Abstract
A detailed conception of intranuclear messenger ribonucleoprotein particle (mRNP) dynamics is required for the understanding of mRNP processing and gene expression outcome. We used complementary state-of-the-art fluorescence techniques to quantify native mRNP mobility at the single particle level in living salivary gland cell nuclei. Molecular beacons and fluorescent oligonucleotides were used to specifically label BR2.1 mRNPs by an in vivo fluorescence in situ hybridization approach. We characterized two major mobility components of the BR2.1 mRNPs. These components with diffusion coefficients of 0.3 ± 0.02 μm2/s and 0.73 ± 0.03 μm2/s were observed independently of the staining method and measurement technique used. The mobility analysis of inert tracer molecules revealed that the gland cell nuclei contain large molecular nonchromatin structures, which hinder the mobility of large molecules and particles. The mRNPs are not only hindered by these mobility barriers, but in addition also interact presumably with these structures, what further reduces their mobility and effectively leads to the occurrence of the two diffusion coefficients. In addition, we provide evidence that the remarkably high mobility of the large, 50 nm-sized BR2.1 mRNPs was due to the absence of retarding chromatin.
Introduction
The delivery of nucleoplasmic mRNA from the transcription site to the cytoplasmic ribosomes is one of the key transport processes in eukaryotic cells. Upon transcription, the nascent mRNA associates with proteins and thus forms messenger ribonucleoprotein particles (mRNPs) (1). The subsequent intranuclear trafficking and nuclear export of mRNPs is regulated (2–6). This regulation presumably has a strong impact on the gene expression outcome, but is poorly understood. In addition, evidence accumulates that RNA itself is directly involved in gene regulation (7). Such effects and functions are traceable by studying intranuclear mRNA dynamics, and therefore intranuclear mRNP trafficking is a topic of intense research (8–19). Over the years, a potent and impressive arsenal of different mRNP labeling and imaging techniques has been developed to perform such studies (20).
In mammalian cells, the interphase nucleus is crowded with both hetero- and euchromatin, affecting the mobility of every macromolecule or macromolecular complex, and of course also that of mRNPs (21). Using the almost pointlike observation volume of a fluorescence correlation spectroscopy (FCS) instrument, Politz et al. (13) showed that mRNP particles can move in cell nuclei almost as fast as in aqueous solution, with diffusion coefficients up to 10 μm2/s (13,14). These researchers argued that such fast diffusion could occur within interchromatin channels (22). In addition, massively reduced diffusion coefficients were also found, likely corresponding to particles retarded due to processing steps, which suggested that complex regulatory mechanisms govern intranuclear mRNA diffusion. Unfortunately, retardation due to nonchromatin structures, which would be of special interest because it would point to functional processes, could not be distinguished from passive crowding effects of the unfolded chromatin itself. Using video microscopy to analyze diffusion on longer timescales and greater spatial domains than possible by FCS, Politz et al. (13) demonstrated that the extension of the intranuclear observation field has a significant impact on the obtained effective diffusion coefficient (14), making it difficult to interpret the mobility data with respect to mRNP processing and binding: whereas FCS measures the diffusion within a confocal volume <1 μm3, techniques like fluorescence recovery after photobleaching or photoactivation measure the effective diffusion over a field of many square microns. In this work, we show that, for longer distances, mobility restrictions exerted by euchromatin will have a great impact on the mobility of relatively bulky mRNPs.
Taken together, studying the specific interaction of mRNP particles with their nuclear environment is quite challenging in mammalian cells. It would be most instructive to visualize single mRNPs on their track through the nucleoplasm and to distinguish mobility restriction due to chromatin obstruction from that caused by specific binding events to nonchromatin structures.
One cell system allowing such studies is that of the Balbiani Ring (BR) mRNPs in the salivary gland cell nuclei from Chironomus tentans. These nuclei are large, and though being in interphase, their chromatin is concentrated in polytene chromosomes, which are surrounded by a vast region of chromatin-free nucleoplasm (23,24). Here exists the unique situation, where mRNP mobility can be studied without the obstruction of chromatin. Several genes coding for salivary proteins are up to 40 kilobases and the size of their mRNA is only slightly reduced by splicing (25). The corresponding mRNPs have a diameter of ∼50 nm and contain internal repetitive sequences. Complementary fluorescently labeled oligonucleotides can specifically bind and label the endogenous BR2.1 mRNPs. Using this experimental system, we previously visualized and tracked the pathways of single BR2.1 mRNPs by single molecule microscopy (17). We observed that BR2.1 mRNPs moved in the chromatin-free nucleoplasm in a discontinuous manner, meaning that the mRNPs could not be sorted into slow and fast fractions; instead, the mode of motion changed along the trajectories. Consequently, the mobility could not be quantified by a single diffusion coefficient, but required the consideration of different mobility components. Thus, three main mobility components were found with D1 = 0.24 μm2/s, D2 = 0.7 μm2/s, and D3 = 4 μm2/s. In addition, at times the particles were retarded down to D0 = 0.015 μm2/s, which corresponded virtually to immobility. However, the exact reason for the discontinuous mobility remained to be elucidated.
In this work we focused on this problem. We performed in-depth studies of BR2.1 mRNP and also of tracer molecule mobility by single molecule tracking and fluorescence correlation spectroscopy (FCS) to understand the intranuclear dynamics of mRNPs on different time- and length scales. The complementary techniques of single molecule tracking and FCS and the comparison of tracer mobility in salivary gland cells and mammalian cells revealed new, to our knowledge, aspects of nucleoplasmic mRNP dynamics and provided a rationale for the occurrence of the different BR mRNP mobility components.
Materials and Methods
Buffer and reagents
Fluorescent RNA probes used for in vivo hybridization of the BR mRNA were from IBA, Göttingen, Germany. The specific 30-mer BR2.1 RNA probe,
and the unspecific 30-mer CTR RNA probe,
were Atto647N-labeled on the 5′ end and were dissolved in water with a concentration of 0.1 nmol/μL.
The specific BR2.1 MB RNA probe,
and the unspecific CTR MB RNA probe,
were Atto-Fluor 647N-labeled on the 5′ end and BHQ2-labeled on the 3′ end. The stem sequence of the MBs is underlined. MBs were dissolved in water with a concentration of 0.16 nmol/μL.
Both singly labeled RNA probes and molecular beacons were synthesized from 2′-O-methyl-RNA-oligonucleotides. An estimate of the number of oligonucleotides that can maximally bind to a single BR2.1 mRNP was given previously (17). Transport buffer was used for the dilution of microinjection probes (20 mM HEPES/KOH (pH 7.3), 110 mM potassium acetate, 5 mM sodium acetate, 2 mM magnesium acetate, 1 mM EGTA, and 2 mM DTT).
Amino-derivatized dextran (molecular mass 500 kDa; Invitrogen, Darmstadt, Germany) was dissolved in 0.1 M NaHCO3, pH 8, and fluorescence-labeled with a fivefold excess of ATTO647 succidinimidyl ester (ATTO-TEC, Siegen, Germany). Labeling reactions were set up at room temperature for 2 h and free dye was removed by gel filtration on a BioRad-P6 desalting column (molecular mass cutoff 6 kDa; BioRad, Munich, Germany). Labeled probes were finally size-fractionated on a Superose 12 column to remove smaller fragments, and were stored at 4°C.
Preparation of salivary glands
Preparation of C. tentans salivary glands was performed as described earlier (17).
Cell culture
HeLa cells stably expressing histone 2B conjugated to the green fluorescent protein (H2B-GFP) were a kind gift from Heinrich Leonhardt (Ludwig-Maximilians-Universität, Munich, Germany). Cells were cultivated in Dulbecco's modified Eagle's Medium (Biochrom, Berlin, Germany) containing 20 mM HEPES with 10% fetal calf serum (HyClone/Perbio, Bonn, Germany) plus 5 mM L-glutamine (Biochrom) and the antibiotics penicillin (100 U/mL) and streptomycin (100 μg/mL) (Biochrom), as well as 100 μg/mL gentamycin (Gibco/Invitrogen, Grand Island, NY). For live cell microscopy, cells were seeded on dishes (MatTek, Ashland, MA) 1–2 days before measurements. All measurements were performed for <40 min at room temperature.
Fluorescence video microscopy
For single molecule microscopy, standard RNA oligonucleotides were diluted 1:400, MB RNA oligonucleotides were diluted 1:200–1:400, and dextrans 1:1000 in transport buffer, and microinjected in freshly prepared salivary gland cell nuclei. A custom-built high-speed fluorescence video microscope based on an inverted microscope was employed (26), which was equipped with an LCI Plan Neofluar 63×/1.3 Imm Korr PH3 water immersion or a 63×/1.4 oil immersion objective (Zeiss, Göttingen, Germany). The frame rate was 100 Hz (10.22-ms acquisition time) or 200 Hz (5.22-ms acquisition time). Identification and tracking of the single-particle signals were accomplished by using Diatrack 3.03 (Semasopht, www.semasopht.com). In cells, tracking was only performed in the nucleoplasm. All further data processing was performed using Origin 8 (OriginLab, Northampton, MA) and ImageJ (http://rsbweb.nih.gov/ij/). Heterogeneous mobility populations can be analyzed by a jump-distance analysis. The probability for a particle starting at a specific position to be encountered within a distance r and width dr after a time t is given as
| (1) |
where D is the diffusion coefficient. The probability distribution function can be approximated by a frequency distribution by counting the jump distances within respective intervals [r, r + dr] traveled by single particles. When particles move discontinuously, the jump-distance distributions cannot satisfactorily be fitted by Eq. 1. Different mobility fractions must be considered by summing up several diffusion terms according to Eq. 1. We used two or three fractions with different diffusion constants
| (2) |
where fj are mobility fractions with different diffusion coefficients Dj and x depends on the number of fractions.
Special care was taken to avoid weighting artifacts in the calculation of the mean-square displacement (MSD). First, it was decided how many time points M the MSD plot should contain. Then, only trajectories detected for M time points or more were selected. Subsequently on this selected data the mth MSD point (m ≤ M) was calculated by averaging the square distances of all particle positions at time point m and time point 0 (trajectory start). Thereby we made sure that each trajectory counted only once for each time point m, independently of the complete trajectory length. Thus, an overweighting of very long and most likely very slow trajectories was avoided (see the Supporting Material for further details). The linear part of the MSD was fitted according to
| (3) |
with the MSD in two dimensions 〈r2〉 and the diffusion coefficient D. Thus, an (initial) diffusion coefficient was calculated.
Fluorescence correlation spectroscopy
FCS measurements were performed using a ConfoCor 1 microscope setup equipped with a water immersion objective (C-Apochromat, 63×, NA 1.2; Zeiss). For calibration of the beam width, Atto647N-maleimide dye molecules (Atto-Tec, Siegen, Germany) with a molecular mass of 870 Da in buffer were used. The diffusion coefficient of Atto655-maleimide dye with a molecular mass of 810 Da has been measured previously with a high precision (27), and a diffusion coefficient of 400 μm2/s was determined. We measured characteristic diffusion times for the two dyes of 63 and 62 μs, respectively. Because both dyes were comparable in molecular mass, we assumed a diffusion coefficient of 400 μm2/s for Atto647N-maleimide as well. The laser power used was ∼5 μW.
For FCS measurements, RNA oligonucleotides were microinjected into freshly prepared salivary gland cell nuclei with a dilution of 1:1000 in transport buffer. A z-scan was recorded to ensure that the measurement was performed inside the nucleoplasm. Data analysis was performed as has been described earlier (28) using FCS ACCESS (Evotec, Hamburg, Germany). An FCS autocorrelation curve was recorded typically within 30–60 s. The theoretical autocorrelation curve for three-dimensional diffusion of up to x mobility components is
| (4) |
where fj are the fractions corresponding to the different diffusion times τD,j, N is the total number of fluorescent molecules, T is the ratio of triplet state, τt is the triplet time, and κ is the axial e−2 beam radius of the laser beam divided by the lateral e−2 beam radius. The κ was determined once a day with a solution of dye in buffer and held constant for in vivo measurements. The κ was usually close to 5. The value τD is related to the diffusion coefficient by
| (5) |
where w is the lateral e−2 beam radius of the laser spot.
Atto647N shows an extended triplet state. This state was visible in every cell measurement we performed, likely due to the long observation time of dye bound to BR mRNPs. Cell measurements were fitted with j = 2 or 3, where one component was related to the extended triplet state with a typical τD of 20–80 μs (data not shown).
Results
Single molecule microscopy of 2′-O-methyl-RNA-labeled BR mRNPs
As previously demonstrated, BR2.1 mRNPs can efficiently and specifically be labeled by fluorescent oligonucleotides, which are complementary to segments of their internal repetitive sequence (17). We showed that single BR2.1 mRNPs can be visualized and found that they move in the nucleoplasm in a discontinuous manner. In the previous approach, we relied on confocal line-scanning microscopy to study BR mRNP motion with a limited frame rate (20 fps) and sensitivity. To improve time resolution and sensitivity, we now performed single-particle tracking experiments with a highly sensitive and fast video microscope. We could reduce the concentration of microinjected oligonucleotides by a factor of 40 and increase the time resolution by a factor of 5, thus minimizing the risk of introducing artifacts and missing fast kinetic events. Furthermore, we only employed fluorescently labeled 2′-O-methyl-RNA-oligonucleotides, which are known to be more stable than DNA-oligonucleotides in the cellular environment (29).
Upon microinjection of fluorescently labeled, specific oligonucleotides, the BR transcription site was immediately stained (Fig. 1 A), demonstrating that the RNA-oligonucleotides virtually instantaneously bind to nascent mRNA transcripts. Not only the nascent mRNPs at the BR were labeled, but also mRNPs in the nucleoplasmic space, which could be visualized and tracked at a single particle level by fluorescence video microscopy at a frame rate of 100 Hz (Fig. 1 B, see Movie S1 in the Supporting Material). We recorded movies revealing the BR mRNP motions in real-time from six cell nuclei from four different glands. A total of 28,861 trajectories comprising 124,172 single jumps from frame to frame were extracted from the movie data as previously described (17). Numerous trajectories showed frequent retardation periods as exemplified in Fig. 1 C, which indicated that the particles moved in a discontinuous manner. The distribution of molecular jumps of the obtained trajectories from frame to frame was determined and analyzed as discussed in previous publications (17,30). The jump distance histogram could not be described by a single diffusion term according to Eq. 1. Instead, as expected, the particles displayed a complex mobility. The histograms could well be described by two major mobility fractions, a slow one with a D1,SPT = 0.3 ± 0.02 μm2/s with a fractional weight of 38 ± 4%, and a faster one with a D2,SPT = 0.73 ± 0.03 μm2/s with a fractional weight of 61 ± 4% (Fig. 2 A). The remaining 1% was due to immobile particles (D = 0.001 μm2/s).
Figure 1.
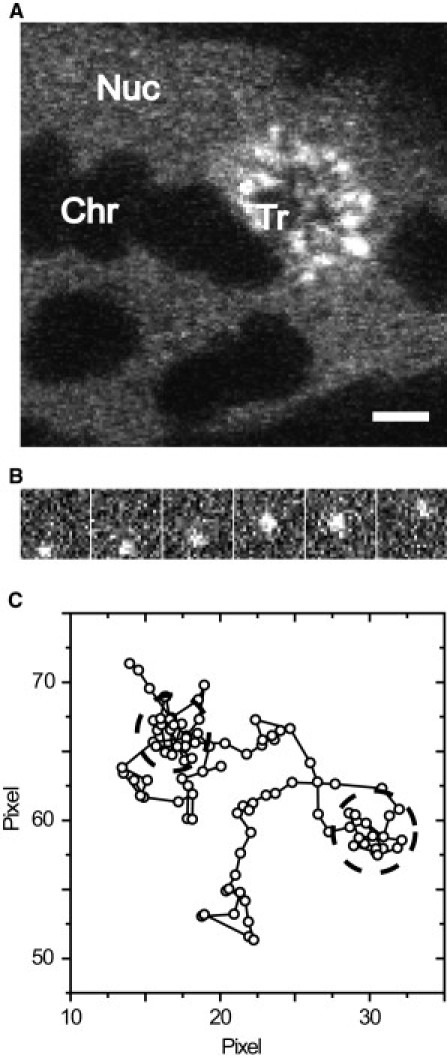
2′-O-methyl-RNA-oligonucleotides were microinjected into salivary gland cell nuclei and analyzed with single particle tracking. (A) Confocal image. Upon microinjection of specific oligonucleotides, the BR2.1 transcription site became visible within a minute. Nuc, nucleoplasm; Chr, polytene chromosome; Tr, transcription site. Bar, 5 μm. (B) Sequence from a typical movie showing every fifth frame. Pixel size is 95.2 nm. (C) Sample trajectory showing discontinuous motion. Pixel size is 95.2 nm.
Figure 2.
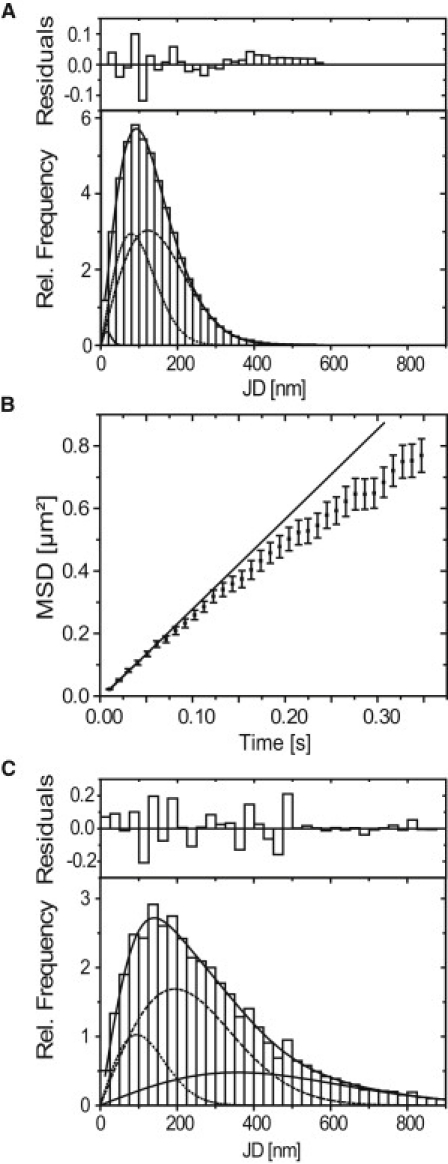
Frame-to-frame jump distance histograms and MSD analysis. The residuals above the jump distance histograms correspond to the overall fit (solid line) due to two or three mobility components. (A) Specific oligonucleotides: D1,SPT = 0.3 ± 0.02 μm2/s with a fractional weight of 38 ± 4% (dots) and D2,SPT = 0.73 ± 0.03 μm2/s with a fractional weight of 61 ± 4% (dashed). The remaining percent to hundred are due to a small fraction with a D of 0.001 μm2/s (solid line, lower left corner). (B) MSD analysis of specific oligonucleotide trajectories with 35 or more jumps. A clearly nonlinear behavior is observed. (Solid line) Linear MSD behavior of the first jumps. The initial diffusion coefficient is 0.71 ± 0.02 μm2/s. (C) Control oligonucleotides: D1,SPT = 0.5 ± 0.1 μm2/s (dots), D2,SPT = 2 ± 0.5 μm2/s (dashed), and D3,SPT = 6.5 ± 2.5 μm2/s (solid). The fractional weights were 17 ± 5%, 54 ± 11%, and 29 ± 14%, respectively.
We already stressed that the three mobility components described above should not be understood as referring to different particles, e.g., as three particle types with different radii. Instead, the different mobility components indicate that the observed particles suffered from retardation, which becomes manifest in the jump distance analysis by deviation from a single diffusion term. It can be demonstrated that the two major components D1,SPT and D2,SPT were found within single trajectories (see the Supporting Material). Finally, it should be noted that the three components of the jumps distance analysis were in agreement with previous results on a quite different timescale from this system (17).
To view the molecular dynamics from a different perspective, we calculated the MSDs for all trajectories with an equal weight for each trajectory (see Materials and Methods). This approach guaranteed that retarded and therefore very long trajectories were not overweighted, which would yield a misleadingly low diffusion coefficient. Despite this approach, a clear deviation from linear behavior indicating anomalous diffusion was found (Fig. 2 B). The results of the jump distance and the MSD analysis were summarized in Tables 1 and 2.
Table 1.
Experimental results
| A1 [%] | D1 [μm2/s] | A2 [%] | D2 [μm2/s] | A3 [%] | D3 [μm2/s] | N | |
|---|---|---|---|---|---|---|---|
| SPT RNA-Oligon. | 38 ± 4 | 0.3 ± 0.02 | 61 ± 4 | 0.73 ± 0.03 | — | — | 6 nuclei, 4 glands |
| SPT CTR | 17 ± 5 | 0.5 ± 0.1 | — | — | 83 ± 15 | 2–6 | 4 nuclei, 3 glands |
| SPT MB∗ | 56 ± 12 | 0.31 ± 0.03 | 44 ± 11 | 0.68 ± 0.08 | — | — | 4 nuclei, 3 glands |
| FCS RNA-Oligon. | 20 ± 4 (12/77) | 0.15 ± 0.05 | 80 ± 2 (77/77) | 0.8 ± 0.05 | 10 ± 1 (7/77) | 11 ± 2 | 9 nuclei, 3 glands |
| FCS CTR | — | — | 13 ± 1 (45/61) | 0.95 ± 0.1 | 73 ± 1 (61/61) | 10 ± 0.4 | 7 nuclei, 3 glands |
N, number of examined nuclei and of different glands, respectively. SPT, single particle tracking; RNA-Oligon., specific 2′-O-methyl-RNA-oligonucleotides; CTR, unspecific 2′-O-methyl-RNA-oligonucleotides; MB, specific 2′-O-methyl-RNA molecular beacons. FCS, fluorescence correlation spectroscopy. Errors are mean ± SE.
See the Supporting Material.
Table 2.
Experimental results
| DMSD [μm2/s] | Linear? | N | Trajectory No. | |
|---|---|---|---|---|
| BR2.1 mRNPs | 0.71 ± 0.02 | No | 6 nuclei, 4 glands | 306 |
| 500 kDa dextrans buffer | 5.3 ± 0.1 | Yes | — | 322 |
| 500 kDa dextranssalivary gland cell nuclei | 2.2 ± 0.04 | No | 11 nuclei, 6 glands | 226 |
| 500 kDa dextransH2B-GFP cell nuclei | 1.1 ± 0.1 | No | 22 cells | 66 |
MSD analysis for oligonucleotide labeled BR2.1 mRNPs and labeled 500 kDa dextrans. A nonlinear MSD behavior is noted. N, number of examined nuclei and of different glands, respectively. D, initial diffusion coefficient, corresponding to the linear part of the MSD plot. Trajectory No., number of trajectories in the MSD plot. Errors are mean ± SE.
Inverse oligonucleotides, which were incapable of binding to BR 2.1 mRNPs, were analyzed in comparable experiments. As expected, the BR transcription site was not marked in these experiments. Unbound fluorescent oligonucleotides diffuse extremely rapidly in the cell nucleus. Assuming a viscosity of 3–5 centipoise (cP) in the nucleus (17), a diffusion coefficient of >10 μm2/s can be expected for unbound oligonucleotides. Molecules moving so fast should not be detectable with a single frame integration time of 10 ms, because the signal would be completely smeared over a large area, resulting in an insufficient signal/noise ratio for detection. Nevertheless, movies were taken after microinjection of the control oligonucleotides, and the same data acquisition conditions and analysis procedures as described above were applied. Movies of four nuclei from three different glands were evaluated. In contrast to expectation, we observed some single molecule signals. However, only 822 trajectories containing a total of 4761 jumps were detected—significantly less than in the case of the specific oligonucleotides. In the corresponding jump distance histogram, three diffusion coefficients—namely D1,SPT = 0.5 ± 0.1 μm2/s, D2,SPT = 2 ± 0.5 μm2/s, and D3,SPT = 6.5 ± 2.5 μm2/s with fractional weights of 17 ± 5%, 54 ± 11%, and 29 ± 14%—were determined (Fig. 2 C). Table 1 summarizes all results. Here, D2,SPT and D3,SPT of the control oligonucleotides were shown together, and designated D3,SPT. The results indicated that at times the control oligonucleotides became visible, although they do not bind to the BR2.1 mRNPs. Only ∼17% of the 4761 jumps for the control oligonucleotides showed a diffusion coefficient in the range of the specific ones (0.5 μm2/s), whereas for the specific oligonucleotides, 99% of the 124,172 jumps were in that range. Presumably, the control oligonucleotides show some nonspecific interactions with intranuclear proteins or particles.
Both the reduction of the microinjected oligonucleotide concentration as well as the increased time resolution enhanced our particle tracking reliability compared to our previous study (17). Due to the reduced concentration, the density of labeled particles was significantly lower than previously, and no extended photobleaching phases were needed to reduce further the particle density. The larger signal-to-signal distance, combined with a higher time resolution, yielding shorter jumps from frame to frame made tracking very reliable. Under these improved conditions, a fast mobility fraction for the specific oligonucleotides was not detectable. In addition, the fastest component (4 μm2/s) detected in our previous study was suspiciously similar to D2,SPT and D3,SPT of the unspecific oligonucleotides. Therefore, we speculate that this component was due to unbound oligonucleotides interacting nonspecifically with intranuclear proteins. Additional experiments using specific and unspecific molecular beacons confirmed this conclusion (see the Supporting Material and Table 1).
FCS of 2′-O-methyl-RNA-labeled BR mRNPs
Compared to single molecule tracking, FCS provides a complementary approach to molecular dynamics. A single FCS measurement reports the molecular dynamics within a limited spatial region of <1 μm3 over a time period of tens of seconds. Therefore, it produces a snapshot of the local dynamics in the confocal FCS volume. Furthermore, FCS is especially suited to detect and quantify fast molecular dynamics. Thus, we applied it here in order to complete our examination of the mRNP dynamics.
2′-O-methyl-RNA oligonucleotides were microinjected into salivary gland cell nuclei and subsequently analyzed with FCS. For specific oligonucleotides, nine nuclei from three different glands were analyzed. Seventy-seven separate FCS measurements were performed. In all measurements a fraction with a diffusion coefficient D2,FCS = 0.8 ± 0.05 μm2/s was found with a mean weight of 80 ± 2%. A corresponding representative data set for a single component is shown in Fig. 3 A (red curve). Furthermore, in 13 out of 77 FCS measurements a second slowly diffusing component was found with D1,FCS = 0.15 ± 0.05 μm2/s (mean weight 20 ± 4%). Fig. 3 B shows a representative FCS curve, which required D1,FCS and D2,FCS for a satisfactory fit.
Figure 3.
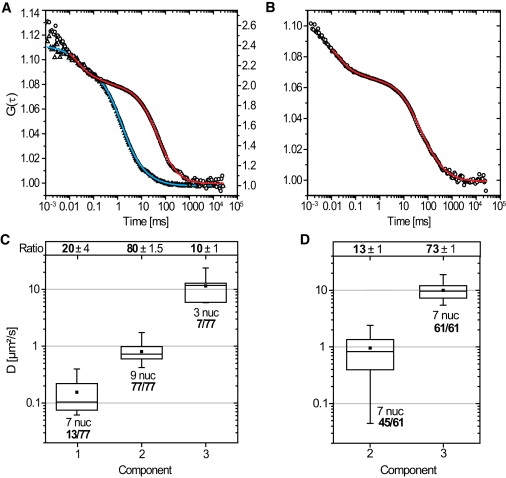
2′-O-methyl-RNA-oligonucleotides were microinjected into salivary gland cell nuclei and analyzed with FCS. (A) Two typical autocorrelation curves for control (triangles, right scale) and specific oligonucleotides (circles, left scale) are shown with their corresponding fits (control, blue; specific, red). The red curve shows only D2,FCS. (B) Autocorrelation curve for specific oligonucleotides, comprising two diffusion coefficients D2,FCS and D3,FCS. (C) Results of the specific oligonucleotides are shown in an overview. (Box plots) Distributions of the diffusion coefficients of the three mobility components, summarized. (Solid square) Average value. (Line) Median value. (Whiskers) Minimum and the maximum value. The box contains 50% of all values obtained. The relative occurrence of the respective component in relation to all 77 FCS measurements was indicated below the boxes. The ratio of the components is shown above. (D) Results for the unspecific oligonucleotides; for a detailed description of the graph, see panel C.
The two mobility fractions characterized by D1,FCS and D2,FCS correspond to the values determined above by SPT and were attributed to oligonucleotides bound to BR mRNPs. A box plot demonstrated that the distribution of D1,FCS was rather broad (Fig. 3 C), indicating the spatial and temporal inhomogeneity of BR mRNP dynamics. It should be noted that only mobility component 2 was detectable in all measurements.
A third small but fast component was seen in seven out of 77 FCS runs, which could be characterized by D3,FCS = 11 ± 2 μm2/s with a mean weight of only 10 ± 1%. This third component was unlikely to be due to BR2.1 mRNPs, because these are expected to diffuse with a maximal diffusion coefficient of ∼D = 2–3 μm2/s inside C. tentans salivary gland cell nuclei, because of their great diameter of 50 nm and the effective nuclear viscosity of ∼3–5 cP. Hence, only significantly smaller particles than BR mRNPs can diffuse with such a high diffusion coefficient. All results were accumulated in Table 1.
Finally, unrelated control oligonucleotides were analyzed in seven nuclei from three different salivary glands yielding a total number of 61 FCS measurements. In every measurement the fast component D3,FCS = 10 ± 0.4 μm2/s was determined, which dominated the dynamics with a mean weight of 73 ± 1% in contrast to the specific oligonucleotides (mean weight 10 ± 1%; see Fig. 3 A, triangles, and Fig. 3 D). In 45 out of 61 measurements, a slow component was also observed with a D2,FCS = 1.0 ± 0.1 μm2/s but with a weight of 13 ± 1% only. Hence, as expected, the unrelated control oligonucleotides showed a large fast mobile fraction, which corresponded to D3,FCS of the specific 2′-O-methyl-RNA oligonucleotides. Yet, as known from the single particle tracking experiments, control oligonucleotides do show some unspecific binding.
In general, the FCS measurements clearly supported the single particle tracking data. Furthermore, we noted that oligonucleotides unspecifically bound to small proteins or mRNPs may be observed in rare cases. Finally, the results of FCS measurements apparently varied with the specific location, where the measurement was performed.
Single molecule tracking of large tracer molecules in cell nuclei
Deviations from linear MSD plots occur in molecular crowded media or when distributions of binding sites exist (31,32). In order to possibly discriminate between these two options for the BR mRNPs, and to understand the reason for the deviation from normal diffusive behavior of the BR2.1 mRNPs, we used large inert tracer molecules to probe the chromatin-free salivary gland cell nucleoplasm. For comparison, similar measurements were performed in the interior of mammalian cell nuclei, which contain unfolded chromatin, and in buffer solution. As tracer molecules, we employed fluorescently labeled dextrans with a molecular mass of 500 kDa labeled by Atto647N.
To begin with, the tracers were examined by single molecule imaging at 200 Hz in buffer solution. A total number of 3184 dextran trajectories was identified and analyzed. As expected, a linear dependence of the MSD as a function of time was obtained, and the analysis by Eq. 3 yielded an average diffusion coefficient of 5.3 ± 0.1 μm2/s, indicating a Stokes radius of 80 nm (Fig. 4, triangles).
Figure 4.
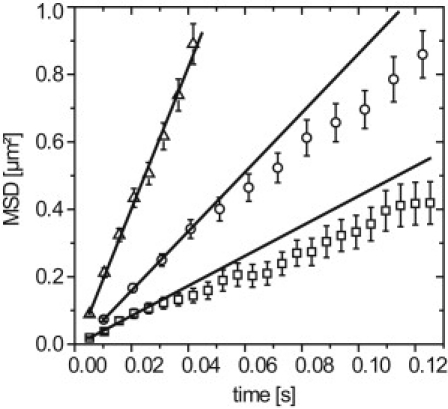
MSD plots for fluorescently labeled 500 kDa dextrans visualized and tracked in buffer (triangles), salivary gland cell nuclei (circles), and nuclei of mammalian H2B-GFP cells (squares). A nonlinear behavior is observed in the salivary gland cell nuclei, and in the mammalian cell nuclei. Errors are mean ± SE.
Next, the fluorescent dextran molecules were microinjected into C. tentans salivary gland cell nuclei (11 nuclei of six different salivary glands). Single molecule imaging and tracking yielded 6673 trajectories. An example data set was shown in Movie S2. From these trajectories, we determined the MSD as a function of time (Fig. 4, circles). Although linear on a short timescale, the MSD clearly deviated from a linear dependence for t ≥ 50 ms, indicating a distance-dependent retardation of the inert tracer. This mobility behavior was principally similar to that of the endogenous BR2.1 mRNPs (Fig. 2 B). Thus, the retardation effects of nonchromatin nuclear components become visible. A straight line fitted to the first four linear points yielded a diffusion coefficient of 2.2 ± 0.04 μm2/s. This value was reduced compared to that in buffer (5.3 ± 0.1 μm2/s) by a factor of 2.4. Assuming a viscosity of ∼1 cP for buffer, a salivary gland nucleoplasmic viscosity of ∼2.4 cP could be deduced, which was in good agreement with a previously determined value (17).
Finally, the dextran tracers were examined inside the cell nuclei of mammalian HeLa cells stably expressing H2B-GFP. In 22 different cell nuclei, a total number of 13,974 dextran trajectories were imaged and analyzed (see Movie S3). Again, the determined MSD versus time plot showed a nonlinear behavior for t ≥ 25 ms (Fig. 4, squares), and from the linear part of the plot a diffusion coefficient of 1.1 ± 0.1 μm2/s was obtained. This value indicated an effective intranuclear viscosity of ∼4.8 cP in these cell nuclei. Very similar results were obtained for mouse C2C12 myoblasts, suggesting that the effect was not a cell-type-specific effect (data not shown). All results of the tracer experiments were summarized in Table 2.
Discussion
The transcription and intranuclear processing of mRNA molecules is a well-organized cellular process. Numerous proteins and protein complexes are involved and contribute to a highly dynamical meshwork of biochemical interactions. In the last years a very dynamic view of intranuclear spatio-temporal processes evolved, and a detailed analysis of intranuclear mRNA trafficking will further improve the understanding of the dynamical nuclear mRNA metabolism. In this study, we focused on the nature of the complex dynamics that was recently uncovered for Balbiani Ring (BR) mRNPs of C. tentans by single particle tracking (17). In contrast to previous work, we employed both fluorescently labeled 2′-O-methyl-RNA oligonucleotides and molecular beacons based on 2′-O-methyl-RNA oligonucleotides in an in vivo fluorescence in situ hybridization approach for visualizing these mRNPs. This approach yielded an excellent signal/noise ratio of the labeled mRNA molecules. In addition, the exploitation of complementary optical techniques (single particle tracking and FCS) used to study the molecular mobility of the probes created a detailed view of native mRNP trafficking, which provided, to our knowledge, new insights into the internal structure of these cell nuclei, where the chromatin is condensed in polytene chromosomes, leaving a large nuclear volume devoid of chromatin.
As seen before, the trajectory analysis revealed two major mobility components, which could be related to BR2.1 mRNP movements, in addition to a small almost immobile fraction. The major components, in the range of D1 = 0.3 ± 0.02 μm2/s and D2 = 0.73 ± 0.03 μm2/s, were detected independently of the labeling approach and measurement technique used. At times a third mobility component of the probe molecules with a diffusion coefficient D3 ≥ 4 ± 1.5 μm2/s was observed. We found here that this component was due to unspecific bound probe oligonucleotides.
Next, we addressed what the cause for the occurrence of two different diffusion components might be. Our data analysis demonstrated that they were not due to particles moving with distinct diffusion coefficients (see the Supporting Material). Instead, the particle trajectories contained phases with both slow and fast diffusion. To understand this behavior, we examined the mRNP dynamics in great detail. In addition to the jump-distance distributions, we also examined the MSD of the particles as a function of time. Thereby we detected a nonlinear dependence of the MSD versus time, with an initial slope indicating a diffusion coefficient of D = 0.7 μm2/s. Generally, a nonlinear MSD behavior is observed, when binding or crowding effects occur (31,33,34). This leads to the question of whether large intranuclear structures exist, which would retard the mRNP motion simply by geometric hindrance (i.e., molecular crowding), and whether mRNPs might bind specifically to such structures.
To answer this question, we examined the dynamics of inert tracer molecules with a diameter close to that of the mRNPs. To this end, we used dextran molecules with a molecular mass of 500 kDa. As expected, these molecules displayed a linear dependence of the MSD on time in solution, yielding a diffusion coefficient of Ddex,sol = 5.3 μm2/s. In contrast, the diffusion of these tracers in mammalian cell nuclei, which are well known for strongly retarding the diffusion of inert molecules, was significantly slowed down (35–37). Inside mammalian cell nuclei we measured a diffusion coefficient of Ddex,mcn = 1.1 μm2/s only, and observed also a clear nonlinear dependence of the MSD on time. This clearly indicated the known molecular crowding inside these nuclei, which was due to the existence of the distributed chromatin (38,39). Finally, we analyzed the diffusion characteristics of the dextran tracer molecules inside a C. tentans salivary gland cell nuclei. We expected a certain reduction of the diffusion coefficient in comparison to solution due to an increased effective viscosity. Indeed, we found a diffusion coefficient Ddex,sgn = 2.2 μm2/s, which corroborated that the effective intranuclear viscosity was ∼3 centipoise (17). Furthermore, we also detected a nonlinear functional dependence of the MSD on time in these nuclei. In chromatin-crowded mammalian cell nuclei, a reduced diffusion coefficient and a nonlinear MSD time dependence was expected for inert molecules, but this was not the case for the salivary gland cell nuclei. Chromatin as cause for molecular crowding and hence for a nonlinear MSD-time relationship could definitely be excluded because chromatin is located in distinct polytene chromosomes here. The observed nonlinearity of the MSD clearly indicated that numerous and extended molecular nonchromatin structures were present, which reduced even the mobility of the inert tracer molecules.
Comparison of the BR mRNP mobility with that of the dextran tracers inside the gland cell nuclei revealed that the mRNPs (D = 0.7 μm2/s) moved slower than the tracers (D = 2.2 μm2/s) although they were smaller in size with 50 vs. 80 nm. This suggested that the mRNPs were not only slowed down due to nonchromatin structures causing a mobility hindrance, but also interacted presumably with these structures causing a even further retardation. Indeed, a simple Monte Carlo simulation demonstrated that a random distribution of binding sites with binding and unbinding times below the imaging time resolution may result in the observed intranuclear dynamics: nonlinear MSD-versus-time plots, and two mobile components in the jump-distance analysis (see the Supporting Material). We consider this to be a probable scenario.
What nonchromatin structures, which hinder both the movements of tracers and mRNPs, and in addition function as interactions sites for mRNPs, might exist in the salivary gland cell nuclei?
Miralles et al. (40) showed by electron tomography that BR mRNPs interacted with certain intranuclear supramolecular nonchromatin complexes, so-called fibrogranular clusters (FGCs), which presumably correspond functionally to interchromatin granule clusters in mammalian cells. Therefore we assume that the structures, which reduce mRNP mobility as both hindrances and binding sites, are identical with FGCs, and we speculate that mRNPs in mammalian cells also interact with such nonchromatin structures, leading to a structural model as described in Fig. 5.
Figure 5.
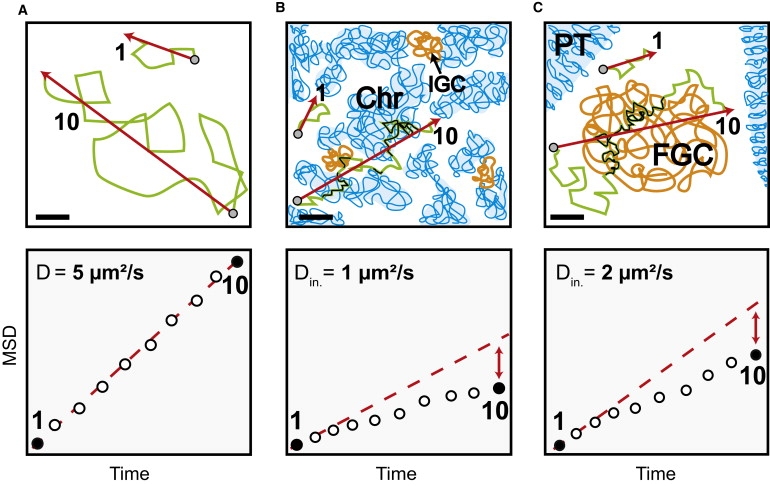
Impact of the local nuclear environment on the mobility of large particles and corresponding MSD-time plots (lower lane). (A) Determination of MSD in buffer. (Lower lane) The MSD is linear due to Brownian motion. (B) Mobility in mammalian cell nuclei. The unfolded chromatin (Chr) and interchromatin granule clusters reduce particle mobility on long timescales (t = 10), but not on short timescales (t = 1), when particles diffuse within interchromatin channels with an initial diffusion coefficient due to the effective nucleoplasmic viscosity. At long times the traveled distance is reduced due to frequent collisions with mobility barriers or unspecific and specific binding events (black sections of the trajectories) due to unfolded chromatin and interchromatin structures leading to a nonlinear MSD-time dependence (lower lane). (C) Mobility in salivary gland cell nuclei. The interphase chromatin is compacted into polytene chromosomes (PT) leaving vast regions devoid of chromatin. The diffusion on short timescales (t = 1) is due to the effective local viscosity as in mammalian cell nuclei. (Lower lane) The particle mobility at long times and large spatial scales (t = 10) shows a nonlinear MSD-time dependence. Because this cannot be due to chromatin it implies the presence of another extended nuclear structure influencing the mobility (black trajectory parts) of endogenous molecules (BR2.1 mRNPs) and inert tracer particles (500 kDa dextrans). Presumably, these structures correspond to FGCs (40). Initial diffusion coefficients for 500 kDa dextrans are shown in the lower lane. Bars, 2 μm.
Altogether, our experiments revealed that mRNP mobility is intrinsically complex even within cell nuclei that have a seemingly simple intranuclear structure. Simple diffusion defined by a single diffusion coefficient and a linear MSD behavior does not exist, even when chromatin is absent. Instead, a view of mRNPs engaged in numerous subsequent molecular interactions with special nonchromatin structures, which not only hinder the free motion of large molecules and nuclear particles by presenting mobility restrictions but also function as interaction partners, is emerging.
Acknowledgments
U.K. acknowledges financial support by the German Research Foundation under grant No. Ku 2474/7-1, and S.T. gratefully acknowledges support by the National Institutes of Health under grant No. MH079197.
Supporting Material
References
- 1.Dreyfuss G., Kim V.N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 2.Iglesias N., Stutz F. Regulation of mRNP dynamics along the export pathway. FEBS Lett. 2008;582:1987–1996. doi: 10.1016/j.febslet.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 3.Kohler A., Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 4.Luna R., Gaillard H., Aguilera A. Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma. 2008;117:319–331. doi: 10.1007/s00412-008-0158-4. [DOI] [PubMed] [Google Scholar]
- 5.Misteli T. Cell biology: nuclear order out of chaos. Nature. 2008;456:333–334. doi: 10.1038/456333a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore M.J. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 7.Sharp P.A. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Braga J., McNally J.G., Carmo-Fonseca M. A reaction-diffusion model to study RNA motion by quantitative fluorescence recovery after photobleaching. Biophys. J. 2007;92:2694–2703. doi: 10.1529/biophysj.106.096693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calapez A., Pereira H.M., Carmo-Fonseca M. The intranuclear mobility of messenger RNA binding proteins is ATP dependent and temperature sensitive. J. Cell Biol. 2002;159:795–805. doi: 10.1083/jcb.200203046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishihama Y., Funatsu T. Single molecule tracking of quantum dot-labeled mRNAs in a cell nucleus. Biochem. Biophys. Res. Commun. 2009;381:33–38. doi: 10.1016/j.bbrc.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Molenaar C., Abdulle A., Dirks R.W. Poly(A)+ RNAs roam the cell nucleus and pass through speckle domains in transcriptionally active and inactive cells. J. Cell Biol. 2004;165:191–202. doi: 10.1083/jcb.200310139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mor A., Suliman S., Shav-Tal Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 2010;12:543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- 13.Politz J.C., Browne E.S., Pederson T. Intranuclear diffusion and hybridization state of oligonucleotides measured by fluorescence correlation spectroscopy in living cells. Proc. Natl. Acad. Sci. USA. 1998;95:6043–6048. doi: 10.1073/pnas.95.11.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Politz J.C., Tuft R.A., Singer R.H. Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr. Biol. 1999;9:285–291. doi: 10.1016/s0960-9822(99)80136-5. [DOI] [PubMed] [Google Scholar]
- 15.Politz J.C., Tuft R.A., Pederson T. Rapid, diffusional shuttling of poly(A) RNA between nuclear speckles and the nucleoplasm. Mol. Biol. Cell. 2006;17:1239–1249. doi: 10.1091/mbc.E05-10-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shav-Tal Y., Darzacq X., Singer R.H. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siebrasse J.P., Veith R., Kubitscheck U. Discontinuous movement of mRNP particles in nucleoplasmic regions devoid of chromatin. Proc. Natl. Acad. Sci. USA. 2008;105:20291–20296. doi: 10.1073/pnas.0810692105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tadakuma H., Ishihama Y., Funatsu T. Imaging of single mRNA molecules moving within a living cell nucleus. Biochem. Biophys. Res. Commun. 2006;344:772–779. doi: 10.1016/j.bbrc.2006.03.202. [DOI] [PubMed] [Google Scholar]
- 19.Vargas D.Y., Raj A., Tyagi S. Mechanism of mRNA transport in the nucleus. Proc. Natl. Acad. Sci. USA. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyagi S. Imaging intracellular RNA distribution and dynamics in living cells. Nat. Methods. 2009;6:331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- 21.Wachsmuth M., Caudron-Herger M., Rippe K. Genome organization: balancing stability and plasticity. Biochim. Biophys. Acta. 2008;1783:2061–2079. doi: 10.1016/j.bbamcr.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 22.Albiez H., Cremer M., Cremer T. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 2006;14:707–733. doi: 10.1007/s10577-006-1086-x. [DOI] [PubMed] [Google Scholar]
- 23.Daneholt B. Packing and delivery of a genetic message. Chromosoma. 2001;110:173–185. doi: 10.1007/s004120000127. [DOI] [PubMed] [Google Scholar]
- 24.Daneholt B. Assembly and transport of a premessenger RNP particle. Proc. Natl. Acad. Sci. USA. 2001;98:7012–7017. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wieslander L. The Balbiani ring multigene family: coding repetitive sequences and evolution of a tissue-specific cell function. Prog. Nucleic Acid Res. Mol. Biol. 1994;48:275–313. doi: 10.1016/s0079-6603(08)60858-2. [DOI] [PubMed] [Google Scholar]
- 26.Siebrasse J.P., Grunwald D., Kubitscheck U. Single-molecule tracking in eukaryotic cell nuclei. Anal. Bioanal. Chem. 2007;387:41–44. doi: 10.1007/s00216-006-0763-0. [DOI] [PubMed] [Google Scholar]
- 27.Müller C.B., Loman A., Enderlein J. Precise measurement of diffusion by multi-color dual-focus fluorescence correlation spectroscopy. Europhys. Lett. 2008;83:46001. [Google Scholar]
- 28.Magde D., Elson E.L., Webb W.W. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 29.Molenaar C., Marras S.A., Tanke H.J. Linear 2′ O-Methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.17.e89. E89–E89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kues T., Peters R., Kubitscheck U. Visualization and tracking of single protein molecules in the cell nucleus. Biophys. J. 2001;80:2954–2967. doi: 10.1016/S0006-3495(01)76261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dix J.A., Verkman A.S. Crowding effects on diffusion in solutions and cells. Annu. Rev. Biophys. 2008;37:247–263. doi: 10.1146/annurev.biophys.37.032807.125824. [DOI] [PubMed] [Google Scholar]
- 32.Richter K., Nessling M., Lichter P. Macromolecular crowding and its potential impact on nuclear function. Biochim. Biophys. Acta. 2008;1783:2100–2107. doi: 10.1016/j.bbamcr.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 33.Saxton M.J. A biological interpretation of transient anomalous subdiffusion. I. Qualitative model. Biophys. J. 2007;92:1178–1191. doi: 10.1529/biophysj.106.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saxton M.J. A biological interpretation of transient anomalous subdiffusion. II. Reaction kinetics. Biophys. J. 2008;94:760–771. doi: 10.1529/biophysj.107.114074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukacs G.L., Haggie P., Verkman A.S. Size-dependent DNA mobility in cytoplasm and nucleus. J. Biol. Chem. 2000;275:1625–1629. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- 36.Seksek O., Biwersi J., Verkman A.S. Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J. Cell Biol. 1997;138:131–142. doi: 10.1083/jcb.138.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bancaud A., Huet S., Ellenberg J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 2009;28:3785–3798. doi: 10.1038/emboj.2009.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dross N., Spriet C., Langowski J. Mapping eGFP oligomer mobility in living cell nuclei. PLoS ONE. 2009;4:e5041. doi: 10.1371/journal.pone.0005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorisch S.M., Wachsmuth M., Rippe K. Histone acetylation increases chromatin accessibility. J. Cell Sci. 2005;118:5825–5834. doi: 10.1242/jcs.02689. [DOI] [PubMed] [Google Scholar]
- 40.Miralles F., Ofverstedt L.G., Visa N. Electron tomography reveals posttranscriptional binding of pre-mRNPs to specific fibers in the nucleoplasm. J. Cell Biol. 2000;148:271–282. doi: 10.1083/jcb.148.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


