Abstract
In ClpXP and ClpAP complexes, ClpA and ClpX use the energy of ATP hydrolysis to unfold proteins and translocate them into the self-compartmentalized ClpP protease. ClpP requires the ATPases to degrade folded or unfolded substrates, but binding of acyldepsipeptide antibiotics (ADEPs) to ClpP bypasses this requirement with unfolded proteins. We present the crystal structure of Escherichia coli ClpP bound to ADEP1 and report the structural changes underlying ClpP activation. ADEP1 binds in the hydrophobic groove that serves as the primary docking site for ClpP ATPases. Binding of ADEP1 locks the N-terminal loops of ClpP in a β-hairpin conformation, generating a stable pore through which extended polypeptides can be threaded. This structure serves as a model for ClpP in the holo-enzyme ClpAP and ClpXP complexes and provides critical information to further develop this class of antibiotics.
Keywords: ClpP, ADEP, ClpAP, ClpXP, Axial channel, ATP-independent proteolysis
INTRODUCTION
Most intracellular protein degradation is carried out by large multi-subunit complexes belonging to one of four families of ATP-dependent proteases (Gottesman et al., 1997a), including the ClpXP and ClpAP complexes, which degrade a variety of both functional and non-functional proteins in eubacteria and in the major organelles of eukaryotes (Gottesman et al., 1997b; Maurizi, 1992).
The proteolytic core of ClpXP and ClpAP is ClpP, a self-compartmentalized protease that oligomerizes as two stacked heptameric rings enclosing a central chamber containing 14 proteolytic active sites (Wang et al., 1997). Access to the internal chamber is through axial pores in the center of each heptameric ring, and the N-terminal regions of ClpP subunits play a role in controlling substrate entry (Bewley et al., 2006; Gribun et al., 2005; Szyk and Maurizi, 2006). X-ray crystallographic studies show that residues 2-7 line the axial channel, defining a narrow pore 10-12 Å in diameter (Kang et al., 2004). However, the side chain densities of these residues are broken, suggesting that their positions are variable (Bewley et al., 2006; Szyk and Maurizi, 2006; Wang et al., 1997). Residues 8-16 form a loop that extends out from the apical surface of the heptamer. The loops are only partially visible in most structures, suggesting they are also mobile, but in one structure (Bewley et al., 2006) the loops fill the space surrounding the entrance to the axial channel presenting a barrier to substrate entry. The disposition of the loops and the narrowness of the axial pore prevent folded proteins or large polypeptides from directly entering the chamber and severely restrict entry of peptides >5-10 amino acids (Thompson et al., 1994; Woo et al., 1989).
ClpX and ClpA belong to the AAA+ protein family (ATPases associated with various cellular activities). They assemble into hexameric rings that bind both ring surfaces of the ClpP tetradecamer forming a barrel-like holoenzyme complex (Beuron et al., 1998). ClpX and ClpA use ATP hydrolysis to catalyze protein unfolding (Hoskins et al., 1998; Sauer et al., 2004; Singh et al., 1999) and to thread polypeptides into the proteolytic chamber of ClpP (Beuron et al., 1998; Ishikawa et al., 2001; Ortega et al., 2000). The ATPase and protease components also exert allosteric effects on each other. ClpP stabilizes ClpA and ClpX hexamers and inhibits their ATPase activity (Kim et al., 2001) and ClpX and ClpA activate the peptidase activity of ClpP without ATP hydrolysis (Thompson and Maurizi, 1994).
Structural (Bewley et al., 2006; Glynn et al., 2009; Guo et al., 2002; Kim and Kim, 2003; Szyk and Maurizi, 2006; Wang et al., 1997) and biochemical (Kim et al., 2001; Martin et al., 2007, 2008; Singh et al., 2001) data identify two kinds of interactions between ClpP and ClpA or ClpX. The first are stable interactions involving a highly conserved motif, IGF/L, present in loops on the surface of ClpA and ClpX rings. The IGF/L motifs dock into deep hydrophobic pockets on the surface of ClpP. A second interaction involves the ATPase pore-2 loops and the N-terminal loops of ClpP. The ClpP N-terminal loops make direct, though possibly transient, contact with the pore-2 loops of ClpX, which are located near the axial channel proximal to ClpP (Martin et al., 2007, 2008).
Recently, a new class of antibiotics, acyldepsipeptides (ADEPs), was found to activate ClpP in the absence of its cognate ATPases (Brotz-Oesterhelt et al., 2005). ADEPs kill bacterial cells by indiscriminately increasing the activity of ClpP in vivo, redirecting its activity away from its physiological substrates and targeting it to nascent polypeptide chains, resulting in inhibition of cell division and cell death (Kirstein et al., 2009). ADEPs promote dissociation of ClpC/MecA/ClpP complexes purified from Bacillus subtilis and convert the ClpP to an ATP-independent protease capable of degrading unfolded proteins (Brotz-Oesterhelt et al., 2005; Kirstein et al., 2009). While this paper was in preparation, a study with B. subtilis ClpP concluded that ADEP binding caused an increase in the mobility of the N-terminal loops of ClpP (Lee et al., 2010a) and the authors proposed that this increased mobility opens the axial channel and facilitates passage of longer polypeptides into ClpP.
Here, we demonstrate that ADEPs affect the activity and properties of Escherichia coli ClpP in a manner similar to B. subtilis ClpP, but we propose a very different mechanism by which this is accomplished. We show that ADEP1, an ADEP congener purified from Streptomyces hawaiiensis (Brotz-Oesterhelt et al., 2005; Michel and Kastner, 1985), stabilizes the tetradecameric form of ClpP and allows unfolded proteins to be translocated into the degradation chamber. Our crystal structure of E. coli ClpP bound with ADEP1 shows that binding stabilizes the N-terminal region of ClpP, locking the loops in an open conformation that creates a 20 Å diameter axial pore. The rest of the ClpP structure undergoes small structural changes that facilitate the enlargement of the axial pore. Modeling the LGF loop from Helicobacter pylori in place of ADEP1 in the structure indicates that binding of ADEP1 mimics the docking interaction between the Clp ATPases and ClpP. Consequently this structure represents a snapshot of the conformational state of ClpP bound to a Clp ATPase which we propose is the configuration that is ready to accept unfolded substrates.
RESULTS AND DISCUSSION
ADEP1 Activates Protein Degradation by E. coli ClpP
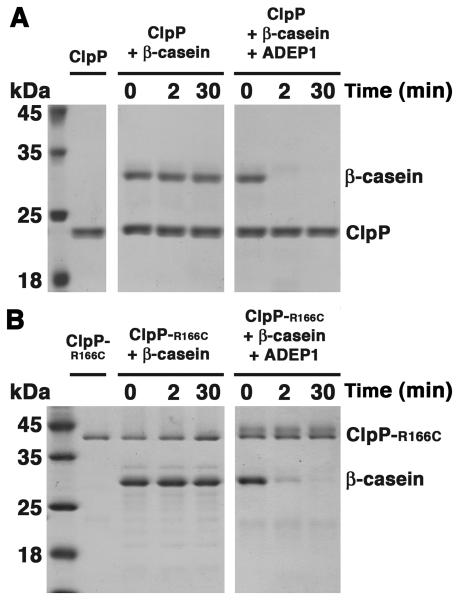
To confirm that ADEPs increase the protease activity of purified E. coli ClpP in the same manner as reported for B. subtilis ClpP, we assayed degradation of a model unfolded protein, β-casein, in the presence and absence of ADEP1. E. coli ClpP alone did not degrade β-casein but cleaved all of the β-casein within 2 min in the presence of ADEP1 (Figure 1A). Degradation in the presence of ADEP1 was comparable to that observed when ClpA and ATP were present (Figure S1A), indicating that ADEP1 renders the ClpP degradation chamber as accessible to unfolded proteins as does ClpA. In agreement with published results (Kirstein et al., 2009), ClpA promoted processive degradation of β-casein, whereas ADEP1-activated ClpP generated numerous partially degraded products (Figure S1A; asterisk).
Figure 1. ADEP Activation of ClpP to Degrade Protein Substrates.
(A) Coomassie brilliant blue 15% SDS-PAGE showing the time course degradation of β-casein by ClpP in the presence and absence of ADEP1. See also Figure S1A.
(B) Time course of the degradation of β-casein by ClpP-R166C, which is a constitutive tetradecamer in the presence and absence of ADEP1. Samples from different time points were resolved in a 15% SDS-PAGE and stained by Coomassie brilliant blue. See also Figure S1B.
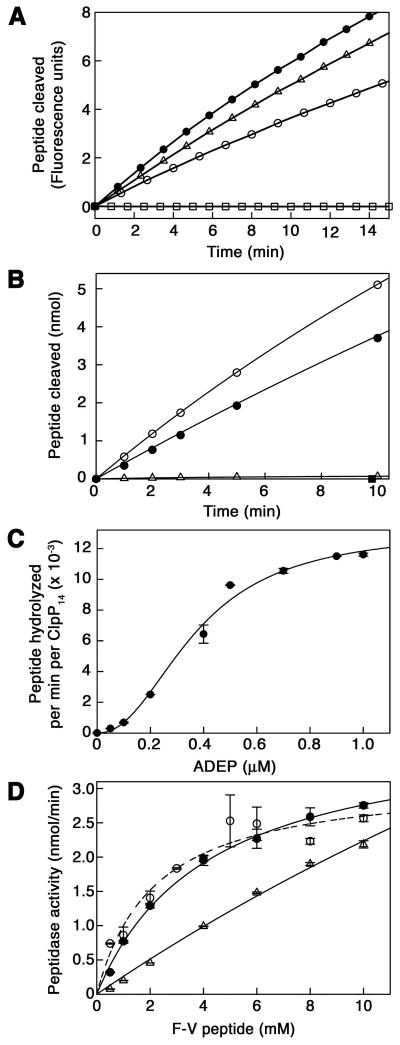
To further evaluate the accessibility of the degradation chamber in the presence of ADEP1, we tested peptidase activity against peptide substrates. The rate of cleavage of the dipeptide, N-Succinyl-Leu-Tyr-7-amido-4-methylcoumarin, was almost unchanged in the presence of ADEP1 (Figure 2A and Supplemental Table 1), suggesting that very small peptides have ready access to the degradation chamber with or without ADEP1. In contrast, the 10-residue peptide, FAPHMALVPV (F-V), which is cleaved at a single specific site by ClpP (Thompson and Maurizi, 1994), was cleaved at least 50 times faster in the presence of ADEP1 (Figure 2B and Supplemental Table 1). Activation of F-V cleavage by ADEP1 was comparable to the allosteric activation seen with ClpA (Figure 2B). ADEP1 also activated cleavage of other peptides, including the 30-residue oxidized insulin β chain (Supplemental Table 1). The enhanced cleavage of longer peptides and the lack of an effect with dipeptides confirm that activation by ADEP1 is primarily due to increased substrate access to the degradation chamber and not to an increase in the catalytic activity of ClpP.
Figure 2. Kinetic Parameters for ADEP Activation of ClpP Peptidase Activity.
(A) Time course of hydrolysis of the fluorogenic peptide N-Succinyl-Leu-Tyr-7-amido-4-methylcoumarin (0.2 mM) by E. coli ClpP (50 μg/ml) in the presence (open circles) and absence (closed circles) of 10 μM ADEP1. Control reactions containing the fluorogenic peptide and ADEP1 but not ClpP (open squares) or ClpP with equivalent amounts of DMSO (open triangles) are also shown. (Arbitrary fluorescence units reflect release of aminomethylcoumarin). See also Supplemental Table 1.
(B) Time course of cleavage of the 10-residue peptide, F-V (1 mM), by ClpP (1 μg/ml) in the presence (open circles) and absence (open triangles) of 10 μM ADEP1. Peptide products released were quantitated by absorbance measurements after reverse phase chromatography. For comparison, ClpP peptidase activity stimulated by ClpA (10 μg/ml) in the presence of 1 mM ATPγS was also measured (closed circles). A control reaction containing only F-V and ADEP1 is also shown (closed squares). See also Supplemental Table 1.
(C) Dependence of ClpP activation on ADEP1 concentration. ClpP (1.0 μg/ml) and F-V (2 mM) were held constant and the concentration of ADEP1 added was varied. Results are the average of two separate experiments.
(D) Substrate-dependence of ClpP peptidase activity in the presence of 10 μM ADEP1 (open circles). ClpP (1.0 μg/ml) and ADEP1 (10 μM) were held constant and the concentration of F-V was varied. For comparison, the substrate-dependence was measured for F-V cleavage by ClpAP (1.0 μg/ml ClpP, 10 μg/ml ClpA) (closed circles) and by ClpP alone (10 μg/ml) (open triangles), which showed no sign of saturation at the highest concentrations used. Results are the average of two separate experiments. Note that the activity with ClpP alone is ~2% of the activity with ADEP1 present.
The saturation curve for ADEP1 activation of F-V cleavage was sigmoidal, with an S0.5 of 0.37 μM and a calculated Hill coefficient of 2.2 ± 0.3 (Figure 2C), reflecting either a slight cooperativity in ADEP1 binding to ClpP or a cooperative allosteric transition involved in the structural changes involved in the activation mechanism.
The concentration dependence for cleavage of F-V in the presence of ADEP1 followed normal Michaelis-Menten kinetics with an apparent Km for F-V of 2.2 ± 0.5 mM (Figure 2D). The Km for F-V is comparable to that obtained for the ClpA-activated cleavage of this peptide (Figure 2D), indicating that the peptide is fully accessible to the active sites and that entry into the chamber is not rate limiting. In contrast, the kinetics of cleavage in the absence of ADEP1 was not saturable because entry into ClpP was rate limiting (Figure 2D).
The similarity between the allosteric activation of peptidase activity produced by ADEP1 and ClpA suggested that they might bind to similar sites on ClpP. Consistent with this interpretation, ADEP1 blocked interaction between ClpP and ClpA (Figure S2). These results confirm that ADEPs preferentially activate ClpP and interfere with regulation by the ATPase components.
ADEP1-induced Protein Degradation is Performed by Tetradecameric ClpP
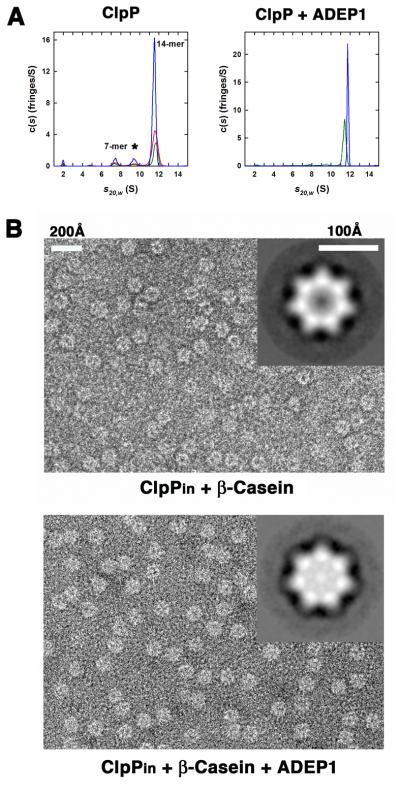
To assess the effect of ADEPs on the oligomeric state of ClpP, we performed sedimentation velocity experiments in the presence and absence of ADEP1 (Figure 3A). The c(s) analysis of ClpP alone showed three species (Figure 3; left panel). The major species, with an average S20,w of 11.58 ± 0.06 S and an estimated molecular mass of 306 ± 10 kDa, corresponds to the ClpP tetradecamer (calculated mass = 301.872 kDa). We observed two minor peaks corresponding to heptamers at 7.52 ± 0.09 S and an additional artifactual species at 9.44 ± 0.06 S (Figure 3A; left panel), arising from the reversible dissociation of the ClpP tetradecamer into smaller species (Schuck, 2000). Addition of ADEP1 produced a single species at 11.77 ± 0.01 S (99% of the loading signal) corresponding to the ClpP tetradecamer and with an estimated molecular mass of 330 kDa (Figure 3A; right panel), revealing that ADEP1 binding enhances rather than disrupts tetradecamer formation. The same results were obtained at two additional ClpP concentrations (data not shown).
Figure 3. ADEP1 Stabilize the ClpP Tetradecamer and Promotes Substrate Translocation into its Degradation Chamber.
(A) Continuous c(s) distributions obtained from sedimentation velocity data collected at 40 krpm for ClpP in the absence of added ADEP1 (left panel). Data were collected at 101 μM (blue), 54 μM (red) and 27 μM (green) of ClpP monomer (left panel). At these concentrations, the ClpP tetradecamer (14-mer) represented the major species at approximately 81% of the total signal. Species formed from the dissociation of the tetradecamer are found at 7.5 S (~ 8% of loading signal, presumed 7-mer) and 9.4 S (~ 8% of loading signal). Traces (~ 2% of loading signal) of ClpP monomer are found at 2.0 S. Similar experiment for ClpP in the presence of added ADEP1 (right panel). Data were collected at approximately 53 μM of ClpP monomer in the presence of 0.7% (v/v) DMSO (green) or 5 equivalents of ADEP1 (blue) dissolved in DMSO. In the presence of ADEP1, data are consistent with the presence of a single ClpP tetradecamer at 11.8 S.
(B) Negative stained electron micrographs comparing ClpPin particles incubated for 2 min with β-casein in the absence (top panel) and presence of ADEP1 (bottom panel). Insets in the micrographs compare top view averages of ~500 particles of ClpPin from each sample. Less stain penetration (brighter) correlates with accumulation of β-casein inside the inner cavity. See also Figure S3.
To rule out that transient formation of heptamers was responsible for ADEP-induced protein degradation, we performed degradation assays with a ClpP mutant, ClpP-R166C, in which pairs of apposing subunits across the tetradecamer interface were cross-linked with 1,11-Bis-maleimidotriethyleneglycol. Cross-linked ClpP-R166C migrates as a dimeric species in SDS gels under reducing conditions (Figure 1B), confirming that the rings are covalently joined and cannot separate under native conditions. ClpP-R166C alone had no protein degrading activity, but it degraded β-casein in the presence of ADEP1 (Figure 1B). Activity of cross-linked ClpP-R166C was similar to that of wild type ClpP with both ADEP1 and ClpA (Figure S1B). These results confirm that proteins do not access the ClpP proteolytic sites as a result of dissociation of the tetradecamer.
ADEP1 Promotes Substrate Translocation into the ClpP Degradation Chamber
To directly visualize translocation of unfolded proteins into the ClpP chamber, we used a chemically inactivated variant of ClpP (ClpPin) that accumulates undegraded substrates translocated by ClpA or ClpX (Ortega et al., 2000; Singh et al., 1999). In negative-stained micrographs, empty ClpPin chambers appear dark because they accumulate the uranyl acetate stain, whereas the presence of protein substrates within the chamber excludes the stain and the chambers remain bright (Ortega et al., 2000). We added an excess of β-casein to ClpPin previously incubated with ADEP1, waited two minutes and imaged the samples. Stain did not penetrate the chamber of most ClpPin molecules treated with ADEP1, while the majority of molecules from a control reaction without ADEP1 showed a stain-penetrated central chamber (Figure 3B). Averaged images obtained from both sets of particles clearly confirmed the difference in stain penetration between treated and untreated populations (Figure 3B; top and bottom panel insets). ADEP1 alone in the absence of β-casein did not prevent stain penetration into the ClpPin chamber (data not shown).
To quantify these observations, we calculated the distribution of the particles with respect to the intensity displayed in an area centered over their digestion chamber. There was a significant shift of particles to higher intensity values when the translocation reaction was performed in the presence of ADEP1 compared to the reactions with no compound (Figure S3A). When the same experiment was conducted using EGFP-SsrA, which is not degraded by ClpP in the presence of ADEP1 (Brotz-Oesterhelt et al., 2005), no significant differences were observed (Figure S3B). These results indicate that ADEP activation of ClpP induces substrate translocation into the degradation chamber.
Structure of E. coli ClpP Bound to ADEP1
To analyze the conformational changes induced in ClpP upon binding of ADEPs, we determined the crystal structure of the ClpP-ADEP1 complex. The structure was solved by molecular replacement using the E. coli ClpP heptamer (PDB ID 1YG6) (Bewley et al., 2006). The N-terminal loops (residues 1-19) and the handles (residues 123-148) were removed from the search model to minimize model bias. Two tetradecamers were found in the asymmetric unit and after rigid body refinement, difference maps showed unequivocal electron density for all ClpP handles and most of the N-terminal loops. ADEP1 molecules were identified at all intra-ring subunit interfaces.
The two tetradecamers in the asymmetric unit are virtually identical and can be superimposed with an r.m.s. deviation of 0.46 Å for 2,592 atoms. In the first tetradecamer (chains A-N), all the N-terminal loops are well defined (Figure S4A), although residues Thr10 to Gly13 engage in minimal interactions and only broken density was seen for this region in half of the N-terminal loops. An apical surface in the second tetradecamer impinges onto the lateral surface of the first tetradecamer, preventing the formation of the β-hairpins for two monomers (R and S) of this ring (Figure S4B). The electron density for the N-terminal loops on the apposing heptamer was also weak, although this ring did not have close crystal contacts that might have precluded formation of the β-hairpins (Figure S4B). Interestingly, the most complete loops in this ring were found in two adjacent monomers (chains X and Y), despite the fact that chain X is the only monomer in which the ADEP1 density is fragmented suggesting lower occupancy at this site. This result supports our model discussed below that organization of the N-terminal loops results from allosteric changes operating through changes in subunit interactions.
The ClpP monomer is virtually identical to previous structures between residues 18 to 193, but the conformation of the N-terminal regions (residues 1-17) varies significantly (Figure 4A and Figure S5A). The tetradecamer is also very similar to other structures, yet not identical. ADEP1 binding causes a rigid-body movement of the monomer resulting in a subtle expansion of the apical surface of the ring and a constriction of the equatorial belt formed by the ClpP handles (Movie S1). This subtle movement (~1.5-2Å) pushes the monomers outwards and locks the N-terminal loops in an open β-hairpin conformation (Figure 4B). Remarkably, all the N-terminal loops adopt an almost identical conformation upon ADEP1 binding with residues 2-7 flush with the channel walls and residues 8-16 rotated away from the axis (Figure 4 and Figure S5B). The best defined N-terminal loops are found in monomers that do not have close crystal contacts around their N-termini (Figure S4A), thereby allowing the hairpin to be properly organized, from which we conclude that this conformation of the axial pore is quite stable. Indeed, the B-factors of the N-terminal loops are comparable to those of the rest of the protein attesting to their stability (Figure S5C).
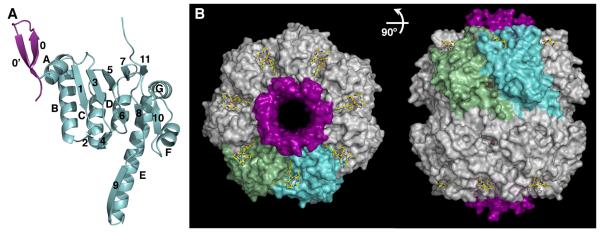
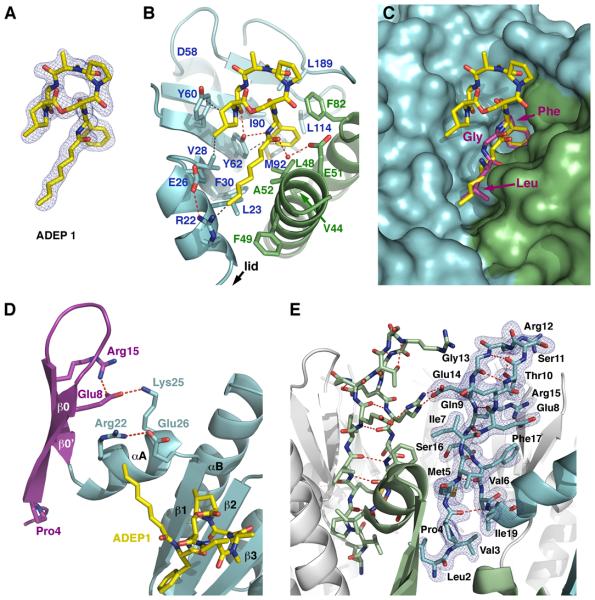
Figure 4. Crystal Structure of the ClpP-ADEP1 Complex.
(A) Ribbon diagram of the ClpP monomer with the N-terminal lid shown in purple and the head domain and handle shown in light blue. The secondary structure motifs are labeled as in (Wang et al., 1997).
(B) Orthogonal views of the ClpP tetradecamer (white surface) bound to ADEP1 (yellow color-coded sticks). The surfaces of two adjacent ClpP monomers are colored light green and blue for reference, and all the N-terminal loops are shown as a purple surface. Two 2-methyl-2,4-pentanediol (MPD) molecules bound to the P1 pocket and the side pores of each ClpP monomer are shown as brown sticks. See also Figure S4, S5, S6 and Movie S1.
ADEP-1 Binding Induces the Change in Conformation of the N-terminal Loops
ADEP1 binds to a hydrophobic pocket on the outer edge of the apical surface of ClpP and, while it sits at the interface between adjacent monomers, it interacts more extensively with one monomer (Figure 4B). The bound configuration of ADEP1 and its contacts with E. coli ClpP are consistent with those reported for the complex of ADEP1 with B. subtilis ClpP (Lee et al., 2010a) (Figure 5A and B). As predicted from kinetic studies of a group of ADEP1 congeners (Brotz-Oesterhelt et al., 2005), the phenylalanine, β-methylproline and alanine moieties, as well as the aliphatic tail in ADEP1 are critical for binding, while the N-methylalanine and proline moieties have minimal interactions with the protein (Figure 5B). The phenyl ring fills a hydrophobic cavity defined by residues Tyr62, Ile90, Met92, Leu114 and Leu189 from one monomer and residues Val44, Leu48, and Phe82 from the adjacent monomer. The aliphatic chain lies along a hydrophobic groove defined by residues Arg22, Leu23, Val28, Phe30 and Tyr62 from one monomer and Leu48, Phe49 and Ala52 from the other. Additionally, Tyr62 forms hydrogen bonds with both the phenylalanine and the alanine moieties of the compound. Interaction between ClpP and ADEP1 is further stabilized by van der Waals contacts between the β-methylproline moiety and the side chains of residues Val28, Tyr60 and Tyr62 and water-mediated hydrogen bonds with Glu51 (Figure 5B).
Figure 5. Binding Interactions of ClpP with ADEP1 and Organization of the N-terminal Loops in the ClpP-ADEP1 structure.
(A) Final model of ADEP1 shown as color-coded sticks with the 2|Fo|-|Fc| electron density map contoured at 1.0 σ level.
(B) Detailed interactions between ADEP1 and the residues defining the hydrophobic pocket in ClpP. Adjacent ClpP monomers are shown as ribbon diagrams in light blue and green, with interacting side chains depicted as color-coded sticks and labeled. Hydrogen bonds (red) and van der Waals interactions (grey) are shown as dashed lines.
(C) Superimposition of the conserved LGF motif (consensus sequence IGF) from Helicobacter pylori ClpX (purple color-coded sticks) onto the structure of ClpP bound to ADEP1 shown in the same orientation and color scheme as in (B).
(D) Ribbon diagram of a ClpP monomer with the N-terminal region shown in purple and the head domain shown in light blue. Side chains of residues involved in anchoring the N-terminal lid to the head domain are shown as sticks with the hydrogen bonds depicted as dashed lines (red). ADEP1 is shown as yellow color-coded sticks.
(E) Ribbon diagram of the ClpP heptamer with two adjacent monomers shown in light green and blue, while the rest are shown in white. Two N-terminal lids (residues 1-19) are shown as sticks, with the intra- and inter-molecular hydrogen bonds maintaining the β-hairpin structure depicted as red dash lines. The 2|Fo|-|Fc| electron density map around one of the lids is shown in blue and contoured at 1.0 σ level.
Binding of ADEP1 widens the interface between adjacent ClpP monomers by about 1 Å, altering the orientation of helix α1 (Ile19-Glu26) and displacing the side chains of Glu26 and Arg22, which are hydrogen bonded in our structure (Figure 5D). This rearrangement disrupts interactions between Tyr20 and the neighboring monomer, and in turn, displaces Phe17, which can now interact with Val6 to form the base of the β-hairpin. Stability of the hairpin is further enhanced by polar interactions between Glu8 and Arg15 and between Gln9 and Glu14 (Figure 5D and E), as well as intermolecular interactions between Arg15 and Glu14 from adjacent monomers (Figure 5E).
Pro4 and Glu8 anchor the β-hairpin in this open conformation (Figure 5D), providing a rationale for the invariance of these residues in the 1XΦΦPΦΦΦE8 consensus sequence of the N-terminus within the ClpP family (Kang et al., 2004). Pro4 causes a kink in the main chain and projects the tip of the loop towards the solvent. The side chain of Glu8 latches the β-hairpin to the globular domain of ClpP through polar interactions with the side-chains of Arg22 and Lys25 (Figure 5D and E), thereby locking the orientation of the N-terminal region relative to the head domain. Interestingly, mutations in Glu8 (Bewley et al., 2009) and Arg 22 (Lee et al., 2010b) are reported to cause significant defects in ClpX-activated degradation by ClpP. Glu14 and Arg15 are also important to stabilized the close conformation of the axial channel and restrict the access to the digestion chamber of peptides longer than 10 amino acids. These residues are also important for the interaction with ClpA and ClpX (Bewley et al., 2006; Lee et al., 2010b) and provide the axial channel with broad translocation specificity. These data support our proposal that the open conformation of the β-hairpin found in the ClpP-ADEP1 structure resembles that adopted when ClpP is bound to Clp ATPases.
Stabilization of the N-terminal Loops Creates a Structured Axial Pore
The structure of the N-terminal region has been a matter of debate. None of the previous studies unequivocally traced all the residues of the N-terminal sequence; however, they all concurred that this region is mobile in “apo” forms of ClpP. Indeed, B-factors of the N-terminal region in previous structures are significantly higher than those in the remainder of the molecule, indicating the increased flexibility of the N-terminal loops (Figure S5C).
Variations of the β-hairpin conformation seen in our structure have been observed in other ClpP structures (Figure S5A) (Bewley et al., 2006; Gribun et al., 2005; Kang et al., 2004; Kim and Kim, 2008; Szyk and Maurizi, 2006). In the E. coli ClpP structure reported by Bewley et al. (2006) (PDB ID: 1YG6), one heptamer had seven nearly complete loops in various orientations that essentially closed off the axial channel, while the opposite heptamer had disordered N-terminal loops that left the properties of the channel in question. In a mutant form of Streptococcus pneumoniae ClpP (Gribun et al., 2005), a rigid body movement of the hairpin dramatically narrowed the axial pore. In every case, the lumen of the pore delimited by the N-terminal regions is not bigger than 12 Å, a diameter that could only accommodate the passage of unfolded single-chain polypeptides. In our structure the N-termini of the ClpP ring are retracted further from the lumen defining a pore of 20 Å (Figure 4B and Figure S5A and B), large enough to allow the entry of two or three polypeptide chains from disulfide-cross-linked substrate dimers but sufficient to restrict passage of folded proteins (Burton et al., 2001).
In our structure, the N-terminal loops adopt an open conformation only when they are not engaged in crystal contacts (Figure S4A, chains A-N), whereas they are disordered in monomers that have close packing contacts in their N-terminal region (Figure S4B, chains R and S). In the recently reported structures of ADEP1 and ADEP2 bound to B. subtilis ClpP, the N-terminal loops are not visible, leading the authors to conclude that binding increases the flexibility of this region (Lee et al., 2010a). In contrast to our structure, contacts between the apical surfaces of B. subtilis ClpP do mediate crystal packing, leaving virtually no space for the N-terminal loops (Figure S4C). Therefore, the increased flexibility of the N-terminal region could likely be a consequence of the crystal packing that prevented the N-terminal loops from adopting the 〈-hairpin conformation found in the structure of E. coli ClpP bound to ADEP1.
To assay whether the ClpP loops are less flexible in solution after treatment with ADEP1, we used limited protease digestion to probe the accessibility of residues in the loop. ClpP is generally resistant to proteolysis, but Staphylococcus aureus protease V8 cleaves Glu8 and Glu14 in the N-terminal loop of “apo” ClpP, indicating that this region is accessible. When we performed limited proteolysis of ClpP in the presence of ADEP1, cleavage was significantly slower than in the absence of ADEP1 (Figure S6). These data support our conclusion that the N-terminal loops become less flexible as a result of ADEP1 binding.
ClpP-ADEP1 Structure Provides a Model for the ClpX/ClpA Bound State of ClpP
ADEP1 binds in the hydrophobic pocket deemed important for the interaction with ClpX and ClpA (Kim et al., 2001; Martin et al., 2007). We found that the conserved LGF motif in the crystal structure of the monomer of Helicobacter pylori ClpX (PDB ID 1UM8) (Kim and Kim, 2003) can be directly superimposed onto the aliphatic tail and the phenylalanine moiety of ADEP1 without steric hindrance between ClpP and other portions of ClpX (Figure 5C). In the structure of a hexameric form of E. coli ClpX (Glynn et al., 2009), the IGF loops were disordered, but superposition of the H. pylori ClpX monomer onto the hexamer suggests that the LGF loop would have to twist by at least 90° to accommodate axial alignment of the ClpX and ClpP rings and proper orientation of the LGF motifs in the hydrophobic pockets (data not shown). Such movement is feasible, because the regions immediately preceding and following the LGF loop (helices α8 and α9, respectively) in H. pylori ClpX exhibit B-factors that are much higher than the rest of the protein (Kim and Kim, 2003). Our finding that residues 1-7 remain fixed within the axial channel in the open ClpP state are also in concert with a recent cryo-electron microscopy structure of ClpP in complex with ClpA (Effantin et al., 2010), which showed residual density within the lumen of the open channel in the ClpAP complex. Collectively, the data favor the model that ADEP1 mimics the conserved IGF motif binding to the hydrophobic pocket of ClpP, and consequently our structure provides a platform to delineate the conformational changes induced in ClpP upon binding of the ATPase components.
ADEPs do not eliminate the requirement for the ClpA or ClpX to unfold structured substrates prior to degradation by ClpP; however, an immediate question arising from our study is whether the ability of ClpP to take up unfolded proteins is sufficient to account for degradation of proteins unfolded by ClpA and ClpX complexed with ClpP. In ClpXP and ClpAP complexes, the ATPase mediates substrate selection and unfolding, and current models suggest that they also actively translocate proteins into ClpP (Martin et al., 2008; Singh et al., 2000). An alternative possibility is that the Clp ATPases translocate unfolded proteins into the vestibule between the ATPase and ClpP and that the unfolded proteins rapidly diffuse through the expanded axial channel in ClpP rather than being actively driven into the chamber. While ADEP-activated ClpP can make only one or two cuts in the time ClpAP completely degrades proteins, processive cleavage by ADEP-activated ClpP might be prevented because the cleaved protein is free to diffuse away. In the ClpAP and ClpXP complexes, the ATPase prevents the partially cleaved unfolded protein from escaping and diffusion into the ClpP chamber would continue. Studies with proteasomes have suggested that some unfolded proteins can trigger gate opening even to the extent of allowing folded domains to which they are fused to be degraded in the absence of an ATP-dependent unfoldase (Liu et al., 2003). Thus, to what extent translocation into the degradation chamber of ClpP or proteasomes is energy-dependent in the context of the holoenzyme complex remains an open question.
Maximal opening of the axial pore is obtained when the N-terminal region of all seven ClpP monomers adopts the well-defined conformation described in our structure of ClpP with ADEP1 bound (Figure 4B). This seven-fold symmetry of the pore is achieved by binding one molecule of ADEP1 to each one of the seven hydrophobic clefts in the heptameric ring (Figure 4B). However, in ClpAP or ClpXP complexes, the seven hydrophobic pockets of ClpP cannot be bound simultaneously by the six IGF/L loops in the ATPase hexamer. In principle, only two or three IGF/L loops might interact simultaneously with hydrophobic clefts in ClpP without major conformational changes affecting the positions or orientations of the loops. Nonetheless, mutational data indicate that at least five, and most favorably six, IGF loops are needed for formation of stable complexes between ClpX and ClpP (Martin et al., 2007). It is possible that the asymmetry in the ClpX hexamer (Glynn et al., 2009) enables additional loops to be positioned for interaction, and the flexibility of the polypeptides flanking the IGF loops noted earlier should contribute as well (Kim and Kim, 2003). The sigmoidal response of ClpP peptidase activity to ADEP1 concentration hints at some degree of cooperativity, which could involve a concerted induction of the extended conformation in the remaining loops after some threshold number of the sites are occupied. Interaction of the first few IGF/L loops might trigger conformational changes in the respective N-terminal loops, which in turn promote changes in other subunit interfaces or interactions between adjacent N-terminal regions. The extended conformation of the loop in subunit b, with its low ADEP1 occupancy, might be attributed to allosteric effects from adjacent occupied subunits.
Model for the Activation of ClpP by ADEPs and Implications of this Study
In conclusion, this work provides a description of the molecular mechanism of activation of the proteolytic activity of ClpP by ADEP1 (Figure 6). The compound docks into the hydrophobic clefts located on the apical surface of each ClpP ring. This interaction locks the N-terminal region of ClpP into a well-structured conformation that opens an axial pore of ~20 Å diameter. This effect removes normal regulatory constraints on ClpP allowing uncontrolled access to unfolded proteins. This study also constitutes an initial foray into a structural understanding of the communication between the ATPase and protease components in the ClpXP and ClpAP complexes. However, in ClpXP complexes, the N-terminal loop of ClpP makes at least transient contact with the pore-2 loop of ClpX (Martin et al., 2007). Such an interaction might further stabilize the extended β-hairpin observed in the current structure. As the pore loops of ClpX and ClpA are dynamic and responsive to changes in nucleotide states, such interactions probably also provide a mechanism by which the size or shape of the proximal portion of the ClpP channel would remain dynamic, which might allow a broader range of translocating substrates to be accommodated. Nevertheless, given the inherent difficulty of obtaining co-structures of these complexes, the characterization of ClpP, ClpX, and ClpA bound to small molecules that mimic interactions between these proteins will remain a promising approach to understand the allosteric communication between the Clp ATPases and ClpP.
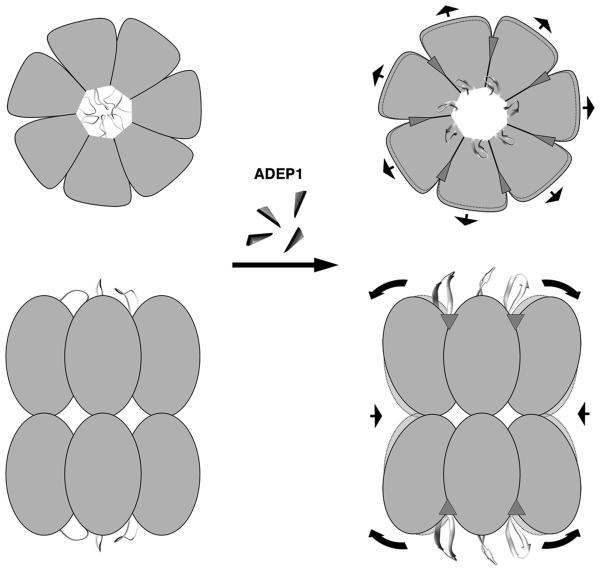
Figure 6. Model for the Activation of ClpP by ADEPs.
Schematic representations of a top and a side-view of ClpP in the absence and presence of ADEP1 are shown in the left and right hand panels, respectively. The N-terminal regions of the ClpP monomers in the absence of ADEP1 are shown in multiple conformations representing the flexible nature of this region and the 12 Å diameter pore that they delimitate. This small diameter pore restricts the passage of protein substrates to the digestion chamber. ADEP1 molecules, represented as small triangles, dock into the seven hydrophobic clefts located on the apical surface of each ClpP ring. Upon binding, ADEP1 locks the ClpP N-terminal loops in a β-hairpin conformation retracting these loops from the lumen and generating a stable pore of 20 Å diameter through which extended polypeptides can be threaded into the degradation chamber. ADEP1 binding also triggers an outward movement of the ClpP head domain causing a subtle expansion of the apical surface of the ring. Simultaneously, the equator of the tetradecamer formed by the ClpP handle domains slightly contracts as a result of the rigid body movement of the ClpP monomers. The arrows indicate the direction of these movements and the areas delimitated by dotted lines represent the ClpP structure before ADEP1 binding and are shown for reference.
EXPERIMENTAL PROCEDURES
Isolation of ADEP1 from Streptomyces hawaiiensis
ADEP1 was purified to at least 95% homogeneity from the fermentation broth of Streptomyces hawaiiensis NRRL 15010 according to (Michel and Kastner, 1985) with minor modifications. This strain was obtained from the US Agricultural Research Service Culture Collection (NRRL). Amberlite XAD16 and Diaion HP-20 resins were purchased from Sigma-Aldrich (St. Louis, MO) and all other chemicals and solvents for the purification were from Fisher Scientific (Pittsburgh, PA) and associated providers. The final compound preparations were verified by mass spectrometry (MS) and by nuclear magnetic resonance (NMR) analysis.
Protein Expression and Purification
To over express ClpA, ClpP and the ClpP-R166C mutant, the pBAD33-ClpA, pAT9a-ClpP and pET3d-ClpP-R166C plasmids were transformed into E. coli BL21 (DE3) competent cells. Over expression and purification of these proteins was performed as described previously (Maurizi et al., 1994). To cross-link ClpP-R166C, the purified protein (500 μg/ml in 50 mM Tris/HCl (pH 7.5), 0.2 M KCl, and 10% (v/v) glycerol) was incubated with 20 μM of the bifunctional reagent, BM(PEG)3 (Pierce) for 30 min on ice. The unreacted reagent was removed by passing the protein over Sephadex G25 column in the same buffer. The extent of cross-linking (>90%) was confirmed by SDS-PAGE. ClpPin was prepared by treating purified ClpP with carbobenzoxy-Leu-Tyr-chloromethylketone (Singh et al., 1999). Peptidase and protease activity of ClpPin was measured before and after the treatment. Over expression and purification of EGFP-SsrA with a N-terminal His-tag was done according to previous studies (Iwanczyk et al., 2007).
Peptidase and Protease Activity Assays
Peptidase activity of ClpP in the presence and absence of ADEP1 was assayed at 37 °C using N-Succinyl-Leu-Tyr-7-amido-4-methylcoumarin and oxidized insulin B chain (both from Sigma) or the peptide, FAPHMALVPV (F-V). Assay mixtures (50 μl) contained 100 mM Tris/HCl, pH 8.0 with 0.1 M KCl and reactions were initiated by addition of ClpP as indicated. Loss of substrate or appearance of products was assayed according to published protocols (Maurizi et al., 1994; Thompson and Maurizi, 1994). When substrates were present at subsaturating concentrations, assays times were limited to permit <10% cleavage in order to maintain initial rate conditions.
Protease assays were assembled in 100 μL reactions containing 4.6 μM of ClpP monomer in 50 mM Tris/HCl (pH 7.5), 0.2 M KCl, and 1 mM DTT. ClpP was incubated on ice for 2 min with a 5 molar excess of ADEP1 added from a stock solution in DMSO. Control reactions contained an equivalent amount of DMSO as a control. Reactions were started by adding 9 μM of bovine β-casein (Sigma). ClpA-dependent reaction conditions were the same as described previously with α-casein in place of β-casein (Thompson and Maurizi, 1994). All reactions mixtures were incubated at 37°C and quenched by addition of hot SDS sample buffer. Proteins were resolved by SDS-PAGE on 15% polyacrylamide gels and visualized by staining with Coomassie Brilliant Blue.
Analytical Ultracentrifugation
Sedimentation velocity experiments were conducted at 20.0°C on a Beckman Coulter Proteome XL-I analytical ultracentrifuge using the Rayleigh interference detection optics. ClpP samples purified in 50mM Tris/HCl (pH 7.5) and 0.2M KCl were studied at loading concentrations ranging from 101 to 27 μM of ClpP monomer. Samples containing ClpP and ADEP1 were loaded at concentrations 53, 14 and 7 μM of ClpP monomer. The ADEP compounds were dissolved in pure DMSO at high concentration (40 mM) and added to the sample such that at least 5 stoichiometric equivalents of ADEP1 per ClpP monomer were present. All samples were loaded into 2-channel, 12 mm path length sector shaped cells (400 μL) and 50 scans were acquired at approximately 7-min intervals and rotor speeds of 40 krpm. Data were analyzed in SEDFIT 11.9b (Schuck, 2000) in terms of a continuous c(s) distribution. The solution density ρ and viscosity η were calculated using the program SEDNTERP 1.2 (Cole et al., 2008). The partial specific volume of ClpP was also calculated using SEDNTERP. The c(s) analyses were carried out using an s range of 0 to 15 with a linear resolution of 150 and confidence levels (F-ratio) of 0.68. In all cases, reasonable fits were observed with root mean square deviations ranging from 0.0284 – 0.0044 fringes. Sedimentation coefficients were corrected to standard conditions at 20.0°C in water, s20,w.
Electron Microscopy
To visualize the effect of ADEP1 in the translocation of β-casein and EGFP-SsrA into ClpPin using electron microscopy, reactions were assembled in 50μL of 50mM Tris/HCl (pH 7.5), 0.2M KCl, and 20mM MgCl2 by adding 0.93 μM of ClpPin monomer previously incubated on ice with 5 molar excess of ADEP1 or equivalent amount of DMSO. Respective reactions were started by adding equimolar amount of β-casein or EGFP-ssrA and at the indicated time points, 10 μL were taken and applied on grids for negative staining.
All samples were applied by floating a 10 μL drop to carbon-coated grids previously glow discharged and negatively stained with 2% uranyl acetate. Specimens were observed in a JEOL 2010F electron microscope operated at 200 kV. Images were collected at 50,000x with a dose of ~10 electrons/Å2 and a defocus of 2.7 μm. All images were recorded on Kodak SO-163 films, scanned on a Nikon super COOLSCAN 9000 ED at 6.35 mm/pixel and averaged 2x to produce data at 2.54 Å/pixel. Particles were extracted interactively from the digitized electron micrographs using the Boxer (EMAN) program (Ludtke et al., 1999). Two-dimensional averages were obtained using cross-correlation based methods using the Xmipp software package (Scheres et al., 2008).
Crystallization, Data collection and Structure Determination
The ClpP-ADEP1 complex was assembled by mixing ClpP (20 mg/mL in 0.01 M MES pH 6.5 and 0.2 M NaCl) with ADEP1 (in 100% DMSO) at 1:2 ratio. Crystals of the complex were grown in 25-35% (v/v) MPD and 0.1 M sodium acetate at pH 5. A complete data set diffracting to 1.9 Å was collected at the X25 beam line (NSLS, Brookhaven National Laboratory). Data were indexed, processed and merged using HKL2000 (Table 1) (Otwinowski and Minor, 1997). Initial phases were determined by molecular replacement using PHASER (McCoy et al., 2007). Two complete ClpP tetradecamers were found in the asymmetric unit. Refinement and model building were done using standard protocols in phenix.refine and COOT (Afonine et al., 2005; Emsley and Cowtan, 2004). The restraints for the ADEP1 molecule were generated using the PRODRG server and phenix.elbow (Moriarty et al., 2009; Schuttelkopf and van Aalten, 2004). Over 96% of the residues in the final model are found in the most favored regions of the Ramachandran plot and none in the disallowed regions. Figures depicting molecular structures were generated using PyMol (DeLano, 2002).
Table 1.
Data Collection and Refinement Statistics
|
Data Collection and Processing
| |
| Wavelength (Å) | 0.9795 |
| Unit Cell (Å, °) | a= 93.3, b= 121.2, c= 276.2, β=91.4° |
| Resolution (Å) | 30-1.9 (1.93-1.9) |
| Space Group | P21 |
| Total Reflections | 3046626 |
| Unique Reflections | 472053 |
| Mean I/σ(I) | 27.0 (3.0) |
| Rmerge (%) | 6.5 (40.3) |
| Completeness (%) | 98.3 (84.0) |
| Redundancy | 6.5 (4.6) |
|
| |
|
Refinement | |
| Reflections (work) | 471,961 |
| Reflections (test) | 5,212 |
| Atoms refined | 46,555 |
| Solvent atoms | 3,651 |
| Rwork/Rfree (%) | 20.1/17.0 |
| Rmsd in bond lengths (Å) | 0.007 |
| Rmsd in bond angles (°) | 1.269 |
Values in parentheses refer to the highest resolution shell.
SIGNIFICANCE.
Identification of the self-compartmentalized ClpP protease, a key bacterial enzyme for maintenance of cellular protein homeostasis, as the target for the new class of antibiotics acyldepsipeptides (ADEPs) has been heralded as a major advance in the search for new drug leads. However the molecular mechanism of ADEP activity has been elusive. ClpP forms a tetradecamer that encloses a central hollow chamber containing the proteolytic sites. ClpA and ClpX ATPases unfold and translocate proteins into the degradation chamber through axial pores gated by the N-terminal region of ClpP. Here, we presents the crystal structure of Escherichia coli ClpP complexed with ADEP1 and reveals the activation mechanism of the enzyme by this antibiotic. We show that binding of ADEP1 bypasses the requirement for the ATPases by locking the N-terminal loops of ClpP in a β-hairpin conformation that defines a 20 Å diameter axial pore. This conformational change allows unfolded proteins to enter the degradation chamber. In addition, we found that binding of ADEP1 mimics the interaction of ClpP with its cognate ATPases, ClpX or ClpA. Consequently, our structure provides the first snapshot of the conformation of ClpP bound to a Clp ATPase in a configuration that accepts translocated substrates. Given the inherent difficulty of obtaining co-structures of ClpAP and ClpXP complexes, our manuscript shows that the characterization of ClpX, ClpA and ClpP bound to small molecules is a promising approach to understand the allosteric communication between these proteins. Finally, the fundamental understanding of the mode of interaction of ADEP1 with ClpP presented here can now be used to further explore the ADEP chemical scaffold for the development of more efficient antibiotics and to further exploit ClpP as a new antibiotic target.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr. Walid A. Houry for providing the pAT9a-ClpP plasmid. We are in debt with Dr. M. Junop and the staff at the X25 beam line (BNL, NSLS) for assistance during data collection. This work was supported by a grant from the Canadian Institutes of Health Research to JO (MOP-82930) and GDW (MT-14981), by a Canada Research Chair in Antibiotic Biochemistry (to GDW) and an Early Researcher Award to AG (Ontario Ministry of Research Innovation). JO and AG are supported by the New Investigators program from CIHR. This work was also supported by the intramural research program of the Center for Cancer Research, NCI (MRM and EJ) and the National Institute of Diabetes and Digestive and Kidney Diseases (RG), NIH, Bethesda, MD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Atomic coordinates and structure factors of ClpP-ADEP1 have been deposited in the Protein Data Bank (accession code 3MT6).
SUPPLEMENTAL INFORMATION
Supplemental Data include six figures, one table and one movie that can be found with this article online.
REFERENCES
- Afonine PV, Grosse-Kunstleve RW, Adams PD. phenix.refine. CCP4 Newsletter. 2005;42 contribution 8. [Google Scholar]
- Beuron F, Maurizi MR, Belnap DM, Kocsis E, Booy FP, Kessel M, Steven AC. At sixes and sevens: characterization of the symmetry mismatch of the ClpAP chaperone-assisted protease. J Struct. Biol. 1998;123:248–259. doi: 10.1006/jsbi.1998.4039. [DOI] [PubMed] [Google Scholar]
- Bewley MC, Graziano V, Griffin K, Flanagan JM. The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes. J Struct. Biol. 2006;153:113–128. doi: 10.1016/j.jsb.2005.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley MC, Graziano V, Griffin K, Flanagan JM. Turned on for degradation: ATPase-independent degradation by ClpP. J Struct. Biol. 2009;165:118–125. doi: 10.1016/j.jsb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotz-Oesterhelt H, Beyer D, Kroll HP, Endermann R, Ladel C, Schroeder W, Hinzen B, Raddatz S, Paulsen H, Henninger K, et al. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 2005;11:1082–1087. doi: 10.1038/nm1306. [DOI] [PubMed] [Google Scholar]
- Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. Embo J. 2001;20:3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JL, Lary JW, T PM, Laue TM. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 2008;84:143–179. doi: 10.1016/S0091-679X(07)84006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphic Systems. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- Effantin G, Maurizi MR, Steven AC. Binding of the CLP-A unfoldase opens the axial gate of CLP-P peptidase. J. Biol. Chem. 2010;139:14834–14840. doi: 10.1074/jbc.M109.090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Maurizi MR, Wickner S. Regulatory subunits of energy-dependent proteases. Cell. 1997a;91:435–438. doi: 10.1016/s0092-8674(00)80428-6. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Wickner S, Maurizi MR. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997b;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- Gribun A, Kimber MS, Ching R, Sprangers R, Fiebig KM, Houry WA. The ClpP double ring tetradecameric protease exhibits plastic ring-ring interactions, and the N termini of its subunits form flexible loops that are essential for ClpXP and ClpAP complex formation. J. Biol. Chem. 2005;280:16185–16196. doi: 10.1074/jbc.M414124200. [DOI] [PubMed] [Google Scholar]
- Guo F, Maurizi MR, Esser L, Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J. Biol. Chem. 2002;277:46743–46752. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- Hoskins JR, Pak M, Maurizi MR, Wickner S. The role of the ClpA chaperone in proteolysis by ClpAP. Proc. Natl. Acad. Sci. U S A. 1998;95:12135–12140. doi: 10.1073/pnas.95.21.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Beuron F, Kessel M, Wickner S, Maurizi MR, Steven AC. Translocation pathway of protein substrates in ClpAP protease. Proc. Natl. Acad. Sci. U S A. 2001;98:4328–4333. doi: 10.1073/pnas.081543698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanczyk J, Damjanovic D, Kooistra J, Leong V, Jomaa A, Ghirlando R, Ortega J. The Role of the PDZ Domains in Escherichia coli DegP Protein. J. Bacteriol. 2007;189:3176–3185. doi: 10.1128/JB.01788-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Maurizi MR, Thompson M, Mueser T, Ahvazi B. Crystallography and mutagenesis point to an essential role for the N-terminus of human mitochondrial ClpP. J. Struct. Biol. 2004;148:338–352. doi: 10.1016/j.jsb.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Kim DY, Kim KK. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J. Biol. Chem. 2003;278:50664–50670. doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- Kim DY, Kim KK. The structural basis for the activation and peptide recognition of bacterial ClpP. J. Mol. Biol. 2008;379:760–771. doi: 10.1016/j.jmb.2008.04.036. [DOI] [PubMed] [Google Scholar]
- Kim YI, Levchenko I, Fraczkowska K, Woodruff RV, Sauer RT, Baker TA. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat. Struct. Biol. 2001;8:230–233. doi: 10.1038/84967. [DOI] [PubMed] [Google Scholar]
- Kirstein J, Hoffmann A, Lilie H, Schmidt R, Rubsamen-Waigmann H, Brotz-Oesterhelt H, Mogk A, Turgay K. The antibiotic ADEP reprogrammes ClpP, switching it from a regulated to an uncontrolled protease. EMBO Mol. Med. 2009;1:37–49. doi: 10.1002/emmm.200900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BG, Park EY, Lee KE, Jeon H, Sung KH, Paulsen H, Rubsamen-Schaeff H, Brotz-Oesterhelt H, Song HK. Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nat. Struct. Mol. Biol. 2010a;17:471–478. doi: 10.1038/nsmb.1787. [DOI] [PubMed] [Google Scholar]
- Lee ME, Baker TA, Sauer RT. Control of Substrate Gating and Translocation into ClpP by Channel Residues and ClpX Binding. J. Mol. Biol. 2010b;399:707–718. doi: 10.1016/j.jmb.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CW, Corboy MJ, DeMartino GN, Thomas PJ. Endoproteolytic activity of the proteasome. Science. 2003;299:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Distinct static and dynamic interactions control ATPase-peptidase communication in a AAA+ protease. Mol. Cell. 2007;27:41–52. doi: 10.1016/j.molcel.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat. Struct. Mol. Biol. 2008;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizi MR. Proteases and protein degradation in Escherichia coli. Experientia. 1992;48:178–201. doi: 10.1007/BF01923511. [DOI] [PubMed] [Google Scholar]
- Maurizi MR, Thompson MW, Singh SK, Kim SH. Endopeptidase Clp: ATP-dependent Clp protease from Escherichia coli. Methods Enzymol. 1994;244:314–331. doi: 10.1016/0076-6879(94)44025-5. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel KH, Kastner RE. US patent 4492650 A54556 antibiotics and process for production thereof. 1985
- Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega J, Singh SK, Ishikawa T, Maurizi MR, Steven AC. Visualization of substrate binding and translocation by the ATP- dependent protease, ClpXP. Mol. Cell. 2000;6:1515–1521. doi: 10.1016/s1097-2765(00)00148-9. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW, Sweeet RM, editors. Methods in Enzymology. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, et al. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat. Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc. Natl. Acad. Sci. U S A. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Guo F, Maurizi MR. ClpA and ClpP remain associated during multiple rounds of ATP-dependent protein degradation by ClpAP protease. Biochemistry. 1999;38:14906–14915. doi: 10.1021/bi991615f. [DOI] [PubMed] [Google Scholar]
- Singh SK, Rozycki J, Ortega J, Ishikawa T, Lo J, Steven AC, Maurizi MR. Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis. J. Biol. Chem. 2001;9:9. doi: 10.1074/jbc.M103489200. [DOI] [PubMed] [Google Scholar]
- Szyk A, Maurizi MR. Crystal structure at 1.9A of E. coli ClpP with a peptide covalently bound at the active site. J. Struct. Biol. 2006;156:165–174. doi: 10.1016/j.jsb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Thompson MW, Maurizi MR. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J. Biol. Chem. 1994;269:18201–18208. [PubMed] [Google Scholar]
- Thompson MW, Singh SK, Maurizi MR. Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli. Requirement for the multiple array of active sites in ClpP but not ATP hydrolysis. J. Biol. Chem. 1994;269:18209–18215. [PubMed] [Google Scholar]
- Wang J, Hartling JA, Flanagan JM. The structure of ClpP at 2.3 A resolution suggests a model for ATP- dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- Woo KM, Chung WJ, Ha DB, Goldberg AL, Chung CH. Protease Ti from Escherichia coli requires ATP hydrolysis for protein breakdown but not for hydrolysis of small peptides. J. Biol. Chem. 1989;264:2088–2091. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.