer [10 mM Tris–HCl (pH 8.0), 400 mM LiCl], and once with water.
Immunoprecipitated proteins were analyzed by SDS-PAGE on 12–14% gels and transferred to BioTrace™ PVDF membranes (Pall Corporation, Port Washington, NY, USA). After blocking with skimmed milk, membranes were probed with anti-FLAG M2 monoclonal antibody (Sigma-Aldrich, St Louis, MO, USA). After washing, membranes were incubated with alkaline phosphatase-labeled secondary antibody. Antigen–antibody complexes were detected by hydrolysis of nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate.
Generation of transgenic torenia with modified class B genes
The coding region of TfGLO was amplified using primer sets of Torenia GLO1 1F and Torenia GLO1 over R primers for TfGLO overexpression and Torenia GLO1 1F and Torenia GLO1 SRDX R primers for TfGLO suppression, respectively (Supplementary Table S4). Amplified fragments were cloned into SmaI sites of p35SSG and p35SSRDXG vectors (Mitsuda et al. 2005) to produce p35S::TfGLO and p35S::TfGLO-SRDX plasmids, respectively. After sequence confirmation, the region corresponding to each transgene was transferred to the pBCKH plant binary vector using the Gateway system (Invitrogen). For generation of transgenic plants containing 35S::TfDEF and 35S::TfDEF-SRDX, the coding region of TfDEF was amplified using primer sets of Torenia DEF1 1F and Torenia DEF1 over R primers for TfDEF overexpression and Torenia DEF1 1F and Torenia DEF1 SRDX R primers for TfDEF suppression, respectively (Supplementary Table S4). The p35S::TfDEF and p35S::TfDEF-SRDX plasmids were constructed in an identical manner as described above for TfGLO plasmid, and each transgene was transferred to the pBCKH plant binary vector.
Binary vectors were introduced in Agrobacterium tumefaciens strain EHA105 by electroporation. After infection of torenia leaf discs by Agrobacterium, the transgenic torenia were screened and regenerated following the procedure described by Aida et al. (2000a).
Expression analysis by RT-PCR
Total RNA was prepared by TRIzol (Invitrogen). The cDNA was synthesized from total RNA using a cDNA synthesis kit (Toyobo) and used to amplify the transcripts. The RT-PCR reaction was performed using KOD Plus 2 (Toyobo). The sequence of each specific primer is described in Supplementary Table S3. The quality and quantity of each cDNA sample were checked using the TfACT3 gene (AB330989) as an internal control. Three independent transgenic plants were used for expression analysis.
Scanning electron microscopy
Fresh tissues were prepared from greenhouse-grown plants and immediately observed under a scanning electron microscopy (SEM; VE-7000) (Keyence Co., Osaka, Japan) without fixing.
Extraction and quantification of anthocyanins and flavones
Anthocyanins and flavones were extracted and quantified using the procedure described by Aida et al. (2000b), with some modifications. Fresh torenia petals (approximately 0.1 g fresh weight for each sample) were incubated for 20 h with 2–3 ml 10% (v/v) acetic acid to extract anthocyanins and flavones. The anthocyanin and flavone samples were analyzed by HPLC using a LiChrospher 100 RP-18 column (4 × 250 mm, Kanto Chemical, Tokyo). Eluted components were monitored at 530 nm for anthocyanins and at 340 nm for flavones. Relative quantities of anthocyanins and flavones in the samples were determined from the peak areas of the chromatogram.
Results
Phylogenetic analysis of torenia class B genes
To examine the function of torenia class B genes, TfGLO and TFDEF in planta, we first investigated whether TfGLO and TfDEF proteins contain characteristic domains specific to individual GLO/PI or DEF/AP3 proteins. These two class B proteins contained both MADS and K domains (Fig. S1), which are usually conserved in MADS proteins (Yang et al. 2003a; Yang and Jack 2004). Furthermore, the PI motif was conserved on the C terminus of TfGLO, and the PI motif-derived sequence and the euAP3 motif were conserved on the C terminus of TfDEF (Fig. S1; Kramer et al. 1998). Next, we examined whether the two class B genes could actually be classified into GLO/PI or DEF/AP3 by phylogenetic analysis. Many class B genes have been found in the plant kingdom, and they have been classified into GLO/PI or DEF/AP3 according to their amino acid sequences and functions in flowering (Vandenbussche et al. 2004; for review Soltis et al. 2007). DEF/AP3, in particular, has been further classified into euAP3, TM6, and paleoAP3 lineages (Kramer et al. 1998). The euAP3 proteins in core eudicots contain PI-derived and euAP3 motifs including TfDEF (Fig. S1), while TM6 proteins in core eudicots and paleoAP3 proteins in basal eudicots, magnoliids, and monocots contain distinct PI-derived and paleoAP3 motifs (Kramer et al. 1998; Vandenbussche et al. 2003). Phylogenetic analysis further confirmed that TfGLO and TfDEF were classified as expected into GLO/PI and euAP3 lineage of DEF/AP3, respectively (Fig. 1). TfGLO is homologous with A. majus GLO (AmGLO; 79.6%) and P. hybrida GLO (PhGLO2; 63.0%) and TfDEF is similar to AmDEF (79.9%) and PhDEF (74.8%). Based on these results, we confirmed the genes as torenia class B genes and analyzed their functions.
Fig. 1.
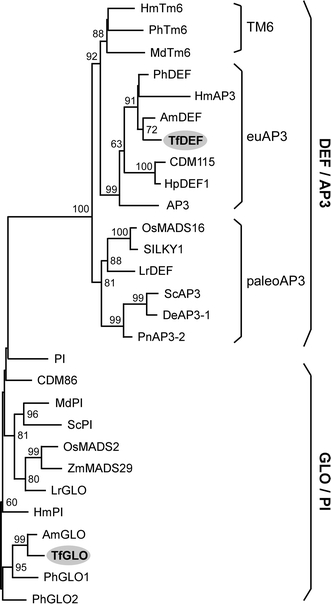
Phylogenetic relationship among class B genes. The neighbor-joining tree was generated based on amino acid sequences with coding regions of class B genes, and 10,000 bootstrap samples were generated to assess support for the relationships. TfDEF and TfGLO were classified into euAP3 and GLO/PI families, respectively. Origins and accession numbers of each protein used here are described in Supplementary Table S1. Local bootstrap probabilities (in percentages) are indicated near the branching points, and values below 50 are omitted
Interaction between TfGLO and TfDEF
In A. thaliana, GLO/PI and DEF/AP3 have been reported to interact directly with each other in vitro (Goto and Meyerowitz 1994; Riechmann et al. 1996a) via the K domain (Yang et al. 2003a). Therefore, to confirm their characteristic heterodimerization activity, we examined whether TfGLO and TfDEF also interact with each other. We produced the recombinant proteins Trx-TfGLO, TfGLO-FLAG, Trx-DEF, and TfDEF-FLAG for this analysis. Trx-TfGLO and Trx-TfDEF proteins were synthesized in bacteria, and the purified proteins were detected as a single band by SDS-PAGE analysis (Fig. 2A). The Trx antibody recognized a protein of approximately 49 kDa that corresponded to the predicted molecular masses of Trx-GLO and Trx-DEF (45.6 and 46.0 kDa, respectively, Fig. 2A), indicating the quality of synthesized proteins and specific recognition by the antibody. TfGLO-FLAG and TfDEF-FLAG proteins were produced as C-terminal FLAG-fusion proteins by in vitro transcription and translation. The FLAG antibody recognized a protein that corresponded to the predicted molecular masses of TfGLO-FLAG and TfDEF-FLAG (27.9 and 28.2 kDa, respectively, Fig. 2B), thus indicating the quality of synthesized proteins and specific recognition by the FLAG antibody. After mixing TfGLO and TfDEF fusion proteins, anti-Trx antibody was added for precipitation, and the resulting immunoprecipitation was detected by immunoblot analyses using anti-FLAG antibody (Fig. 2C). The Trx-TfGLO protein co-precipitated with the TfDEF-FLAG protein and the Trx-TfDEF protein also co-precipitated with the TfGLO-FLAG protein, indicating that the two proteins interact with each other in vitro. These results indicated that TfGLO and TfDEF retain the activity necessary to form heterodimers as reported in A. thaliana (Goto and Meyerowitz 1994; Riechmann et al. 1996a).
Fig. 2.
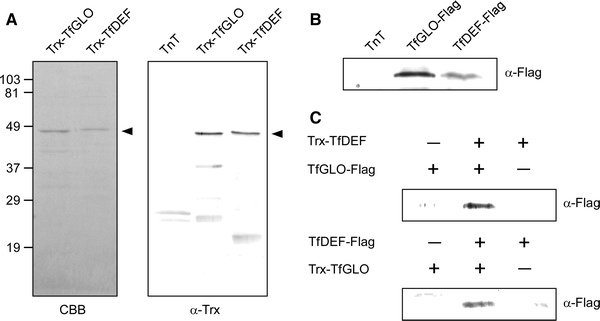
Co-immunoprecipitation of TfGLO and TfDEF proteins. A Recombinant proteins of Trx-TfGLO and Trx-TfDEF were produced in E. coli and purified by nickel affinity chromatography. Purified proteins (0.2 μg) were separated on SDS-PAGE gel and stained with Coomassie Brilliant Blue (left; CBB). The positions of molecular mass markers are indicated on the left. The mass unit is kDa. Production of Trx fusion proteins was confirmed by immunoblot analysis using anti-Trx antibody (right; α-Trx). B TfGLO- and TfDEF-FLAG was produced by in vitro transcription/translation using rabbit reticulocyte lysate (TnT). Production of FLAG-fusion proteins was confirmed by immunoblot analysis using anti-FLAG antibody (α-FLAG). C Co-immunoprecipitation of TfGLO and TfDEF. Protein mixtures were precipitated with α-Trx, and the precipitated proteins were detected by α-FLAG
Unique phenotypes of floral organs in transgenic plants with class B transgenes
To understand the biological functions of TfGLO and TfDEF, we generated four types of transgenic plants, TfGLO- and TfDEF-overexpressing and TfGLO- and TfDEF-suppressed plants. The CaMV 35S promoter (35S) was used to overexpress TfGLO and TfDEF (Supplementary Fig. S2). To suppress the function of endogenous TfGLO and TfDEF, we used an artificial dominant repression domain of 12 amino acids (SRDX; Hiratsu et al. 2003) because Southern gel blot analysis suggested that TfDEF exists as at least two copies while TfGLO exists as one copy in torenia (data not shown). When genes of interest fuse to SRDX, transcription activators are converted to dominant repressors, and they suppress expression of target genes overcoming the activity of endogenous and functionally redundant transcription factors (Hiratsu et al. 2003). The transgenic phenotypes generated by this technology resemble those generated by loss of function of transcriptional activators, even if there are endogenous and functionally redundant transcription factors (Hiratsu et al. 2003; Mitsuda and Ohme-Takagi 2009). SRDX was fused to the C terminus of TfGLO and TfDEF to express their chimeric repressors. In this study, expression of TfGLO, TfGLO-SRDX, TfDEF, and TfDEF-SRDX transgenes was confirmed by RT-PCR analysis (Supplementary Fig. S2).
While no phenotypic change was observed in petals of 35S::TfGLO plants (Fig. 3A-c and A-d), ectopic accumulation of anthocyanin was observed in sepals (Fig. 3A-e and A-f). Anthocyanin accumulation coincided with transgene expression at 81.8% (Supplementary Table S5). In 35S::TfGLO-SRDX plants, distinctive serrations on the petal margin (Fig. 3A; red arrowhead) were primarily observed, and they coincided with transgene expression at 75.0% (Supplementary Table S5), while no phenotypic alteration was observed in sepals (Fig. 3B-i and B-j). In 35S::TfDEF plants, petals and sepals showed no phenotypic change (Fig. 3B-c, B-d, B-e, and B-f). However, in 35S::TfDEF-SRDX plants, partially decolorized petals were primarily observed (Fig. 3B-g and B-h; red arrowhead), and they coincided with transgene expression at 68.4% (Supplementary Table S5), while no clear phenotypic alteration was observed in sepals (Fig. 3B-i and B-j). Both TfGLO- and TfDEF-SRDX transgenic plants exhibited no phenotypic change in stamens including the number of stamens per flower and shape of anthers. Although the length of filaments seemed to shorten, this would be because the whole flower size of TfGLO- and TfDEF-SRDX plants shrunk (data not shown).
Fig. 3.
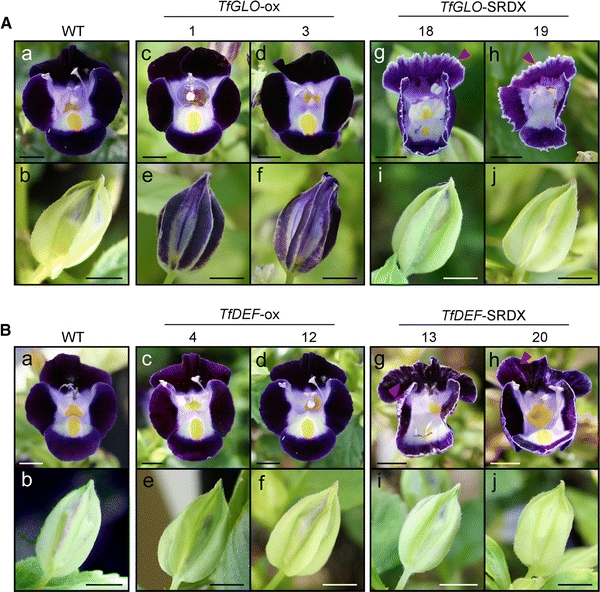
Phenotypes of transgenic torenia with modified torenia class B genes. A Photographs of TfGLO transgenic torenia. Photographs of the flower (a) and flower bud (b) in wild-type plants are shown. Photographs of flowers (c and d) and flower buds (e and f) in two independent TfGLO-overexpressing plants and flowers (g and h) and flower buds (i and j) in two independent TfGLO-SRDX plants, respectively. B Photographs of TfDEF transgenic torenia plants. Photographs of the flower (a) and flower bud (b) in wild-type plants. Photographs of flowers (c and d) and flower buds (e and f) in two independent TfDEF-overexpressing plants and flowers (g and h) and flower buds (i and j) in two independent TfDEF-SRDX plants are shown. Scale bar 5 mm
From the viewpoint of petal pigmentation, sepals of TfGLO-overexpressing plants, but not of TfDEF-overexpressing plants, were suggested to be partially converted to petals. Furthermore, TfGLO- and TfDEF-suppressed plants showed altered petal shapes compared to wild-type plants; however, phenotypes of both plants were distinct from each other. Next, we promoted TfGLO and TfDEF research focused on sepals and petals using transgenic torenia.
Scanning electron microscopy analysis of floral phenotypes in transgenic plants with class B transgenes
To further confirm whether sepals of TfGLO-overexpressing plants would partially convert to petals, we performed SEM analysis using transgenic plants. Because the tip of flower buds in TfGLO-overexpressing plants sometimes showed petal-like morphological changes (Fig 4A-a; black arrowhead), we first observed cell shapes at this position on both adaxial and abaxial surfaces. Cells at the tip of sepals of TfGLO-overexpressing plants were corn-shaped on both surfaces (Fig. 4B-a and B-b). The shapes of surface cells of sepals of TfGLO-overexpressing plants were similar to those of petals (Fig. 4B-c and B-d; observed position was blue arrowhead in Fig. 4A-b) but not to those of the tip of sepals (Fig. 4B-e and B-f; observed position was blue arrowhead in Fig. 4A-c) in wild-type plants. In the middle region of sepals of TfGLO-overexpressing plants (Fig. 4A-a; red arrowhead), cells on the adaxial surface contained both puzzle-shaped cells (Fig. 4C-a; black arrowhead) similar to those of wild-type sepals and cells like in the process of change to corn-shaped cells (Fig. 4C-a; red arrowhead). The upper middle regions of sepals of TfGLO-overexpressing plants mostly contained corn-shaped cells similar to those of petals and were not observed to have sepal-like, puzzle-shaped cells on the adaxial surface (Fig. 4C-b) as observed on the adaxial surface of sepals of wild-type plants (Fig. 4D). Although wing parts of TfGLO-overexpressing plants accumulated a large amount of anthocyanins, cells on the abaxial surface showed puzzle-shaped cells like sepals of wild-type plants (Fig. S3). Sepals of TfDEF-overexpressing plants also showed puzzle-shaped cells on the adaxial surface similar to those of sepals of wild-type plants (Fig. S4). These results further suggest that sepals of TfGLO-overexpressing plants were partially converted to petals.
Fig. 4.
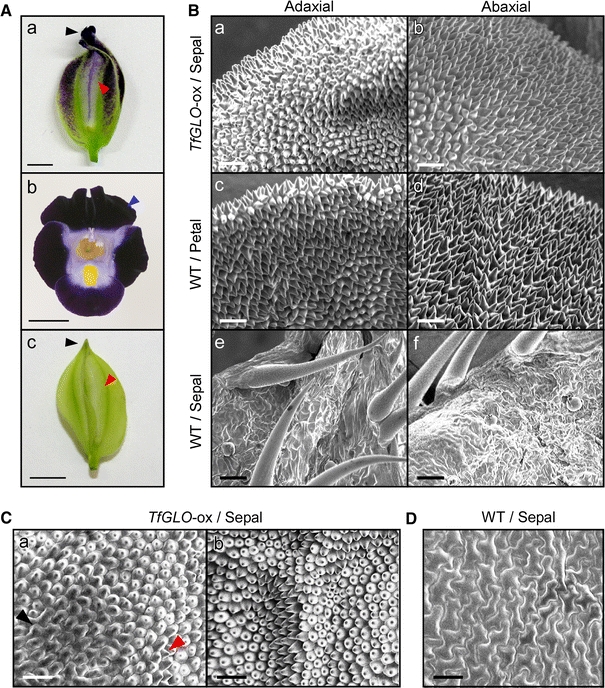
Overexpression of TfGLO gene in torenia. A A flower bud of 35S::TfGLO transgenic plants (TfGLO-ox; a). A flower (b) and flower bud (c) of wild-type plants. Scale bar 5 mm. B First and second whorls of wild-type plants and second whorls of TfGLO-ox plants viewed by scanning electron microscopy. Adaxial (a) and abaxial (b) surfaces of the tip of a flower bud with 35S::TfGLO indicated by black arrowhead in Fig. 4A–a. Adaxial (c) and abaxial (d) surfaces of petals of wild-type indicated by blue arrowhead in Fig. 4A–b. Adaxial (e) and abaxial (f) surfaces of the tip of a wild-type flower bud indicated by black arrowhead in Fig. 4A–c. Scale bar 50 μm. C The adaxial surface of sepals of TfGLO-ox plants indicated by red arrowhead in Fig. 4A–a. Middle parts of sepals of TfGLO-ox plants (a) partially contained sepal-type cells (black arrowhead), but cells of upper parts (b) were mostly altered to petal-like cells. Scale bar 50 μm. D The adaxial surface of sepals of wild-type plants indicated by red arrowheads in Fig. 4A–c. Scale bar 50 μm
We next observed petal cells in TfGLO- and TfDEF-suppressed plants to confirm whether the cell shapes would change or not. Petal cells in wild-type plants showed corn-shaped cells both on abaxial and adaxial surfaces (Fig. 5a, b). While petals of TfGLO-SRDX plants showed distinctive serrated regions, cells on both adaxial and abaxial surfaces were mainly rounded at the tip and changed to short, corn-shaped cells (Fig. 5c, d; blue arrowhead) when compared to wild-type petal cells. Furthermore, cells on the adaxial surface sometimes contained smooth cells (Fig. 5c; red arrowhead) and those on the abaxial surface clearly showed sepal-like cells (Fig. 5d; red arrowhead). Petals of TfGLO-SRDX plants in the middle region, outside of the serrated region, showed rounded cells at the tip that changed to short corn-shaped cells on both the adaxial and abaxial surfaces (Fig. 5e, f; blue arrowhead). In TfDEF-SRDX plants, petals of the decolorized regions contained some sepal-like cells that were arranged in tandem on both abaxial and adaxial surfaces (Fig. 5e, f; red arrowhead). Furthermore, other petal cells (Fig. 5e, f) including those in the middle region, which were outside the decolorized regions (Fig. 5g, h), showed almost the same cell shapes as those observed in wild-type petals on both surfaces. These results suggest that suppression of TfGLO and TfDEF functions causes partial conversion of petal cells to sepal cells.
Fig. 5.
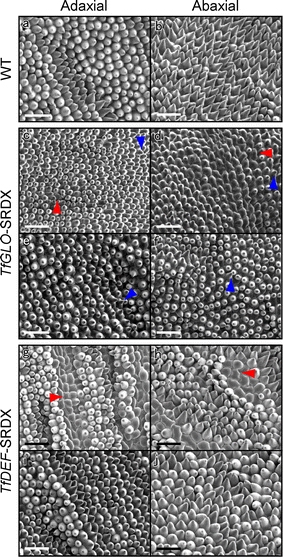
Second whorls of wild-type, TfGLO- and TfDEF-suppressed plants viewed by scanning electron microscopy. Adaxial (a) and abaxial (b) surfaces of petals of wild-type torenia. Adaxial (c and e) and abaxial (d and f) surfaces of petals of TfGLO-SRDX plants. Petals of the distinctive serrated region (c and d) and the middle region (e and f) outside the serrated region in TfGLO-SRDX plants. Adaxial (g and i) and abaxial (h and j) surfaces of petals of TfDEF-SRDX plants. Petals of partial decolorized region (g and h) and the middle region (i and j) outside the decolorized region in TfDEF-SRDX plants. Blue arrowheads indicate petal cells that have lost the characteristics of petal cells, i.e., without corn-shaped cells. Red arrowheads indicate petal cells that showed sepal-like cells. Scale bar 50 μm
Expression analyses of putative downstream genes of class B genes in transgenic plants
In A. thaliana, expression of downstream genes was analyzed by microarray analysis in ap3 or pi mutants (Zik and Irish 2003). To understand functions of TfGLO and TfDEF in the regulation of downstream gene expression, we isolated some putative class B gene-regulated genes from torenia based on the results of microarray analysis in A. thaliana (Zik and Irish 2003), and then examined their expression using transgenic plants. We attempted to isolate several types of downstream genes of class B genes in torenia, and were able to isolate the putative ortholog genes of AtXEG, AtLKR, and AtCAB, which were down-regulated in ap3 or pi mutants and up-regulated in AP3/PI overexpressing plants (Zik and Irish 2003). The isolated genes from torenia encoded the following seven candidate genes: TfXEG1, TfXEG2, TfXEG3, TfLKR, TfCAB1, TfCAB2, and TfCAB3. First, expression of TfGLO, TfDEF, and the above putative class B gene-regulated genes were examined using 10 different organs of wild-type plants (Fig. 6). TfGLO transcript was detected in immature petals, mature petals, and stamens, while TfDEF transcript was unexpectedly detected in sepals and carpels in addition to petals and stamens. The expression pattern of three TfXEG genes was similar in petals, but their transcripts were detected differently in other organs. TfLKR transcript was detected in immature petals, mature petals, and stamens. The expression pattern of three TfCAB genes was similar, and their transcripts were detected strongly in vegetative organs, except roots and carpels.
Fig. 6.
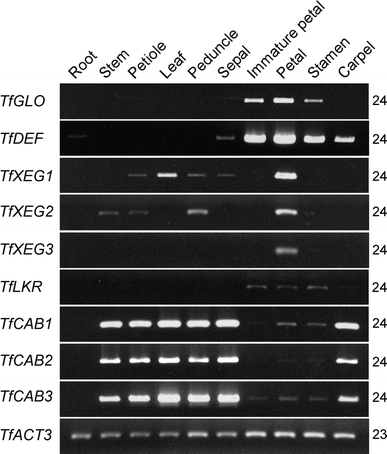
Organ-specific expression of class B genes and putative downstream genes of class B genes. Ten torenia organs—root, stem, petiole, leaf, peduncle, sepal, immature petal, petal, stamen, and carpel—were used for expression analysis of each gene. The torenia actin gene TfACT3 was used as an internal control. PCR cycles are indicated on the right side of each column
Next, we examined expression of these genes in petals and sepals of transgenic torenia by RT-PCR. Endogenous TfGLO and TfDEF were up-regulated in 35S::TfGLO and down-regulated in 35S::TfGLO-SRDX transgenic plants (Fig. 7A). Furthermore, TfXEG1, 2, and 3 were also up-regulated in 35S::TfGLO and down-regulated in 35S::TfGLO-SRDX plants as expected from previous reports on A. thaliana (Zik and Irish 2003). Expression of TfLKR was also up-regulated in 35S::TfGLO plants, while that of TfLKR was not influenced in 35S::TfGLO-SRDX plants. Unexpectedly, expression of TfCAB1, 2, and 3 was also not influenced in 35S::TfGLO plants; however, it was up-regulated in 35S::TfGLO-SRDX plants. In petals of 35S::TfDEF-SRDX plants, expression of endogenous TfDEF was down-regulated, while that of TfGLO remained unchanged (Fig. 7B). Expression of TfXEGs and TfCABs in TfDEF transgenic plants was similar to that in TfGLO plants. Expression of TfLKR in TfDEF plants was different from that in TfGLO plants, and this expression was up-regulated only in 35S::TfDEF-SRDX plants. These results of expression of three TfXEG genes coincided with those of expression observed in previous reports on A. thaliana, while results of expression of TfLKR and TfCABs were different (Zik and Irish 2003).
Fig. 7.
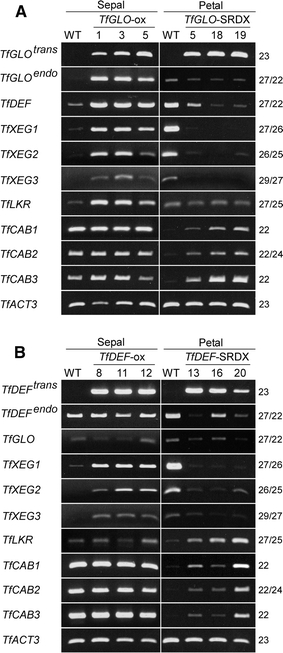
RT-PCR analysis of putative downstream genes of class B genes in transgenic torenia. Expression of putative downstream genes was analyzed using transgenic torenia with A 35S::TfGLO and 35S::TfGLO-SRDX genes and B 35S::TfDEF and 35S::TfDEF-SRDX genes. TfACT3 was used as an internal control. PCR cycles are indicated on the right side of each column. Trans, transgene; endo, endogenous gene
Regulation of anthocyanin biosynthesis-related gene expression in transgenic plants
To further confirm whether sepals of 35S::TfGLO plants that exhibited distinct anthocyanin accumulation (Figs. 3A and 4A-a) obtain characteristics of petals, we examined the influence of modified TfGLO and TfDEF transgenes on expression of anthocyanin biosynthesis-related genes by RT-PCR. Because no anthocyanin biosynthesis-related gene had been isolated from torenia used in this study, we isolated 10 genes (Supplementary Fig. S5), including TfANS, TfCHI, TfCHS1 and TfCHS2, TfDFR, TfF3H, TfF3′H1 and TfF3′H2, TfF3′5′H, and Tf3GT, and then examined their expression in 10 organs of wild-type plants (Fig. 8). Expression of these 10 genes was preferentially found in sepals and immature petals, but was stronger in the latter. Transcripts of most genes were not detected in vegetative organs such as roots, stems, petioles, and leaves, except that of TfF3′H2, which was detected in stems.
Fig. 8.
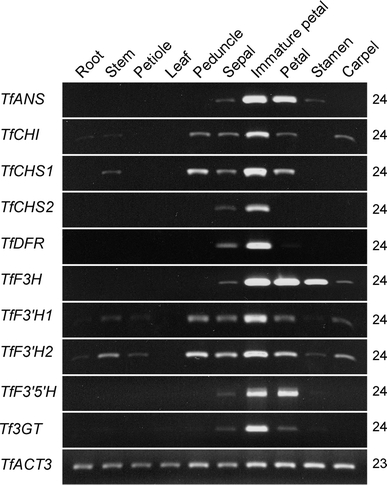
Organ-specific expression of anthocyanin biosynthesis-related genes. Ten torenia organs were used for expression analysis of anthocyanin biosynthesis-related genes. All anthocyanin biosynthesis-related genes were similarly expressed in immature petals. TfACT3 was used as an internal control. PCR cycles are indicated on the right side of each column
To examine expression of anthocyanin biosynthesis-related genes in transgenic plants, we performed RT-PCR using immature petals of addition to sepals of 35S::TfGLO plants that ectopically accumulated anthocyanins. The 10 genes were up-regulated in sepals of 35S::TfGLO plants and down-regulated in immature petals of 35S::TfGLO-SRDX plants (Fig. 9A). On the other hand, all genes showed distinctive expression patterns in 35S::TfDEF plants and were classified into three types (Fig. 9B). The first type including TfCHI, TfCHS1, and TfCHS2 showed no alteration in expression. The second type including TfANS, TfDFR, TfF3H, TfF3′5′H, and Tf3GT showed down-regulated expression, and the third type including TfF3′H1 and TfF3′H2 showed up-regulated expression. The expression patterns in petals of 35S::TfDEF-SRDX plants were also diverse, and the genes were classified into two types. The first type including TfCHI and TfCHS1 showed no alteration in expression, while the second type comprising the remaining genes showed down-regulated expression in immature petals. These results indicated that TfGLO and TfDEF act differently on anthocyanin biosynthesis, while the two class B genes function synergistically.
Fig. 9.
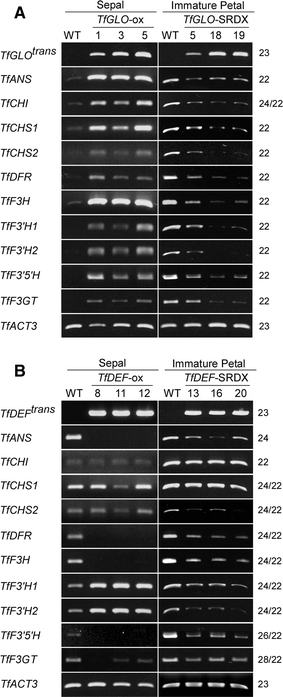
RT-PCR analysis of anthocyanin biosynthesis-related genes. RT-PCR analysis of anthocyanin biosynthesis-related genes using transgenic plants with A 35S::TfGLO and 35S::TfGLO-SRDX genes and B 35S::TfDEF and 35S::TfDEF-SRDX genes. TfACT3 was used as an internal control. PCR cycles are indicated on the right side of each column. Trans, transgene
Accumulation of anthocyanins and flavones in sepals of TfGLO-overexpressing plants
Next, we investigated anthocyanin and flavone compositions in sepals of 35S::TfGLO plants to elucidate whether the same types of pigments were accumulated as those in wild-type petals. Anthocyanins and flavones were extracted and quantified by HPLC, following the procedure described by Aida et al. (2000b). We used petals and sepals of wild-type plants and those of 35S::TfGLO and 35S::TfDEF plants as samples (Fig. 10). In petals of wild-type plants, five anthocyanin peaks were identified with regard to absorbance maximum ( λ max) as delphinidin 3,5-diglucoside (238, 274, and 522 nm), cyanidin 3,5-diglucoside (238, 278, and 514 nm), petunidin 3,5-diglucoside (240, 276, and 524 nm), peonidin 3,5-diglucoside (240, 278, and 516 nm), and malvidin 3,5-diglucoside (240, 276, and 526 nm) (Aida et al. 2000b; Fig. 10A). Five types of anthocyanins were identified in sepals of 35S::TfGLO plants, and their composition was similar to that in petals of wild-type plants. Peonidin 3,5-diglucoside accumulated to 72% of the level found in wild-type plants, and the other anthocyanins accumulated to 28–48% of the levels found in wild-type plants. No anthocyanin was detected in sepals of wild-type or 35S::TfDEF plants.
Fig. 10.
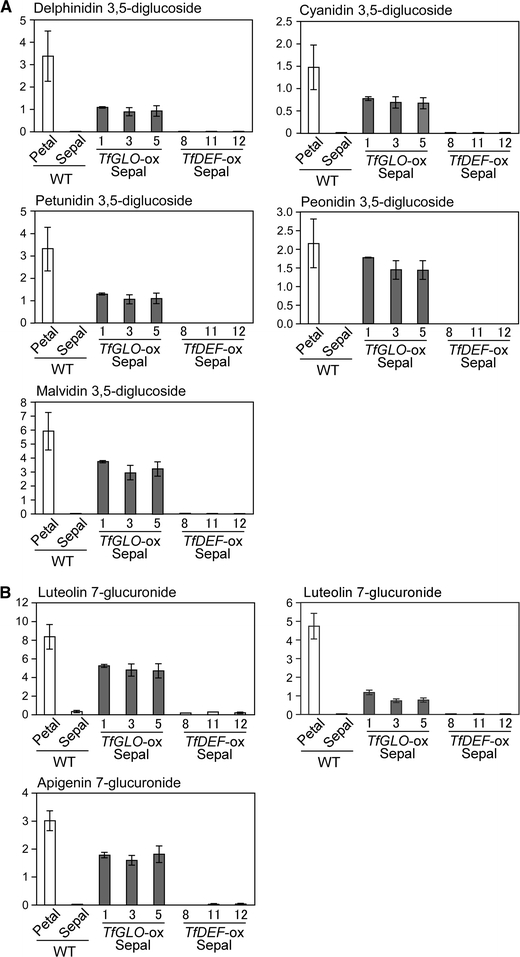
Quantification of anthocyanins and flavones. Torenia anthocyanins were quantified using petals and sepals of wild-type plants and sepals of transgenic plants with 35S::TfGLO or 35S::TfDEF. In these organs, five anthocyanins (A) and three flavones (B) were detected by HPLC. The horizontal axis represents the mean of three independent experiments with standard deviation
Three major and several minor peaks for flavones in petals of wild-type plants were observed. Peaks were identified with regard to λ max as luteolin 7-glucoside (257, 266, and 348 nm), luteolin 7-glucuronide (256, 266, and 348 nm), and apigenin 7-glucuronide (268 and 338 nm) (Aida et al. 2000b; Fig. 10B). Three types of flavones were identified in sepals of 35S::TfGLO plants and their composition was similar to that in petals of wild-type plants. Luteolin 7-glucoside and apigenin 7-glucuronide accumulated to ca. 60% of the levels in wild-type plants, and luteolin 7-glucuronide accumulated to 19% of the wild-type levels. Flavones were hardly detected in sepals of wild-type and 35S::TfDEF plants. These results elucidate that sepals of 35S::TfGLO plants accumulate identical types of pigments as petals, further suggesting that sepals were partially converted to petals.
Discussion
In this study, we demonstrated individual behaviors of TfGLO and TfDEF in floral organ development using transgenic torenia. TfGLO-overexpressing plants showed accumulation of anthocyanins and flavones in sepals (Figs. 3A, 10). TfGLO- and TfDEF-suppressed plants showed distinctive serrations and partial decolorization in petals, respectively (Fig. 3A, B). Furthermore, sepals of TfGLO-overexpressing plants showed petal-like cells (Fig. 4), and petals of TfGLO- and TfDEF-suppressed plants partially showed sepal-like cells (Fig. 5) on both adaxial and abaxial surfaces. These results indicate that the torenia class B genes individually perform important functions in floral organ development.
Sepals of TfGLO-overexpressing plants were thought to acquire the property of petals because they accumulated anthocyanins similar to petals (Fig. 3A-e and A-f). SEM analysis also revealed that surface cells of sepals of TfGLO-overexpressing plants were petal-like corn shaped (Fig. 4). In addition, we confirmed by HPLC that sepals accumulated anthocyanins and flavones, which were similar to those accumulated in petals of wild-type plants (Aida et al. 2000b) (Fig. 10). This type of data has not been reported for other plant species that do not accumulate anthocyanins in petals, such as A. thaliana. Genes in which transcripts were detected in petals but not in sepals were also up-regulated in sepals of TfGLO-overexpressing plants (Figs. 7, 9). Because the expression level of the endogenous TfDEF gene, the product of which interacted with TfGLO (Fig. 2), was weak in sepals (Fig. 6), the TfGLO transgene synergistically promoted conversion of sepals to petal-like organs. In contrast, overexpression of the TfDEF transgene was not sufficient to convert sepals to petals (Fig. 3B-e, B-f, and S4). This may be because expression of TfGLO is intrinsically very low and below the detection level in sepals (Fig. 6). Conversion of sepals of TfGLO-overexpressing plants to petals was incomplete. In fact, sepals remained as sepal shapes (Fig. 3A) and accumulated about half the amount of anthocyanins and flavones present in wild-type petals (Fig. 10). However, we assume that sepals of TfGLO-overexpressing plants were in a transition state from sepals to petals, and co-overexpression of TfDEF and TfGLO could have converted them to petals.
We assume that petals of TfGLO- and TfDEF-SRDX transgenic plants partially converted to sepals. Petals of these torenia transgenic plants apparently did not convert to sepals as reported in A. thaliana in which petals of ap3 and pi mutants morphologically changed to sepals (Bowman et al. 1989; Bowman et al. 1991). However, we believe our results were basically similar to those of ap3 and pi mutants in A. thaliana. Indeed, morphological alterations were observed in petals of TfGLO- and TfDEF-SRDX plants (Fig. 3); furthermore, the petal cells on both adaxial and abaxial surfaces partially but clearly converted to sepal-like cells (Fig. 5). Genes for which transcripts were detected in petals but not in sepals, such as TfXEGs and anthocyanin biosynthesis-related genes, were also down-regulated in petals of TfGLO- and TfDEF-suppressed plants (Figs. 7, 9). Petals of TfGLO- and TfDEF-SRDX plants would also be in a transition state from petals to sepal-like organs as observed in ap3 and pi mutants. These results indicate that petals of TfGLO- and TfDEF-suppressed transgenic plants partially showed characteristics of sepals and suggested that the torenia class B genes have common functions in floral organ development as shown in A. thaliana.
In A. thaliana, stamens of ap3 and pi mutants converted to carpels (Bowman et al. 1989; Bowman et al. 1991). While petals of TfGLO- and TfDEF-suppressed plants were partially converted to sepals, stamens did not seem to obtain the characteristics of carpels. Although it is possible that an invisible phenotype, such as carpel-specific gene expression, has been changed in stamens of the transgenic plants, we think that TfGLO- and TfDEF-SRDX could not disrupt stamen development. In torenia, the flower structure is unique in which stamens generally attach to the middle region of petals; however, petals of TfGLO- and TfDEF-SRDX plants apparently retained their shape (Fig. 3) and were only partially converted to sepals at the cell level (Fig. 5). Because suppression of petal development was incomplete in TfGLO- and TfDEF-SRDX plants, we could not observe morphological conversion in stamens of these transgenic plants. Therefore, we anticipate that co-suppression of TfGLO and TfDEF would achieve stronger suppression of TfGLO/TfDEF activities and affect stamen development in transgenic plants. Furthermore, the 35S promoter may also drive TfGLO- and TfDEF-SRDX insufficiently to repress stamen development. Activity of the 35S promoter was different in each organ and was low in some organs and tissues (Mitsuhara et al. 1996; Shirasawa-Seo et al. 2005). Utilization of their own promoters may provide a more accurate understanding of the functions of torenia class B genes in floral development, especially in stamens.
We isolated three TfXEG, one TfLKR, and three TfCAB downstream genes regulated by TfGLO and TfDEF (Fig. 4) according to the microarray data in A. thaliana (Zik and Irish 2003). Among them, we assumed that TfXEG genes may be regulated directly by TfGLO and TfDEF because their expression coincided with that of TfGLO and TfDEF in transgenic plants. This is because class B genes function as transcriptional activators as shown in analysis of A. thaliana in which the AP3/PI heterodimer directly binds CArG motifs of the AP3 promoter, thus resulting in activation of the promoter (Tilly et al. 1998). The A. thaliana XEG gene is down-regulated in ap3 or pi mutants and up-regulated in AP3/PI-overexpressing plants (Zik and Irish 2003). In our case, the three TfXEG genes were also up-regulated in TfGLO- and TfDEF-overexpressing plants and down-regulated in TfGLO- and TfDEF-suppressed plants (Fig. 5). In this study, we further isolated a promoter of TfXEG2 and determined the sequence as a representative gene which may be regulated directly by torenia class B genes (Fig. S6). The TfXEG2 promoter contained one candidate sequence for CC(A/T)6GG CArG-like motif (Riechmann et al. 1996b; Tilly et al. 1998) and two C(A/T)8G-type CArG motifs (Tang and Perry 2003). TfXEG2 did not include the reported typical sequences of CC(A/T)6GG-type CArG motifs, but MADS proteins have been reported to also bind incomplete CC(A/T)6GG-type CArG-box-like sequences (Riechmann et al. 1996b; Hill et al. 1998; Tilly et al. 1998). Because there are several CArG and CArG-like motifs on TfXEG2 promoters, gel mobility shift analysis using these putative CArG motifs and chromatin immunoprecipitation analysis with TfGLO and TfDEF proteins will help clarify this question in future analyses. While expression of TfLKR and TfCABs did not coincide with overexpression and suppression of TfGLO and TfDEF in transgenic plants, these genes appeared to be affected by certain factors as a result of modified torenia class B genes.
Anthocyanin biosynthesis-related genes were similarly regulated in TfGLO plants (Fig. 9A). In contrast, their expression patterns in TfDEF transgenic plants were complicated, and those in TfDEF-overexpressing and TfDEF-suppressed plants were divided into two and three types, respectively (Fig. 9B). We assume that the complex expression patterns were primarily derived from three reasons that are complicatedly related each other. The first reason is broad-ranging expression of TfDEF in not only petals and stamens but also sepals and carpels (Fig. 6). In fact, expression of class B genes in some plant species was found in not only petals and stamens but also sepals (for review Soltis et al. 2007). Although expression of class B genes of A. thaliana was not observed in sepals (Jack et al. 1994), transcripts of TfGLO were detected in only petals and stamens as expected, and the transcripts of TfDEF were detected in sepals and carpels (Fig. 6). Because of broad expression of TfDEF, the TfDEF protein may interact in various combinations with four class E proteins, known as SEPALLATA (SEP1–4; Pelaz et al. 2000, Ditta et al. 2004), which is described below and are different from TfGLO. The broad expression of TfDEF would influence regulation of anthocyanin biosynthesis-related genes, thereby leading to the complex expression that was not observed in petals of TfGLO transgenic plants. The second explanation is the interaction of class B genes with class E genes. In A. thaliana, four class E genes, SEP1–4, have been identified (Pelaz et al. 2000; Ditta et al. 2004) and reported to interact with class B genes (Yang et al. 2003b; Xu et al. 2006; Leseberg et al. 2008). The binding abilities of PI and AP3 with SEP1, SEP2, and SEP3 are different from each other (Yang et al. 2003b; Yang and Jack 2004). Therefore, the binding ability of TfDEF with various torenia SEP proteins may also be different from that of TfGLO. Third, the existence of unidentified torenia DEF and/or TM6 genes that uniquely form heterodimers and bind different CArG motifs from TfDEF is possible. In Petunia hybrida, duplicated class B genes have been identified, such as PhDEF, PhTM6, PhGLO1, and PhGLO2 (Vandenbussche et al. 2004). ThGLO1 and PhGLO2 function redundantly in petal and stamen development, whereas the function of PhDEF seems to be different from that of PhTM6, which is most likely not involved in petal development (Vandenbussche et al. 2004). In fact, we have confirmed by Southern blot analysis that TfDEF exists as at least two copies in the torenia genome (data not shown) and even more unidentified DEF/AP3 genes, which were not detected by this analysis, may also exist. Because the genomic sequence has not been determined and ESTs in torenia have not been fully characterized, we cannot confirm for certain the presence of unidentified DEF/AP3 genes in torenia at present. However, such unidentified DEF/AP3 genes may have a unique influence on expression of anthocyanin biosynthesis-related genes from TfDEF/TfGLO exhibiting different expression patterns in floral organs and/or different interactions in combination with SEPs, thereby conferring possible complex regulation of anthocyanin biosynthesis-related genes.
We aim to develop biotechnological techniques that can control floral traits such as flower color, petal shape, and colorations. However, in ornamental flowers, more information is required with regard to floral development and flower formation to control morphological traits. Therefore, we focused our study on individual functions of class B genes of horticultural torenia and demonstrated the functional divergence in TfGLO and TfDEF using transgenic plants. On the other hand, class B proteins form heterodimers to cooperatively function in floral development in planta (Goto and Meyerowitz 1994; Riechmann et al. 1996a). Therefore, we would like to further study on the function of class B genes using transgenic plants with co-overexpressing or co-suppressed TfGLO and TfDEF in future analyses. These studies would contribute not only to the understanding of the functions of torenia class B genes, but also to the ways in which various floral traits of torenia and many other ornamental flowers can be manipulated.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Dr. Masayoshi Nakayama for instructions regarding pigment analysis, Ms. Satoko Ohtawa for generating and maintaining transgenic torenia, and Ms. Yasuko Taniji and Ms. Yoshiko Kashiwagi for assisting in the molecular biology work and maintaining the plants. This work was supported by the Promotion of Basic Research Activities for Innovative Biosciences from Bio-oriented Technology Research Advancement Institution.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Aida R, Shibata M. Transgenic Torenia fournieri Lind. (torenia) In: Bajaj YPS, editor. Transgenic crops III vol 48 biotechnology in agriculture and forestry. Berlin: Springer Verlag; 2001. pp. 294–305. [Google Scholar]
- Aida R, Kishimoto S, Tanaka Y, Shibata M. Modification of flower color in torenia (Torenia fournieri Lind.) by genetic transformation. Plant Sci. 2000;153:33–42. doi: 10.1016/S0168-9452(99)00239-3. [DOI] [Google Scholar]
- Aida R, Yoshida K, Kondo T, Kishimoto S, Shibata M. Copigmentation gives bluer flowers on transgenic torenia plants with the antisense dihydroflavonol-4-reductase gene. Plant Sci. 2000;160:49–56. doi: 10.1016/S0168-9452(00)00364-2. [DOI] [PubMed] [Google Scholar]
- Becker A, Theissen G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol. 2003;29:464–489. doi: 10.1016/S1055-7903(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Bey M, Stüber K, Fellenberg K, Schwarz-Sommer Z, Sommer H, Saedler H, Zachgo S. Characterization of antirrhinum petal development and identification of target genes of the class B MADS box gene DEFICIENS. Plant Cell. 2004;16:3197–3215. doi: 10.1105/tpc.104.026724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- de Martino G, Pan I, Emmanuel E, Levy A, Irish VF. Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell. 2006;18:1833–1845. doi: 10.1105/tpc.106.042978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol. 2004;14:1935–1940. doi: 10.1016/j.cub.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Fukui Y, Tanaka Y, Kusumi T, Iwashita T, Nomoto K. A rationale for the shift in colour towards blue in transgenic carnation flowers expressing the flavonoid 3′, 5′-hydroxylase gene. Phytochemistry. 2003;63:15–23. doi: 10.1016/S0031-9422(02)00684-2. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- Hill TA, Day CD, Zondlo SC, Thackeray AG, Irish VF. Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development. 1998;125:1711–1721. doi: 10.1242/dev.125.9.1711. [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313X.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 2009;60:1081–1095. doi: 10.1111/j.1365-313X.2009.04064.x. [DOI] [PubMed] [Google Scholar]
- Jack T, Fox GL, Meyerowitz EM. Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell. 1994;76:703–716. doi: 10.1016/0092-8674(94)90509-6. [DOI] [PubMed] [Google Scholar]
- Katsumoto Y, Fukuchi-Mizutani M, Fukui Y, Brugliera F, Holton TA, Karan M, Nakamura N, Yonekura-Sakakibara K, Togami J, Pigeaire A, Tao GQ, Nehra NS, Lu CY, Dyson BK, Tsuda S, Ashikari T, Kusumi T, Mason JG, Tanaka Y. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 2007;48:1589–1600. doi: 10.1093/pcp/pcm131. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Tanaka H, Shiba T, Mii M, Tsujimoto H. Genome size, karyotype, meiosis and a novel extra chromosome in Torenia fournieri, T. baillonii and their hybrid. Chromosome Res. 2006;14:665–672. doi: 10.1007/s10577-006-1077-y. [DOI] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell. 2007;19:473–484. doi: 10.1105/tpc.106.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Dorit RL, Irish VF. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics. 1998;149:765–783. doi: 10.1093/genetics/149.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leseberg CH, Eissler CL, Wang X, Johns MA, Duvall MR, Mao L. Interaction study of MADS-domain proteins in tomato. J Exp Bot. 2008;59:2253–2265. doi: 10.1093/jxb/ern094. [DOI] [PubMed] [Google Scholar]
- Ma H. The unfolding drama of flower development: recent results from genetic and molecular analyses. Genes Dev. 1994;8:745–756. doi: 10.1101/gad.8.7.745. [DOI] [PubMed] [Google Scholar]
- Matsui K, Tanaka H, Ohme-Takagi M. Suppression of the biosynthesis of proanthocyanidin in Arabidopsis by a chimeric PAP1 repressor. Plant Biotechnol J. 2004;2:487–493. doi: 10.1111/j.1467-7652.2004.00094.x. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Ohme-Takagi M. Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 2009;50:1232–1248. doi: 10.1093/pcp/pcp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Hiratsu K, Todaka D, Nakashima K, Yamaguchi-Shinozaki K, Ohme-Takagi M. Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol J. 2006;4:325–332. doi: 10.1111/j.1467-7652.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Umemura Y, Ikeda M, Shikata M, Koyama T, Matsui K, Narumi T, Aida R, Sasaki K, Hiyama T, Higuchi Y, Ono M, Isuzugawa K, Saitoh K, Endo R, Nishihara M, Yamamura S, Yamamura T, Terakawa T, Ohtsubo N, Ohme-Takagi M. FioreDB: a database of phenotypic information induced by the chimeric repressor silencing technology (CRES-T) in Arabidopsis and floricultural plants. Plant Biotechnol. 2008;25:37–43. [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, Ueno K, Mochizuki A, Tanimoto H, Tsugawa H, Otsuki Y, Ohashi Y. Efficient promoter cassettes for enhanced expression of foreign gene in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 1996;37:39–59. doi: 10.1093/oxfordjournals.pcp.a028913. [DOI] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD. The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol. 2005;8:122–128. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Narumi T, Aida R, Niki T, Nishijima T, Mitsuda N, Hiratsu K, Ohme-Takagi M, Ohtsubo N. Chimeric AGAMOUS repressor induces serrated petal phenotype in Torenia fournieri similar to that induced by cytokinin application. Plant Biotechnol. 2008;25:45–53. [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. MADS domain proteins in plant development. Biol Chem. 1997;378:1079–1101. doi: 10.1515/bchm.1997.378.10.1079. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Krizek BA, Meyerowitz EM. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA. 1996;93:4793–4798. doi: 10.1073/pnas.93.10.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Wang M, Meyerowitz EM. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 1996;24:3134–3141. doi: 10.1093/nar/24.16.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Rijpkema AS, Gerats T, Vandenbussche M. Evolutionary complexity of MADS complexes. Curr Opin Plant Biol. 2007;10:32–38. doi: 10.1016/j.pbi.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Aida R, Niki T, Yamaguchi Y, Narumi T, Nishijima T, Hayashi Y, Ryuto H, Fukunishi N, Abe T, Ohtsubo N. High-efficiency improvement of transgenic torenia flowers by ion beam irradiation. Plant Biotechnol. 2008;25:81–89. [Google Scholar]
- Shikata M, Ohme-Takagi M. The utility of transcription factors for manipulation of floral traits. Plant Biotechnol. 2008;25:31–36. [Google Scholar]
- Shirasawa-Seo N, Sano Y, Nakamura S, Murakami T, Gotoh Y, Naito Y, Hsia CN, Seo S, Mitsuhara I, Kosugi S, Ohashi Y. The promoter of Milk vetch dwarf virus component 8 confers effective gene expression in both dicot and monocot plants. Plant Cell Rep. 2005;24:155–163. doi: 10.1007/s00299-005-0917-0. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Ma H, Frohlich MW, Soltis PS, Albert VA, Oppenheimer DG, Altman NS, dePamphilis C, Leebens-Mack J. The floral genome: an evolutionary history of gene duplication and shifting patterns of gene expression. Trends Plant Sci. 2007;12:358–367. doi: 10.1016/j.tplants.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Tang W, Perry SE. Binding site selection for the plant MADS domain protein AGL15: an in vitro and in vivo study. J Biol Chem. 2003;278:28154–28159. doi: 10.1074/jbc.M212976200. [DOI] [PubMed] [Google Scholar]
- Theissen G, Saedler H. Plant biology floral quartets. Nature. 2001;409:469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- Tilly JJ, Allen DW, Jack T. The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development. 1998;125:1647–1657. doi: 10.1242/dev.125.9.1647. [DOI] [PubMed] [Google Scholar]
- Vandenbussche M, Theissen G, Van de Peer Y, Gerats T. Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res. 2003;31:4401–4409. doi: 10.1093/nar/gkg642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M, Zethof J, Royaert S, Weterings K, Gerats T. The duplicated B-class heterodimer model: whorl-specific effects and complex genetic interactions in Petunia hybrida flower development. Plant Cell. 2004;16:741–754. doi: 10.1105/tpc.019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM. Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell. 2004;16:1314–1326. doi: 10.1105/tpc.021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple CJ, Ciceri P, Padilla CM, Ambrose BA, Bandong SL, Schmidt RJ. Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development. 2004;131:6083–6091. doi: 10.1242/dev.01523. [DOI] [PubMed] [Google Scholar]
- Xu Y, Teo LL, Zhou J, Kumar PP, Yu H. Floral organ identity genes in the orchid Dendrobium crumenatum. Plant J. 2006;46:54–68. doi: 10.1111/j.1365-313X.2006.02669.x. [DOI] [PubMed] [Google Scholar]
- Yang Y, Jack T. Defining subdomains of the K domain important for protein–protein interactions of plant MADS proteins. Plant Mol Biol. 2004;55:45–59. doi: 10.1007/s11103-004-0416-7. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fanning L, Jack T. The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 2003;33:47–59. doi: 10.1046/j.0960-7412.2003.01473.x. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xiang H, Jack T. Pistillata-5, an Arabidopsis B class mutant with strong defects in petal but not in stamen development. Plant J. 2003;33:177–188. doi: 10.1046/j.1365-313X.2003.01603.x. [DOI] [PubMed] [Google Scholar]
- Zik M, Irish VF. Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell. 2003;15:207–222. doi: 10.1105/tpc.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


