Summary
In the developing spinal cord, regional and combinatorial activities of Hox transcription factors are critical in controlling motor neuron fates along the rostrocaudal axis, exemplified by the precise pattern of limb innervation by more than fifty Hox-dependent motor pools. The mechanisms by which motor neuron diversity is constrained to limb-levels are however not well understood. We show that a single Hox gene, Hoxc9, has an essential role in organizing the motor system through global repressive activities. Hoxc9 is required for the generation of thoracic motor columns and in its absence neurons acquire the fates of limb-innervating populations. Unexpectedly, multiple Hox genes are derepressed in Hoxc9 mutants, leading to motor pool disorganization and alterations in the connections by thoracic and forelimb-level subtypes. Genome-wide analysis of Hoxc9 binding suggests this mode of repression is mediated by direct interactions with Hox regulatory elements, independent of chromatin marks typically associated with repressed Hox genes.
Introduction
Hox transcription factors have conserved roles in shaping the body plans of animals and function as major determinants of morphological and cellular diversity along the rostrocaudal axis (McGinnis and Krumlauf, 1992). In the vertebrate hindbrain and spinal cord Hox genes are thought to be essential in defining the identity and synaptic specificity of neurons required for vital behaviors such as respiration and locomotion (Dasen and Jessell, 2009; Trainor and Krumlauf, 2000). An early step in the assembly of motor circuits is the establishment of precise connections between motor neurons (MNs) and their peripheral targets, requiring the generation of hundreds of distinct subtypes. Hox genes are particularly important for the specification of MNs involved in limb coordination and differentiate these diverse populations from those necessary for other motor functions. While the regional specialization of MNs appears to be established through Hox combinatorials and additional lineage specific factors (Dalla Torre di Sanguinetto et al., 2008; Jessell, 2000; Shirasaki and Pfaff, 2002), the precise mechanisms by which Hox-dependent subtypes are generated within discrete areas of the spinal cord are not fully understood.
More than half of the 39 chromosomally clustered Hox genes are expressed by MNs (Dasen et al., 2005), yet little is known with respect to the mechanisms underlying one prominent feature of their patterns within the CNS – the restriction of a majority of Hox genes to limb levels. Early in development Hox expression is controlled by gradients of retinoic acid (RA), fibroblast growth factors (FGFs), and Wnts which determine the initial spatial profile of Hox transcription in neural progenitors along the rostrocaudal axis (Bel-Vialar et al., 2002; Liu et al., 2001; Nordstrom et al., 2006). In general, the induction of a Hox gene is linked to its position along the chromosome: genes located at the more 5' end of a cluster are expressed more posteriorly and are induced by progressively higher levels of FGF, and this action is opposed by paraxial mesoderm-derived RA which induces 3' genes (Bel-Vialar et al., 2002; Liu et al., 2001). The sequential activation of Hox genes by signaling gradients defines anterior expression limits (Bel-Vialar et al., 2002), and these boundaries are thought to be maintained by the actions of polycomb group (PcG) repressive complexes which restrict Hox expression through repressive chromatin modifications (Deschamps et al., 1999; Soshnikova and Duboule, 2009). At posterior regions many Hox genes are however initially coexpressed in neuronal progenitors (Bel-Vialar et al., 2002; Deschamps et al., 1999), and only as cells differentiate begin to display mutually exclusive domains of expression (Dasen et al., 2003). Defining the steps which link the early induction of Hox genes to their expression and function during MN differentiation is critical in elucidating how diverse subtypes are generated.
One mechanism thought to shape the final pattern of Hox expression in the CNS involves cross regulatory interactions between Hox proteins and Hox genes. In the developing hindbrain the restricted pattern of Hox expression within rhombomeres is regulated by autoregulatory and feed forward transcriptional cascades (Tumpel et al., 2009). In spinal MNs Hox expression appears to be defined through cross-repressive interactions occurring soon after MNs are born, presumably acting to prevent the generation of neurons with an ambiguous Hox code (Dasen et al., 2003; Dasen et al., 2005). However, several questions relating to the workings of the MN Hox network remain unresolved: 1) Do Hox repressive interactions function simply to sharpen molecular boundaries between neuronal subtypes? 2) Is the high density of Hox genes expressed at limb levels established through regulation of certain Hox genes en masse? 3) Are the repressive interactions mediated by direct binding of Hox proteins to Hox regulatory elements? 4) How would loss of a Hox repressor affect MN identity and patterns of connectivity? Addressing these issues has been challenging due to the redundancies between Hox genes and the inherent difficulty in identifying DNA target sites.
Progress towards understanding how Hox genes contribute to the diversification of neuronal subtypes has emerged through examination of the programs controlling two aspects of MN differentiation - the specification of columnar and pool subtypes. Distinct groups of Hox genes operate at each of these early phases of MN differentiation. The establishment of a MN columnar identity directs axons toward broad target fields including limb, axial, and body wall muscles as well as neurons in the sympathetic chain (Landmesser, 2001). At brachial and lumbar levels of the spinal cord, Hox6 and Hox10 proteins initiate the molecular programs that specify the lateral motor column (LMC) fates and ensure that these subtypes are generated in registry with the position of their limb targets (Dasen et al., 2003; Shah et al., 2004; Tarchini et al., 2005; Wu et al., 2008). Within LMC neurons, the activities of nearly two dozen Hox genes are required to generate the ~50 motor pool subtypes targeting specific muscles in the limb (Dasen et al., 2005). In contrast to limb levels, intervening thoracic levels of the spinal cord contain relatively few Hox-dependent subtypes (Dasen et al., 2005), a possible reflection of the reduced number and variety of synaptic targets (Gutman et al., 1993; Prasad and Hollyday, 1991; Smith and Hollyday, 1983). Thoracic MNs express Hox9 proteins (Liu et al., 2001) and contain columns projecting towards hypaxial muscles and sympathetic chain ganglia, and these populations appear to be relatively homogeneous in molecular profile.
Further insight into the role of the Hox network in MN differentiation has emerged from the analysis of mice lacking the transcription factor FoxP1, a putative cofactor required for deployment of Hox programs in spinal MNs. Each Hox-dependent step of MN diversification relies on FoxP1 activity, as in its absence segmentally restricted columnar and pool subtypes fail to be specified, Hox controlled molecular programs are lost, and MNs revert to an ancestral state (Dasen et al., 2008; Rousso et al., 2008). As a consequence, the normal topographic relationship between MN position and peripheral connectivity is dissolved and limb-level motor axons appear to select their targets at random (Dasen et al., 2008). The columnar and pool specific patterns of Hox expression are unaffected by Foxp1 mutation, indicating that Hox repressive activities are preserved. These observations suggest that FoxP1 functions within the context of a preexisting Hox code, established through cross-repressive interactions, and engages this network to selectively activate downstream columnar and pool specific programs.
Genetic evidence supporting a repression-based strategy in the control of Hox profiles in the CNS has been mostly indirect, due to the presumed functional compensation (Maconochie et al., 1996; McIntyre et al., 2007) amongst the large numbers of Hox genes expressed by MNs. Nevertheless, we initiated a systematic analysis of MN differentiation in Hox mutants, based on the assumption that removal of individual or multiple Hox genes would clarify their role in MN specification and allow a more definitive assessment of the significance of Hox cross-repressive interactions. We find that a single Hox gene, Hoxc9, is required for the generation of thoracic MN subtypes, is essential for organizing the MN topographic map, and acts as a key repressor of the forelimb-level Hox network. We provide evidence that Hoxc9 represses anterior Hox genes through direct interactions at Hox loci, while more posterior Hox genes are silenced by a distinct mechanism. Our studies indicate that Hoxc9 has a central role in patterning neuronal fates within the spinal cord through its activities as a global repressor of multiple Hox genes, and in generating a permissive zone for the Hox network to specify diverse subtypes.
Results
Loss of Thoracic Motor Neuron Columnar Subtypes in Hoxc9 Mutants
To better understand how Hox repressor activities contribute to the diversification of motor neurons in mouse we initiated an analysis of the expression patterns and loss of function phenotypes for 10 of the Hox4–9 paralogs (Hoxc4, c5, c6, c8, c9, a5, a6, a7, a9, d9) expressed at brachial and thoracic levels of the spinal cord. Because of the profound phenotype of Hoxc9 mutants, and observed in an ENU-induced Hoxc9 mutation (K. Liem et al., in preparation), we focus here on the roles of Hox9 genes. Studies in chick implicate Hox9 paralogs in controlling the molecular identity of columnar subtypes generated at thoracic levels (Dasen et al., 2003), in particular MNs that innervate sympathetic chain ganglia and occupy the preganglionic motor column (PGC). To determine whether Hox9 genes function in PGC specification, we analyzed the expression of each of the four Hox9 genes finding that Hoxa9, Hoxc9, and Hoxd9 are expressed in ventral spinal cord at embryonic day (e) 11.5, while Hoxb9 was excluded from postmitotic MNs (Figure S1A). Hoxa9 and Hoxd9 were expressed by MNs extending from thoracic to upper lumbar regions while Hoxc9 expression was largely restricted to thoracic levels (Figure S1A).
We next characterized the expression of molecular markers for early aspects of MN identity and columnar differentiation in Hoxa9, Hoxd9, and Hoxc9 mutant mice, focusing on the impact of loss of Hox9 activity on MN generation and PGC differentiation at e11.5. Features of MN class identity, such as expression of the homeodomain proteins Isl1/2 and Hb9 as well as the cholinergic marker vesicular acetylcholine transferase (VAchT) were not reduced in Hoxa9 and Hoxd9 mutants (Figure S1B and data not shown), while in Hoxc9 mutants the number of thoracic MNs was increased by ~30% (Figure 1A–1D and 1G). We next examined the expression of two markers that distinguish PGC neurons from other thoracic MN subtypes - neuronal nitric oxide synthase (nNOS) and phospho(p)-Smad1/5/8. In Hoxc9 mutants expression of nNOS and p-Smad1/5/8 were not detected at any age examined (e11.5 – e13.5) (Figure 1H–1K) while in Hoxa9 and Hoxd9 mutants expression of these genes were unaltered (Figure S1B).
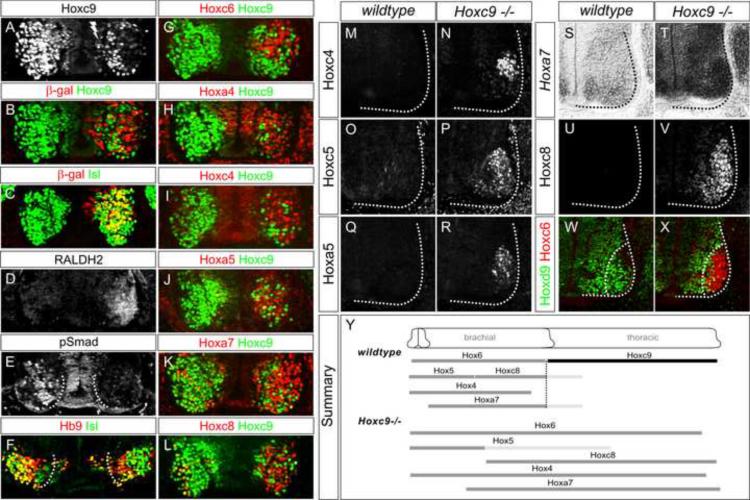
Figure 1. Transformation of Columnar Identities in Hoxc9 Mutants.
(A–B) Loss of Hoxc9 protein at thoracic levels in Hoxc9 mutants. Sections show ventral right quadrant of e11.5 spinal cord. (C–F) Expression of VAchT and the number of Lhx3+Hb9+ MMC MNs are grossly normal in Hoxc9 mutants. (G) Quantification of MN columnar subtypes (n>3 mice, error bars represent SEM). In Hoxc9 mutants total MN number at thoracic levels is increased ~30%, approximating limb-level numbers (data not shown). (H–K) Loss of nNOS and pSmad expression in Hoxc9 mutants. (L–O) In the absence of Hoxc9, the number of Isl1/2+ Hb9+ MNs is reduced and Er81 is not detected. (P–U) Ectopic Hoxc6, RALDH2, and FoxP1high MNs at thoracic levels in Hoxc9 mutants. (V–W) Schematic representation of thoracic MN columnar subtypes in wildtype and Hoxc9 mutants. Motor neuron markers for profiling are shown. (X) Quantification of FoxP1+Hoxc6+ and FoxP1+RALDH2+ LMC MNs along the rostrocaudal axis at e11.5. Results show cell counts for one embryo that are typical of n>5 animals. FoxP1 counts represent ventral lateral MNs that express high levels. (Y) Summary of MN columnar transformations in Hoxc9 mutants.
We next assessed how the loss of Hoxc9 affected the specification of two additional motor columns present at thoracic levels: hypaxial motor column (HMC) and median motor column (MMC) neurons. The HMC is selectively generated at thoracic levels, projects to intercostal and abdominal muscles and is characterized by coexpression of Hb9 and Isl1 and the ETS domain protein Er81 (Cohen et al., 2005; Dasen et al., 2008). In Hoxc9 mutants the number of Hb9+Isl1+ cells was significantly reduced and thoracic expression of Er81 was not detected (Figure 1G and 1L–1O). Neurons in the MMC are a Hox-independent population present at all rostrocaudal levels of the spinal cord, project to axial muscles, and coexpress the LIM homeodomain factors Lhx3 and Hb9 (Arber et al., 1999; Tsuchida et al., 1994). In Hoxc9 mutants the number of Lhx3+Hb9+ MNs was unchanged at all levels, indicating MMC identity is preserved (Figure 1E–1G). Together these observations indicate that Hoxc9 activity is specifically required for the emergence of molecular features for two thoracic-specific motor columns (PGC and HMC) but is dispensable for early aspects of MN identity and specification of MMC neurons.
Thoracic Motor Neurons Acquire an LMC Identity in the Absence of Hoxc9
What are the fate(s) of thoracic MNs which have lost Hoxc9? Hox9 genes have been implicated in restricting Hox6 paralog gene expression to brachial levels and determining the domain in which forelimb-innervating lateral motor column (LMC) neurons are generated (Blackburn et al., 2009; Dasen et al., 2003). In Hoxc9 mutants we detected ectopic expression of Hoxc6 mRNA and protein throughout thoracic spinal cord extending to the boundary between caudal thoracic and rostral lumbar levels (Figure 1P–Q, 1X, S4I–S4J). We next examined whether, as a consequence of Hoxc6 derepression, genes normally restricted to brachial LMC neurons are induced at thoracic levels. At limb levels LMC neurons are characterized by the expression of retinaldehyde dehydrogenase-2 (RALDH2) and high levels of FoxP1 (Dasen et al., 2008; Sockanathan and Jessell, 1998). The normal brachial expression of LMC markers was unaffected by Hoxc9 mutation (Figure S1C). In contrast, analysis of Hoxc9 mutants revealed ectopic RALDH2+ and FoxP1high MNs throughout the thoracic domain of Hoxc6 expression (Figure 1R–1W, 1X, S2). At lumbar levels MNs did not ectopically express Hoxc6 and the position of Hox10+ LMC neurons was preserved (Figure S1D and data not shown).
At limb levels, activation of FoxP1 and RALDH2 initiates a program of MN “divisional” specification, which controls the dorsoventral projection patterns of motor axons in the limb. This program is characterized by the selective expression of homeodomain factors, where medial division LMC MNs express high levels of Isl1 and lateral MNs high levels of Hb9 and Lhx1 (Kania and Jessell, 2003; Tsuchida et al., 1994). In Hoxc9 mutants this divisional pattern of homeodomain expression and MN settling was present at thoracic levels (Figure 1M, S1E). In addition a pattern of EphA4 guidance receptor expression similar to lateral LMC MNs was induced (Figure S1E). Thus, in the absence of Hoxc9 two thoracic-specific columns are lost, Hoxc6 is derepressed in all thoracic segments, and MNs acquire the columnar and divisional fates of forelimb-level LMC neurons. At a molecular level the spinal cord comprises two continuous columns of LMC and MMC neurons extending from cervical to lumbar levels (Figure 1Y).
Consequences of Columnar Transformation on Axonal Projection Patterns
To further examine the impact of switching the columnar identity of thoracic MNs we assessed potential axonal connectivity defects in Hoxc9 mutants. We bred Hoxc9 mice to a transgenic line (Hb9∷GFP mice) in which all motor axons are labeled with GFP (Arber et al., 1999) and analyzed PGC, HMC, and MMC projection patterns. Three major projection pathways are followed by thoracic MNs, corresponding to the three prominent columnar subtypes: MMC neurons project dorsally to axial muscles, HMC ventrolaterally to body wall muscles, and PGC ventromedially to sympathetic chain ganglia. We observed a profound reduction in axonal projections towards the sympathetic chain in Hoxc9−/−; Hb9∷GFP mice, consistent with a loss of PGC fate (Figures 2C–2F and S3). In contrast, motor axon projections towards limb and axial muscles were normal in Hoxc9 mutants indicating LMC and MMC trajectories are preserved (Figure 2A–2D). Thus Hoxc9 is required for both the molecular identity and establishing the peripheral connectivity of PGC MNs.
Figure 2. Altered Motor Axon Projection Patterns in Hoxc9 Mutants.
(A–D) Vibratome sections showing motor axon projections in wildtype and Hoxc9 mutant embryos at e13.5. (A–B) Axonal projections at brachial levels in wildtype and Hoxc9 mutants. Projections to limb (LMC) and axial muscles (MMC) are preserved. (C–D) In Hoxc9 mutants, axonal projections to sympathetic chain ganglia (scg) are significantly reduced at e13.5 (arrows). See also Figure S3. Vibratome sections show GFP+ motor axons in green, Isl1/2+ scg and dorsal root ganglion (drg) neurons in red. (E–F) Schematic representations of axonal projections of thoracic MNs in wildtype and Hoxc9 mutants. (G–H) The thickness of the intercostal nerves is increased in Hoxc9 mutants (white bars). (I–N) Retrograde labeling of MNs after rhodamine (RhD) injection into intercostal nerves. Ectopic FoxP1high LMC neurons are labeled in Hoxc9 mutants, while Lhx3+ MMC MNs are not labeled.
Although molecular features of HMC identity were lost in Hoxc9 mutants, motor axon projections toward hypaxial muscles were present, and there was a >2-fold increase in the overall thickness of the intercostal nerves (16.6 +/− 0.1 μm in control versus 39.2 +/− 0.7 μm in Hoxc9 mutants at e13.5, n>10) (Figures 2G–2H). Because HMC and LMC neurons are similar in their initial pursuit of a distal and ventral trajectory, we hypothesized that in the absence of an appropriate peripheral target many of the aberrant LMC MNs projected like HMC neurons. To test this idea we injected rhodamine dextran (RhD) conjugates into the intercostal nerves of control and Hoxc9−/− mice and assessed the identity of retrogradely labeled neurons. In wildtype mice all RhD labeled MNs lacked FoxP1 expression while in Hoxc9 mutants labeled neurons expressed high levels of FoxP1 (Figure 2I–2J). None of the RhD-labeled neurons expressed the MMC marker Lhx3 in Hoxc9 mutants, consistent with the preservation of this columnar subtype (Figure 2K–2N). These observations indicate in the absence of Hoxc9 MNs fail to project to the sympathetic chain, and the ectopic LMC neurons follow the route normally taken by HMC neurons (Figure 2E–2F).
Hoxc9 is Cell Autonomously Required for Thoracic Fates and Restricting LMC Identity
Because Hoxc9 is broadly expressed at thoracic levels, including the mesoderm surrounding the neural tube (Figure S4W), and these peripheral tissues are known sources of patterning cues that control Hox profiles and MN fates (Bel-Vialar et al., 2002; Ensini et al., 1998; Liu et al., 2001), we performed experiments to determine whether Hoxc9 is cell autonomously required for PGC and HMC specification and restriction of Hoxc6. To ablate Hoxc9 expression selectively in spinal neurons we electroporated double stranded (ds) RNAs directed against Hoxc9 into stage 14 chick neural tube and examined the effects on Hox expression and columnar fates after 2–3 days of further development. Coelectroporation of Hoxc9 dsRNA with a nuclear LacZ expression plasmid (to mark electroporated cells) led to a significant reduction of Hoxc9 protein in the spinal cord (Figure 3A–3B). Knockdown of Hoxc9 had no effect on markers for early aspects of MN identity nor did it affect expression of Hoxa9, indicating the effect is specific for Hoxc9 (Figure 3C and data not shown).
Figure 3. Cell Autonomous Role of Hoxc9 in MN Fate and Hox Gene Expression.
(A–L) Analysis of Hoxc9 knockdown at thoracic levels after dsRNA electroporations in chick neural tube. Bolt indicates electroporated side. (A) Hoxc9 dsRNA reduces Hoxc9 protein expression. (B) Nuclear LacZ expression plasmid was coelectroporated to mark electroporated cells. Note that the LacZ plasmid labels only a fraction of cells that incorporate the dsRNA. (C) Hoxc9 dsRNA does not affect Isl1/2 expression. (D) Ectopic RALDH2 is detected after thoracic Hoxc9 RNAi. (E) Loss of pSmad expression. (F) The number of Hb9+Isl1/2+ HMC neurons is reduced after Hoxc9 removal. (G–L) Hoxc6, Hoxa4, Hoxc4, Hoxa5, Hoxa7, and Hoxc8 are ectopically expressed or upregulated in cells that have lost Hoxc9. (M–V) Derepression of Hoxc4, Hoxc5, Hoxa5, Hoxa7, and Hoxc8 expression at thoracic levels in Hoxc9 mutants. The normal brachial patterns of Hox genes were intact in Hoxc9 mutants (Figure S4A–S4H). Ectopic Hox5 expression was relatively weak at thoracic levels, possibly due to the presence of Hoxc8 which normally restricts Hox5 genes to rostral brachial MNs (Dasen et al., 2005). (W–X) Loss of Hoxd9 expression in MNs that ectopically express Hoxc6. (Y) Summary indicating brachially-restricted Hox genes that are ectopically expressed or upregulated in Hoxc9 mutants. Light-gray bars indicated reduced protein expression levels.
Consistent with the phenotype observed in mice lacking Hoxc9, after RNAi-mediated Hoxc9 ablation expression of Hoxc6 was detected in thoracic MNs (Figure 3G). Ectopic Hoxc6 expression was found only in neurons which had lost Hoxc9 indicating the effects are cell autonomous. In addition MNs that had lost Hoxc9 expressed LMC molecular determinants (RALDH2), failed to express markers for PGC MNs (pSmad), and there was a reduction of MNs with an HMC molecular profile (Figure 3D–3F). Thus Hoxc9 function is required within MNs for the generation of PGC and HMC neurons and the restriction of LMC fates. These observations suggest that thoracic MNs have the capacity to express Hoxc6 relatively late in development in the absence of changes in peripheral signals. In addition the RNAi experiments rule out the possibility that the alteration in Hoxc6 expression in Hoxc9 mutants is due to changes in cis-regulatory elements within the Hox-c locus.
Hoxc9 as a Global Regulator of Anterior Hox Genes
Within the ~50 motor pools present in brachial LMC neurons the profiles of Hox gene expression are determined through cross-repressive interactions between multiple Hox genes expressed at specific rostrocaudal and intrasegmental levels (Dasen and Jessell, 2009). While the selectivity of these interactions has been studied in LMC neurons, the potential influences of Hox9 proteins on the forelimb Hox network have not been fully explored. In Hoxc9 mutants and RNAi knockdown animals we found, unexpectedly, that all brachially-restricted Hox genes became derepressed at thoracic levels. A total of eight Hox genes, Hoxa4, Hoxc4, Hoxa5, Hoxc5, Hoxa6, Hoxc6, Hoxa7, and Hoxc8 were ectopically expressed or markedly upregulated in thoracic MNs after Hoxc9 removal (Figure 3G–3V, S4I–S4N). Hoxd9 was absent from MNs that expressed anterior Hox genes, while Hoxa9 was retained, suggesting some but not all aspects of thoracic “Hox identity” are eroded (Figure 3W–3X, S4O–S4P). The alterations in Hox profiles also appeared to reflect a broad function of Hoxc9 since in Hoxc9 mutants Hox4–8 genes were derepressed throughout the ventral spinal cord as well as in the surrounding mesoderm (Figure S4Q–S4X). These observations indicate that Hoxc9 is required throughout the embryo for restricting expression of more anterior Hox genes.
Do the observed changes in Hox profiles reflect a specific Hoxc9 function or a more general hierarchical relationship of posterior over anterior Hox genes? To address this question we analyzed additional mutants for Hox derepression within the spinal cord. Hoxa9 and Hoxd9 mutants did not express Hox4–8 genes at thoracic levels, consistent with the lack of changes in columnar fates (data not shown). We also analyzed Hoxa7 and Hoxc8 mutants, two genes expressed at brachial levels and at rostral thoracic regions. We did not observe a significant derepression of Hox4, Hox5 or Hox6 genes at thoracic levels in these mutants (data not shown). The brachial expression pattern of the more anterior Hox gene was similar to wildtype mice along the rostrocaudal axis in single mutants for Hoxc5, Hoxc6, Hoxa6, and Hoxa7 analyzed at e11.5 (data not shown). We conclude that Hoxc9 has a selective role in confining Hox4–Hox8 paralog expression to brachial levels (Figure 3Y).
Hoxc9 Expression is Sufficient to Suppress Limb-Level Hox Profiles and MN Fates
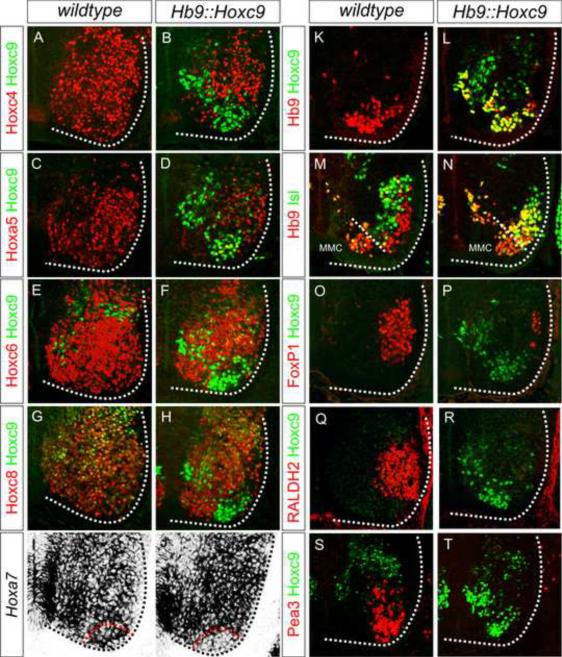
To further explore the repressive influences of Hoxc9, we examined the effects of misexpression in MNs. We used the regulatory sequences of the Hb9 gene to target expression to postmitotic MNs, and performed founder analysis of Hb9∷Hoxc9 mice at e12.5 (Figure 4K–4L). Each of the Hox paralogs expressed by brachial MNs including Hoxc4, Hoxa5, Hoxc6, Hoxa7 and Hoxc8 were repressed or markedly downregulated in Hb9∷Hoxc9 mice, consistent with a broad repressive function of Hoxc9 (Figure 4A–4J). Expression of Hoxc9 did not affect expression of Hoxa9 indicating the influences are specific for a subset of Hox genes (Figure S5E–S5F). The effects also proved to be cell autonomous, as Hox repression was restricted to MNs and appropriate Hox patterns were preserved in ventral interneurons (Figure 4A–4J). Thus Hoxc9 is capable of regulating a subset of Hox genes through repressive functions in MNs.
Figure 4. Hoxc9 Represses Brachial Hox Genes and LMC Identity.
(A–J) Brachial analysis of Hox profiles and motor neuron fates in e12.5 Hb9∷Hoxc9 embryos. Hoxc4, Hoxa5, Hoxc6, Hoxc8, and Hoxa7 expression are repressed or significantly downregulated by Hoxc9 in brachial MNs. Hox expression is preserved in the surrounding ventral interneurons. Red dashed line in panel (J) outlines region where Hoxc9 is misexpressed and corresponding region in control mice (I). (K–L) Hb9+ Hoxc9+ MNs are generated in Hb9∷Hoxc9 transgenic mice. (M–N) The number of Hb9+Isl1/2+ neurons is increased in Hb9∷Hoxc9 transgenic mice. In the absence of a Hox-induced program, MNs appear to remain in a HMC-like ground state. (O–R) Hoxc9 expression in brachial MNs reduces the number of FoxP1+ and RALDH2+ LMC MNs. (S–T) Hoxc9 expression blocks expression of the motor pool marker Pea3.
Previous gain of function studies in chick indicate that Hoxc9 activity prevents LMC specification by repressing Hox6 genes, while its activities in MN progenitors are required for PGC specification (Dasen et al., 2003). Consistent with these observations, postmitotic Hoxc9 expression under Hb9 control was not sufficient to induce PGC fate, and MNs appeared to remain in an HMC-like ground state (Figures 4M–4N, S5A–S5D). In contrast when Hoxc9 was activated in MN progenitors, by breeding mice containing a pCAGGs-loxP-stop-loxP-Hoxc9 cassette to Olig2∷Cre mice, ectopic PGC neurons were detected at brachial levels (Figure S5G–S5L). In Hb9∷Hoxc9 embryos expression of the LMC markers RALDH2 and FoxP1 were lost, and MNs also failed to express the pool marker Pea3, indicating that both Hox-dependent columnar and pool programs are blocked by Hoxc9 (Figure 4O–4T). Thus the absence of Hoxc9 expression from brachial levels appears necessary for MNs to express their appropriate Hox complement and execute their limb-level differentiation programs.
Assessment of the Functional Equivalence of Hox9 Paralogs
The apparent unique role of Hoxc9 in MN organization raises the question of whether the two other Hox9 paralogs expressed by MNs, Hoxa9 and Hoxd9, have a similar capacity to restrict expression of brachial Hox genes. We therefore examined Hoxa9 and Hoxd9 activities by misexpression in the chick neural tube. Previous studies have shown that Hoxa9 can convert LMC MNs to PGC neurons (Dasen et al., 2003), although the influence of Hoxa9 on brachial Hox expression was not assessed. We find that misexpression of Hoxa9 at brachial levels can repress the same group of Hox4–8 genes regulated by Hoxc9 (Figure S5M–S5P). Because Hoxa9 is still expressed in Hoxc9 mutants (Figure S4O–S4P), the absence of functional compensation by Hoxa9 is most likely a reflection of low levels of expression within the spinal cord.
Our gain of function analysis indicates that Hoxd9 is functionally distinct from Hoxa9 and Hoxc9. We find that brachial misexpression of Hoxd9 fails to induce PGC neurons, nor does it inhibit LMC specification (Figure S5Q–S5S). We unexpectedly find that elevating the levels of Hoxd9 at thoracic levels can induce LMC fates (Figure S5T). As with Hoxa9 and Hoxc9 misexpression, anterior Hox genes were repressed after brachial Hoxd9 misexpression suggesting Hoxd9 functions by promoting lumbar over brachial LMC identity (Figure S5U–S5X). In Hoxc9 mutants Hoxd9 expression is lost by MNs (Figure 3W–3X), thereby negating any potential repressive influence of Hoxd9 on the derepressed Hox genes. Taken with the observation that in Hoxa9 and Hoxd9 mutants anterior Hox genes are not derepressed, these data support the notion that Hoxc9 alone has a central role in restricting Hox4–8 gene expression from thoracic levels.
Thoracic Hox Derepression Alters Motor Pool Organization
The combinatorial actions of Hox4–8 genes are critical in the specification of motor pools targeting the forelimb. The expansion of all brachial Hox genes into thoracic levels raises the question of whether the network specifying pools might be preserved in a limbless environment and would generate the appropriate fates for a given transcriptional code. In principle Hox derepression could lead to several outcomes including 1) a scrambling of Hox codes for pool fates, 2) a wholesale shift of pools into the thoracic domain, or 3) the overall expansion of pools from brachial to thoracic levels. We assessed these possibilities by analyzing the expression and connectivity patterns of MNs expressing the transcription factors Pea3 and Scip, which mark pools within caudal LMC regions.
Expansion of the Pea3 motor pool in Hoxc9 mutants
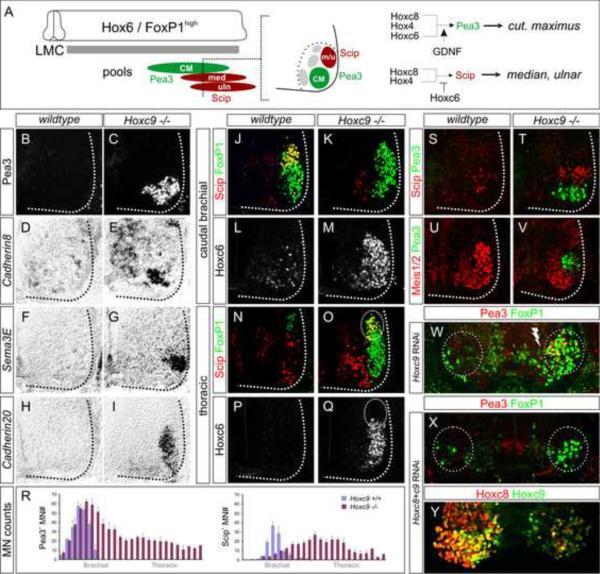
Pea3 expression is initially controlled by a network involving Hox4, Hoxc6, and Hoxc8 activities and marks MNs targeting the cutaneous maximus (CM) muscle (Figure 5A) (Livet et al., 2002). While the normal domain of Pea3 was grossly unaltered in Hoxc9 mutants, Pea3 expression was expanded throughout thoracic levels (Figure 5B–5C, 5R, S6A–S6D). Ectopic Pea3 MNs expressed Hoxc6 and Hoxc8, two proteins implicated in control of Pea3 expression (Figure S6A–S6H). Downstream targets of Pea3, including Cadherin8 and Sema3E, as well as Cadherin20 (Livet et al., 2002) were detected at thoracic levels in Hoxc9 mutants (Figure 5D–5I). Analysis of Hoxc9 RNAi knockdown animals also revealed ectopic Pea3 neurons at thoracic levels (Figure 5W and S6R). These observations indicate that the network controlling Pea3 can operate in the thoracic environment.
Figure 5. Motor Pool Reorganization in Hoxc9 Mutants.
(A) Schematic of the combinatorial Hox codes for motor pools at caudal brachial levels of the spinal cord. Pea3 marks cutaneous maximus (CM) MNs, Scip marks median (med) and ulnar (uln) MNs. Scip MNs are present at the most caudal brachial regions and require exclusion of Hoxc6. (B–I) Multiple markers of the CM pool are detected at thoracic levels in Hoxc9 mutants. The normal brachial patterns were preserved (Figure S6A–S6B and S6I–S6N). (J–M) At caudal brachial levels, upregulation of Hoxc6 expression in Hoxc9 mutants is accompanied by loss of brachial Scip LMC MNs. (N–Q) Altered position of the Scip pool. In Hoxc9 mutants Scip is expressed at thoracic levels. Ectopic Scip neurons are also detected in Hoxc9 RNAi ablated embryos (Figure S6S–S6T). (R) Cell counts for Pea3 and Scip MNs in wildtype and Hoxc9 mutants. Error bars represent n>3 Hoxc9 mutants. (S–T) Ectopic Scip+ and Pea3+ MNs at thoracic levels are clustered normally in Hoxc9 mutants. (U–V) Expression of Meis1/2 is excluded from the Pea3 pool. (W) Pea3 is ectopically expressed at thoracic levels after Hoxc9 RNAi. Bolt: electroporated side. (X) Knockdown of both Hoxc8 + Hoxc9 by dsRNA show that ectopic LMC neurons (FoxP1high) fail to generate ectopic Pea3 at thoracic levels. (Y) Loss of both Hoxc8 and Hoxc9 proteins after coelectroporation of dsRNAs.
We next assessed whether the presence of ectopic Pea3+ MNs in Hoxc9 mutants causes a redirection of motor axons to the CM. We first assessed projections to the CM using whole mount immunohistochemistry, finding that the level of innervation was similar between wildtype and mutant animals (Figure 6A–6B, S7G–S7J). We then performed retrograde tracing assays to ascertain the behavior of the ectopic populations of Pea3 MNs. Injection of RhD into the CM nerve labeled Pea3+ MNs that were confined to the normal brachial domain in Hoxc9 mutants (Figure 6E–6I). Injection into intercostal nerves revealed that the ectopic Pea3 MNs projected along the pathway normally followed by HMC neurons (Figure 6J–6L). This result was unexpected, as Pea3 expression relies on glial-derived neurotrophic factor (GDNF) signaling from the limb (Haase et al., 2002). Analysis of GDNF expression however revealed that in addition to the CM the intercostal mesoderm is a source of GDNF thus providing a permissive context for Pea3 induction (Figure S7A–S7F). In Hoxc9 mutants there is therefore an overall expansion of the Pea3 motor pool with the majority of ectopic MNs targeting inappropriate muscles.
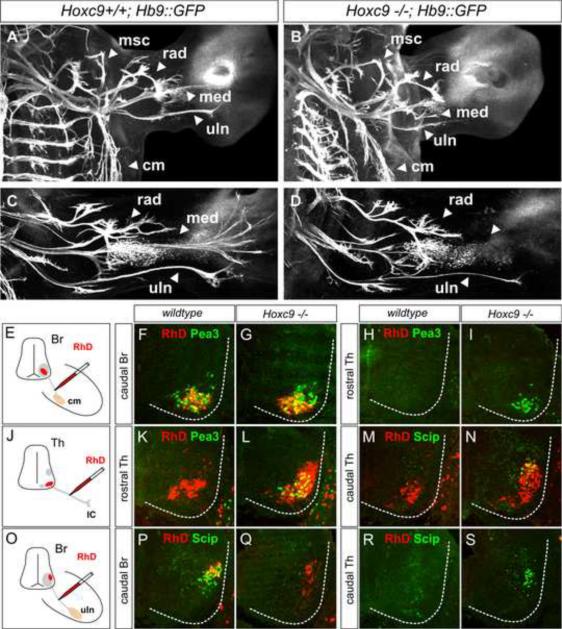
Figure 6. Altered Limb Innervation Patterns in Hoxc9 Mutants.
(A–D) Forelimb innervation in Hoxc9+/+; Hb9∷GFP and Hoxc9−/−; Hb9∷GFP embryos. Motor axons are visualized by whole mount GFP staining. (A–B) At e12.5 both ulnar (uln) and median (med) nerves show a reduction in length in Hoxc9 mutants. Musculocutaneous (mus), radial (rad), and cutaneous maximus (cm) nerves are similar to wildtype in Hoxc9 mutants. See also Figure S7G–S7J. (C–D) At e13.5 the density of ulnar projections are reduced and there is a loss of the distal branch of the median nerve in Hoxc9 mutants. (E–I) Labeled MNs after RhD injection into the CM nerves. RhD labels the normal Pea3 domain at caudal brachial levels in Hoxc9 mutants. (J–N) Ectopic Pea3 and Scip are labeled after RhD injection into the intercostal nerves at thoracic levels in Hoxc9 mutants. (O–S) In Hoxc9 mutants RhD labels scattered Scip− cells at caudal brachial region, but not ectopic Scip+ cells present at thoracic levels after ulnar injection.
Altered pool position and connectivity of Scip MNs in Hoxc9 mutants
We next analyzed the expression of the pool marker Scip which marks MNs projecting along the ulnar and median nerves (Dasen et al., 2005). Scip expression is confined to the most caudal brachial LMC MNs and is controlled by a network requiring Hoxc8, and the late exclusion of Hoxc6 (Figure 5A) (Dasen et al., 2005). We have additionally found that Scip+ MNs express low levels of Hoxc9 (Figure S6O–S6Q), suggesting a possible role in Hoxc6 restriction. Consistent with this idea we observed an upregulation of Hoxc6 at caudal brachial levels in Hoxc9 mutants and the number of Scip+ MNs was reduced (Figure 5J–5M). The loss of Scip MNs was associated with a reciprocal increase in the number of brachial Pea3+ MNs (Figure 5R) consistent with the idea that this pool is specified by a Hoxc6 + Hoxc8 code. Scip+ MNs were detected in Hoxc9 mutants, although this population was shifted to thoracic spinal cord (Figure 5N–5Q, 5R and S6S–S6T), where Hoxc6 levels are apparently reduced in a subset of MNs at the time of pool specification (Figure 5Q).
How does the altered position of Scip MNs affect the pattern of limb innervation? At e12.5 projections along the ulnar and medial nerves were consistently stunted in Hoxc9 mutants (Figure 6A–6B). By e13.5 there was a loss in the distal arbors of the median nerve and the density of ulnar projections was reduced (Figure 6C–6D). We then performed tracing assays to assess the identity of the few neurons projecting into the ulnar nerve and to define the target of ectopic Scip MNs. Ulnar injections of RhD in wildtype mice labeled clusters of LMC neurons that expressed Scip, while injections in Hoxc9 mutants labeled fewer neurons which were scattered and lacked Scip expression (Figure 6O–6Q). Retrograde tracing indicated that, like the ectopic Pea3+ MNs, the aberrant Scip+ neurons project along intercostal nerves (Figure 6M–6N and 6R–6S). Thus in the absence of Hoxc9 there is an erosion of the normal topographic relationship between the identity and projection pattern of motor pools, with the most dramatic effects on the innervation of distal limb muscles.
Characterization of the behavior of ectopic motor pools in Hoxc9 mutants
Additional aspects of the programs controlling MN pool fates were deployed at thoracic levels in Hoxc9 mutants. Pea3 and Scip MNs normally settle in distinct positions, with the Scip pool positioned dorsal to the Pea3 pool. This migratory behavior was retained in the thoracic environment and the ectopic Scip+ and Pea3+ MNs were well clustered (Figure 5S–5T). The specification of these pools requires exclusion of the transcription factor Meis1 (Dasen et al., 2005) and this Hox-dependent program was recapitulated at thoracic levels (Figure 5U–5V). The appearance of ectopic Pea3 and Scip MNs was also dependent on “motor pool” Hox genes, since dual RNAi-mediated knockdown of Hoxc9 and Hoxc8 in chick failed to generate ectopic Pea3 or Scip MNs, although ectopic LMC neurons were still present (Figure 5X–5Y, S6U–S6V). Together these observations indicate that under conditions of Hox derepression, this network is capable of specifying multiple facets of pool identity.
We next considered the possibility that the ectopic LMC pools in Hoxc9 mutants target specific groups of hypaxial muscles. Although the muscle-specific branches of intercostal nerves are too small to inject with tracers individually, we were able to inject at the initial bifurcation that segregates “internal” from “external” HMC axons. Interestingly, we find that injection of internal intercostal nerves in Hoxc9 mutants labeled LMC-like MNs that express Isl1 and lacked Lhx1 (Figure S7K–S7N). In addition all ectopic Scip neurons expressed Isl1, lacked Lhx1, and were labeled after internal intercostal nerve injections (Figure S7O–S7R). These observations suggest the ectopic LMC MNs do not project randomly into hypaxial muscle but may target specific muscle groups.
Hoxc9 Binds Multiple Regions within the Hox-a and Hox-c Clusters
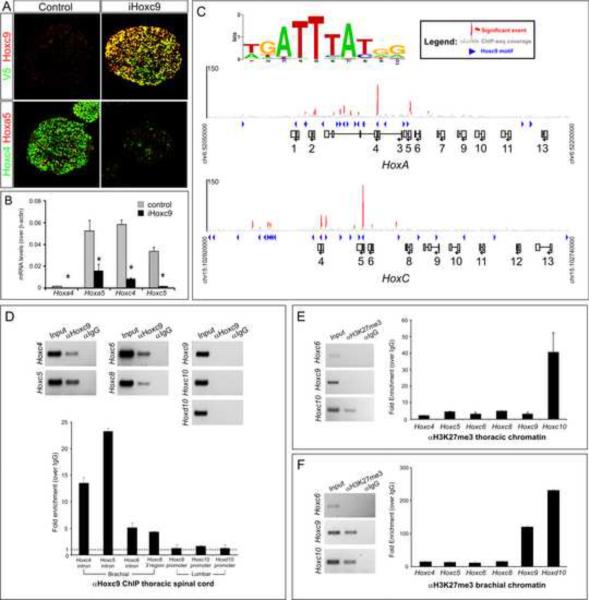
The derepression of a battery of Hox genes in Hoxc9 mutants suggests that the entire set is controlled in a concerted manner. Derepression could be a consequence of Hoxc9 acting on multiple Hox genes or could be a result of derepression of a Hox protein that coordinates brachial Hox gene activation. To determine whether Hoxc9 binds directly to Hox regulatory elements, we used an unbiased approach by taking advantage of an embryonic stem (ES) cell differentiation protocol that recapitulates MN development and allows generation of large quantities of material conducive for biochemical studies (Wichterle et al., 2002). To activate Hoxc9 expression during MN differentiation, epitope (V5) -tagged Hoxc9 was induced as cells become MN progenitors and was maintained until the end of differentiation. We first validated this approach by analyzing the effect of Hoxc9 expression on Hox profiles in ES cell derived MNs which, under standard conditions, are programmed to a rostral cervical (i.e. Hox4+ and Hox5+) identity. Similar to in vivo observations, Hoxc9 induction repressed rostral Hox genes including Hoxc4 and Hoxa5 (Figure 7A–B). Thus Hoxc9 retains its normal repressor function in the context of ES-cell derived MNs.
Figure 7. Genomic Analysis of Hoxc9 Binding at Hox Loci in Motor Neurons.
(A) Immunostaining showing induction of epitope (V5)-tagged Hoxc9 in embryonic bodies represses Hoxc4 and Hoxa5 expression. (B) RT-PCR analysis of Hoxa4, Hoxa5, Hoxc4, and Hoxc5 transcripts in control and Hoxc9 induced (iHoxc9) ES-cell derived MNs. (C) ChIP-seq signal maps for Hoxc9 binding sites within the Hox-a and Hox-c loci. Hoxc9 consensus motifs are indicated by blue arrowheads and significant binding events are shown in red peak. (D) Hoxc9 binds anterior Hox gene regions at thoracic levels in vivo. Top panels show gel images and bottom panels RT-PCR analysis from ChIP assays. Potential binding sites of Hox genes were assessed by ChIP using Hoxc9 specific antibody. Binding of Hoxc9 to the Hoxa7 3' region was not detected, possibly due to reduced sensitivity of in vivo ChIP assay. Error bars represent standard deviation on triplicates. (E) H3K27me3 chromatin status at Hox gene promoters at thoracic levels (F) H3K27me3 status of Hox gene promoters at brachial levels.
We next performed chromatin immunoprecipitation assays followed by sequencing of the enriched DNA fragments (ChIP-seq) to identify potential binding regions within the Hox-c and Hox-a loci. The most over-represented binding motif at enriched sites was similar to the site described for Hox9 paralogs obtained from in vitro studies (Shen et al., 1997) suggesting that tagged Hoxc9 binds to cognate sequences (Figure 7C). Analysis of the location of binding regions indicated that Hoxc9 associates with genomic regions located 3' to the position of Hox9 genes (Figure 7C), including genes derepressed in Hoxc9 mutants. It is of note that the Hox4–6 paralogs, which display mutually exclusive patterns of expression with Hoxc9, contain binding sites situated within the first intron. In contrast both Hoxa7 and Hoxc8, whose expression overlaps with Hoxc9 at rostral thoracic levels, do not contain an intronic binding site, but rather a potential site located more distally (Figure 7C). Certain Hox genes may therefore have evolved differential sensitivities to Hoxc9 repression, with the regulatory sequences retaining conserved positions within Hox loci.
Genome-wide analysis of Hoxc9 binding sites was performed in the context of ES-derived cervical MNs, raising the question of whether a similar occupancy is present at thoracic levels in vivo. We therefore performed ChIP assays on chromatin prepared from e12.5 thoracic spinal cord. We took advantage of the observation that most thoracic spinal neurons, including MNs and interneurons, express Hoxc9 protein and provide a relatively pure population for ChIP analysis (Figure 1A). We found that the majority of the regions identified by ChIP-seq were coimmunoprecipitated with a Hoxc9 antibody when compared to control IgG (Figure 7D). In both assays Hoxc9 was not associated with its own promoter, nor was Hoxc9 associated with the promoter regions of Hoxc10 or Hoxd10 (Figure 7C–7D) in agreement with the finding that these genes are not derepressed in Hoxc9 mutants. These observations additionally suggest a distinct transcriptional mechanism to exclude lumbar Hox10 genes from thoracic spinal cord.
Studies in several systems indicate that Hox gene expression is regulated in part through chromatin modifications at specific lysine residues on histone H3. The repressed state of Hox genes are initiated and maintained by the actions of the polycomb group (PcG) complexes which promote the trimethylation of histone H3 at lysine 27, and subsequent interactions with associated repressor proteins (Schuettengruber and Cavalli, 2009). Using ChIP assays we assessed whether this repressive mark (H3K27me3) is present on brachial Hox genes at thoracic levels. Remarkably, none of the anterior Hox genes which were derepressed in Hoxc9 mutants were associated with high levels of the H3K27me3 mark (Figure 7E). In contrast more posteriorly expressed Hox genes, including Hoxc10 and Hoxd10 were trimethylated at K27 on H3 at thoracic levels (Figure 7E). When H3K27me3 ChIP was performed at brachial levels we found that both Hoxc9 and Hox10 promoters were associated with this repressive mark, suggesting that the exclusion of Hoxc9 from brachial MNs involves histone methylation-dependent silencing (Figure 7F). These observations indicate that different levels of the spinal cord exclude Hox genes by distinct mechanisms and are likely to explain why anterior Hox genes are derepressed at thoracic levels in Hoxc9 mutants while more posterior Hox genes retain their normal expression patterns.
Discussion
Hox genes are essential in the specification of vertebrate CNS cell types, although the strategies used to achieve specific Hox patterns during neuronal differentiation are poorly understood. We have found that a critical step in the transition from the early induction of Hox gene expression to the regionally restricted patterns in MNs is mediated through the actions of a broadly acting Hox gene repressor. These findings may have more general implications for understanding how Hox networks contribute to the diversification of other vertebrate cell types.
Our studies are consistent with the idea that MN diversity is established through a repression-based network, with Hoxc9 functioning as a selective determinant of thoracic columnar fates. In the absence of Hoxc9, motor columns typically associated with respiratory (HMC neurons) and autonomic (PGC) neuronal networks are lost and body wall muscles are appropriated by motor neurons that have acquired the molecular identity of cells involved in limb control. An unexpected finding in our analysis is that Hoxc9 has an additional role in shaping the overall organization of the motor system, by acting as a global repressor of anterior Hox genes and confining the diversity of MN subtypes to limb levels. Genome-wide analysis of Hoxc9 binding suggests that Hox gene repression is mediated through interactions at several loci within the Hox-a and Hox-c clusters. Furthermore, analysis of MN pool disorganization in Hoxc9 mutants provides insights into the strategies used to generate diverse subtypes. We discuss these findings in the context of Hox transcriptional networks and the control of CNS cell type diversification.
A Unique Role for Hoxc9 in Motor Neuron Columnar Fate Specification
While studies in invertebrate systems have established that Hox genes are crucial in the organization of body plans (McGinnis and Krumlauf, 1992), progress towards addressing Hox function in the vertebrate CNS has been thwarted by functional redundancies amongst paralog groups. In a systematic analysis of MN defects in Hox mutants we found that mutation of a single gene, Hoxc9, leads to a remarkably pervasive and fully penetrant phenotype. Hoxc9 mutants lack PGC and HMC neurons with the consequence that all thoracic MNs are transformed into an LMC molecular identity (Figure 8A). This specific activity contrasts with limb levels of the spinal cord, where several Hox genes appear to be necessary to establish the LMC columnar fate. At least three Hox genes, Hoxa10, Hoxc10 and Hoxd10 are required for establishing hindlimb LMC identity (Rousso et al., 2008; Tarchini et al., 2005; Wu et al., 2008) while forelimb LMC specification requires multiple Hox paralogs including Hox6 and Hox8 genes (Dasen et al., 2003; Vermot et al., 2005). In addition the lack of any discernable columnar phenotype in single mutants for Hoxa5, Hoxa6, Hoxa7, Hoxa9, Hoxc4, Hoxc5, Hoxc6, Hoxc8, and Hoxd9 indicates that Hoxc9 has a unique function in shaping the early organization of spinal motor columns.
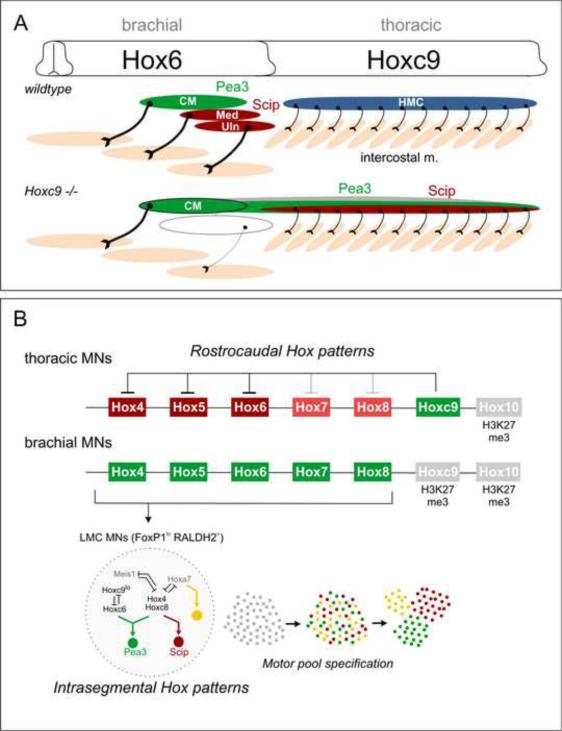
Figure 8. Hox Cross-Repression and Control of Motor Neuron Topography.
(A) Summary of alterations in MN organization and muscle innervation in Hoxc9 mutants. HMC MNs are lost and are transformed to an LMC identity. A subset of ectopic thoracic LMC MNs express Pea3 or Scip and project to intercostal muscle. Other motor pool fates may also be acquired by Hox derepression (indicated in gray). As a consequence of the reduction of Scip+ MNs at caudal brachial levels, median and ulnar nerve projections are profoundly reduced. (B) Hoxc9 is a key repressor of brachial Hox genes. In thoracic MNs, Hoxc9 represses brachial Hox genes by directly binding regulatory regions. The efficacy of Hoxc9 repression appears to be graded, where Hox4–6 genes are strongly repressed, while Hox7–8 gene repression is weaker. Hox10 genes are excluded by the distinct mechanism, likely involving H3K27 methylation-dependent silencing. At brachial levels, an intrasegmental Hox repressor network involving interactions amongst Hox4–8 genes determines pool fate on a cell-by-basis (Dasen et al., 2005). At the brachial-thoracic boundary Hoxc6 and Hoxc8 promote LMC fates, defined by high FoxP1 levels and RALDH2. Low levels of Hoxc9 repress Hoxc6 expression to specify the Scip+ LMC pool whereas MNs maintaining both Hoxc6 and Hoxc8 become Pea3+. Pool clustering occurs after MNs have acquired a specific identity.
In conjunction with previous observations, our findings suggest that Hoxc9 controls the identity of thoracic motor columns through distinct repressive and activator functions. When Hoxc9 is misexpressed in brachial progenitors, presumptive LMC MNs are programmed to a PGC identity, indicating that Hoxc9 has an active role in promoting PGC fate. PGC neurons additionally require specific Hoxc9 activator function, since dominant repressor derivatives fail to respecify LMC MNs, although Hox cross-repressive activities are retained (Dasen et al., 2003). The ability of Hoxc9 to promote PGC fates is likely to be dependent on interactions with the accessory factor FoxP1, since FoxP1 is also required for the specification of PGC MNs (Dasen et al., 2008; Rousso et al., 2008). Hoxc9/FoxP1 interactions may therefore facilitate activities at targets genes that are distinct from the sites repressed by Hoxc9 described in this study.
In contrast the switch of HMC neurons to an LMC fate in Hoxc9 mutants can be attributed to the loss of Hoxc9 repressor function. HMC neurons are normally specified in a Hox-independent manner, as in mice lacking Foxp1 both PGC and LMC MNs are switched to an HMC fate, independent of position or Hox profile (Dasen et al., 2008; Rousso et al., 2008). These observations suggest that the transformation of HMC to LMC MNs in Hoxc9 mutants is due to the derepression of LMC-promoting Hox genes, while the loss of PGC neurons reflects a requirement for Hox activator function. More generally the phenotype of Hoxc9 mutants fits well with a dual functionality for Hox proteins in cell type specification (Li and McGinnis, 1999), through their ability to both activate differentiation programs as well as restrict expression of determinants of other subtypes, even within the same cell.
Strategies for Coordinating Neuronal Diversity with the Periphery
Given the redundancies amongst vertebrates Hox genes why would a single thoracic Hox gene exert a central role in MN organization? Vertebrate species vary widely in the number of thoracic segments, ranging from as few as 6 in frogs to over 300 in certain species of snakes (Dequeant and Pourquie, 2008), and these morphological differences are thought to be shaped by regional Hox gene activities (Wellik, 2009). One possibility is that Hoxc9 is similar to DrosophilaHox genes, by acting as a global determinant of thoracic identity. Hoxc9 function in MNs however does not appear to be associated with the patterning of the thoracic skeletal structures, since these programs are grossly preserved in Hoxc9 mutants (McIntyre et al., 2007). Previous studies have implicated multiple Hox9 paralogs in specifying the regional identity of the lateral plate mesoderm that determines the rostrocaudal position where thoracic segments and limbs form (Cohn et al., 1997). Since Hoxc9 defines the identity of MNs that project into thoracic segments, as well as the position in which limb-innervating MNs are generated, one possibility is that the utilization of a single Hox gene for this purpose allows for a certain degree of adaptability specifically within the motor system, with additional Hox9 genes functioning to coordinately pattern mesoderm-derived structures.
Despite the lack of global morphological changes in Hoxc9 mutants we find that in addition to MNs several Hox genes are derepressed within thoracic mesoderm. Although the significance of this observation is unclear, several studies implicate Hox genes in specifying the precursors that give rise to MN target tissues. In the somites Hox genes have been shown to control the migratory behavior of myogenic precursors that generate the limb musculature (Alvares et al., 2003) while in the limb mesenchyme Hox genes have been implicated in the spatial organization of axonal guidance cues (Burke and Tabin, 1996). These Hox dependent steps in patterning mesodermal derivatives may serve to coordinate the specification of MN subtypes with peripheral signals that help shape motor axon target selection. Thus an additional role of Hoxc9 may be to pattern target regions, by restricting expression of certain Hox genes to forelimb level somitic and lateral plate mesoderm.
Hox Cross-Repression and the Emergence of MN Topographic Maps
Our studies indicate that Hoxc9 acts at an early stage of MN differentiation, by partitioning thoracic and limb-level subtypes, and through restricting Hox4–8 gene expression to brachial LMC MNs. This group of Hox genes has been shown to function as a network to specify the fates of the ~50 motor pools innervating the forelimb (Dasen et al., 2005). We find in Hoxc9 mutants that several downstream aspects of the motor pool Hox network are deployed in thoracic spinal cord, characterized by an expansion of pools expressing Pea3 and Scip, the induction of pool migratory behaviors, and expression of synaptic specificity determinants. As a consequence of Hox derepression there is a loss in the normal topographic relationship between MN position and peripheral target specificity, as most of the ectopic subtypes target inappropriate muscles. Nevertheless these findings are in agreement with a model in which early aspects of the programming of MNs identities, including their columnar, divisional, and pool fates, emerge through a cell intrinsic network, independent of specific signals provided by the limb mesoderm or differentiated muscle.
Analysis of the specification of the motor pool expressing Scip provides additional clues into how the Hox network controls MN diversification. In Hoxc9 mutants we observe a shift of the brachial Scip pool from its normal position, and an erosion of motor axon projections to the distal limb. Two observations suggest the identity of Scip+ MNs requires graded Hoxc9 activity, as opposed to an absolute repressive function used to establish a sharp molecular boundary. In gain and loss of function assays Hoxc9 exhibits repressive activities towards Hoxc8 and Hoxc6, yet Scip neurons express low Hoxc9 levels, retain Hoxc8, and lack Hoxc6. In addition at rostral thoracic levels many MNs coexpress Hoxc9 and Hoxc8 (Liu et al., 2001). These observations indicate that Hoxc9 does not function through a “winner take all” style of cross-repression as occurs during the specification of progenitors along the dorsoventral axis (Briscoe and Ericson, 2001). Similar graded interactions amongst Hox4–8 genes could be involved in the diversification of the ~50 pool fates within the LMC. More generally this strategy for the diversification of MN subtypes could apply to other CNS cell types programmed through networks of transcriptional repressors.
Transcriptional Mechanisms Controlling Hox Patterns in the CNS
Our studies provide insight into the mechanisms through which Hox gene expression boundaries are established during the specification of CNS cell types. The transcriptionally silenced state of Hox genes are maintained in part through the actions of PcG complexes, leading to the trimethylation of histone H3K27 and binding of additional factors which restrict promoter access to activating transcriptional machinery (Schuettengruber and Cavalli, 2009). ChIP analysis of thoracic spinal cord indicates that this mode of Hox repression is not used to silence brachial Hox genes, but rather is mediated through the actions of a single Hox factor. The idea that Hoxc9 directly represses Hox genes is supported by three lines of evidence: 1) Hoxc9 occupies a number of sites in the proximity of repressed Hox genes, 2) loss of Hoxc9 leads to ectopic expression of these same Hox genes at thoracic levels, and 3) misexpression of Hoxc9 represses brachial Hox genes. While Hox genes are known to be negatively regulated by micro- and long noncoding-RNAs (Rinn et al., 2007; Ronshaugen et al., 2005), it is unlikely that Hoxc9 acts through the induction of these regulatory molecules, since dominant-repressor Hoxc9 derivatives display similar repressive activities (Dasen et al., 2003). Although the precise mechanism by which Hoxc9 represses is unresolved, it may include more typical forms of gene regulation, such as selective recruitment of corepressors to be identified.
Hoxc9 mutation does not appreciably affect expression of more posterior lumbar-level Hox genes, raising the question of how they are spatially regulated. Our ChIP analysis of H3K27me3 patterns suggests a distinct mechanism for the restriction of more posterior Hox genes in the spinal cord. We find that at thoracic levels Hox10 promoters contain the H3K27me3 repressive mark, and at brachial levels both Hoxc9 and Hox10 genes are characterized by the presence of this histone modification. The exclusion of more posterior Hox genes in MNs could therefore be mediated by the maintenance of repressive chromatin structure within a Hox cluster. Consistent with this idea, mice bearing mutations in PcG components are characterized by anterior shifts in Hox gene expression, whereas posterior boundaries are maintained (van der Lugt et al., 1996). Thus the mechanisms controlling Hox exclusion at a given segmental level appear to be directionally distinct: recruitment of a Hox protein for repression of anterior Hox genes and silencing of more posterior Hox genes through the actions of PcG complexes (Figure 8B).
Hox Repression in the Assembly of Spinal Neuronal Networks
While our studies have focused on repressive interactions during MN development, it is likely that Hoxc9 and Hox genes in general play a broader role in shaping connections within motor networks. In addition to MNs, Hoxc9 mutation causes a derepression of Hox genes in other cell types, including ventral interneurons. While the role of Hox genes in these diverse classes is unresolved, the local circuits of neurons which coordinate the rhythmic firing patterns of MNs during respiration and locomotion are known to occupy distinct rostrocaudal levels of the spinal cord (Ballion et al., 2001; Kjaerulff and Kiehn, 1996). It is possible that the shared expression of Hox genes in multiple neuronal classes helps establish selective connections in developing motor circuits. Hoxc9 may therefore have a more general role in specifying the regionally restricted subtypes essential for the emergence of motor behaviors through global regulation of neuronal Hox patterns.
Experimental Procedures
Mouse Genetics
The Hox mutant strains are described in McIntyre et al. (2007), the Hb9∷GFP line in Arber et al., (1999). The Hb9∷Hoxc9 construct was generated as described (Dasen et al., 2003) and microinjected into mouse zygotes by standard procedures.
In Ovo Chick Electroporations
Electroporation was performed in chick embryos as described (Dasen et al., 2003). RNA interference was performed using 21-nucleotide dsRNAs (Dharmacon, Option A4). To identify electroporated neurons, siRNAs (suspended in TE to a final concentration 5mg/ml) were combined with a nuclear LacZ expression plasmid (0.5 mg/ml). Target sequence against chick Hoxc9 was: 5'-CGAAGTAGCCCGAGTCCTA-3'. Results for each experiment are representative of at least eight electroporated embryos from three or more independent experiments in which the electroporation efficiency in MNs was >60%.
Chromatin Immunoprecipitation (ChIP) Assays
Brachial and thoracic spinal cords were dissected from e13.5 mouse embryos. Tissues were homogenized in 1.1% formaldehyde using a Dounce B homogenizer. Chromatin was extracted and fragmented to 500–1000 bp by sonication (12 pulses of 10 seconds at 50% amplitude with 50 seconds between pulses). Chromatin extracts (~20 ug) were incubated overnight at 4°C with either specific antibodies or species-matched IgGs. Antibodies used are: rabbit anti-Hoxc9, rabbit anti-H3K27me3 (Upstate). Protein A-agarose (Roche) was added for 3 hours at 4°C and the antibody-protein-DNA complexes were washed 7 times with RIPA and eluted in 1% SDS. DNA-protein decrosslinking was performed overnight at 65°C followed by RNAse and proteinase K treatment at 55°C for 3 hours. DNA was purified using QIAquick columns (Qiagen). Hox regions were amplified using Power Sybr® Green PCR Master Mix (Applied Biosystems) and detected with Mx 3005P real-time PCR apparatus (Stratagene). Fold enrichment were calculated over IgG using the ΔΔCt method: Fold enrichment = 2−(ΔΔCt), where ΔΔCt = (CtIP − CtInput)thoracic − (CtIgG − CtInput)brachial. Primer sequences and details of the Chip-Seq are available in Supplemental Information.
In Situ Hybridization and Immunohistochemistry
In situ hybridization and immunohistochemistry were performed on 16 μm cryostat sections as described (Tsuchida et al., 1994). Whole-mount GFP staining was performed as described (De Marco Garcia et al., 2008) and motor axons were visualized in projections of confocal Z-stacks (500–1000 μm). Antibodies were generated as described (Dasen et al., 2008; Dasen et al., 2005; Liu et al., 2001; Tsuchida et al., 1994). Other antibodies were obtained and used as follows: rabbit anti-nNOS 1:5000 (Cryostar), goat anti-Hoxc6 1:2000 (Santa Cruz), rabbit anti-GFP 1:1000 (Invitrogen). A Hoxc9 antibody was generated in guinea pigs using the peptide sequence: DSLISHENEELLASRFPTKKC.
Retrograde Labeling of Motor Neurons
Retrograde labeling of MNs was performed as described (Dasen et al., 2008). Lysine-fixable dextran-tetramethylrhodamine (RhD, Molecular Probes) was injected into severed muscle-specific nerves of e12.5–e13.5 embryos. To aid in the identification of nerves, we used GFP fluorescence from Hb9∷GFP transgenic mouse embryos, visualized using a MVX10 wide-field fluorescent macroscope (Olympus). Nerves were severed using Oban Bioscissors and RhD was injected onto the cut terminal. Embryos were incubated for 4 to 5 hours in oxygenated F12/DMEM (50:50) solution at 32–34°C and subsequently fixed in 4% paraformaldehyde.
Supplementary Material
Acknowledgements
We thank Tom Jessell, Gord Fishell and members of the lab for comments on the manuscript. Mario Capecchi and Deneen Wellik provided Hox mutant lines, Tom Jessell provided Olig2∷Cre mice, and Silvia Arber provided Pea3 antibodies. We thank NYU transgenic facility for mouse husbandry, and Brian Dynlacht for help with the in vivo ChIP assays. EOM is supported by Damon Runyon Cancer Research Foundation. JD is supported by grants from the Alfred P. Sloan Foundation, Burroughs Welcome Fund, McKnight Foundation, NIH R01 NS062822 and Project ALS. JD is an HHMI Early Career Scientist.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvares LE, Schubert FR, Thorpe C, Mootoosamy RC, Cheng L, Parkyn G, Lumsden A, Dietrich S. Intrinsic, Hox-dependent cues determine the fate of skeletal muscle precursors. Dev Cell. 2003;5:379–390. doi: 10.1016/s1534-5807(03)00263-6. [DOI] [PubMed] [Google Scholar]
- Arber S, Han B, Mendelsohn M, Smith M, Jessell TM, Sockanathan S. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- Ballion B, Morin D, Viala D. Forelimb locomotor generators and quadrupedal locomotion in the neonatal rat. Eur J Neurosci. 2001;14:1727–1738. doi: 10.1046/j.0953-816x.2001.01794.x. [DOI] [PubMed] [Google Scholar]
- Bel-Vialar S, Itasaki N, Krumlauf R. Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes in two distinct groups. Development. 2002;129:5103–5115. doi: 10.1242/dev.129.22.5103. [DOI] [PubMed] [Google Scholar]
- Blackburn J, Rich M, Ghitani N, Liu JP. Generation of conditional Hoxc8 loss-of-function and Hoxc8-->Hoxc9 replacement alleles in mice. Genesis. 2009;47:680–687. doi: 10.1002/dvg.20547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Ericson J. Specification of neuronal fates in the ventral neural tube. Curr Opin Neurobiol. 2001;11:43–49. doi: 10.1016/s0959-4388(00)00172-0. [DOI] [PubMed] [Google Scholar]
- Burke AC, Tabin CJ. Virally mediated misexpression of Hoxc-6 in the cervical mesoderm results in spinal nerve truncations. Dev Biol. 1996;178:192–197. doi: 10.1006/dbio.1996.0210. [DOI] [PubMed] [Google Scholar]
- Cohen S, Funkelstein L, Livet J, Rougon G, Henderson CE, Castellani V, Mann F. A semaphorin code defines subpopulations of spinal motor neurons during mouse development. Eur J Neurosci. 2005;21:1767–1776. doi: 10.1111/j.1460-9568.2005.04021.x. [DOI] [PubMed] [Google Scholar]
- Cohn MJ, Patel K, Krumlauf R, Wilkinson DG, Clarke JD, Tickle C. Hox9 genes and vertebrate limb specification. Nature. 1997;387:97–101. doi: 10.1038/387097a0. [DOI] [PubMed] [Google Scholar]
- Dalla Torre di Sanguinetto SA, Dasen JS, Arber S. Transcriptional mechanisms controlling motor neuron diversity and connectivity. Curr Opin Neurobiol. 2008;18:36–43. doi: 10.1016/j.conb.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Dasen JS, De Camilli A, Wang B, Tucker PW, Jessell TM. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell. 2008;134:304–316. doi: 10.1016/j.cell.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Jessell TM. Hox networks and the origins of motor neuron diversity. Curr Top Dev Biol. 2009;88:169–200. doi: 10.1016/S0070-2153(09)88006-X. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Liu JP, Jessell TM. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425:926–933. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Dequeant ML, Pourquie O. Segmental patterning of the vertebrate embryonic axis. Nat Rev Genet. 2008;9:370–382. doi: 10.1038/nrg2320. [DOI] [PubMed] [Google Scholar]
- Deschamps J, van den Akker E, Forlani S, De Graaff W, Oosterveen T, Roelen B, Roelfsema J. Initiation, establishment and maintenance of Hox gene expression patterns in the mouse. Int J Dev Biol. 1999;43:635–650. [PubMed] [Google Scholar]
- Ensini M, Tsuchida TN, Belting HG, Jessell TM. The control of rostrocaudal pattern in the developing spinal cord: specification of motor neuron subtype identity is initiated by signals from paraxial mesoderm. Development. 1998;125:969–982. doi: 10.1242/dev.125.6.969. [DOI] [PubMed] [Google Scholar]
- Gutman CR, Ajmera MK, Hollyday M. Organization of motor pools supplying axial muscles in the chicken. Brain Res. 1993;609:129–136. doi: 10.1016/0006-8993(93)90865-k. [DOI] [PubMed] [Google Scholar]
- Haase G, Dessaud E, Garces A, de Bovis B, Birling M, Filippi P, Schmalbruch H, Arber S, deLapeyriere O. GDNF acts through PEA3 to regulate cell body positioning and muscle innervation of specific motor neuron pools. Neuron. 2002;35:893–905. doi: 10.1016/s0896-6273(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kania A, Jessell TM. Topographic motor projections in the limb imposed by LIM homeodomain protein regulation of ephrin-A:EphA interactions. Neuron. 2003;38:581–596. doi: 10.1016/s0896-6273(03)00292-7. [DOI] [PubMed] [Google Scholar]
- Kjaerulff O, Kiehn O. Distribution of networks generating and coordinating locomotor activity in the neonatal rat spinal cord in vitro: a lesion study. J Neurosci. 1996;16:5777–5794. doi: 10.1523/JNEUROSCI.16-18-05777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmesser LT. The acquisition of motoneuron subtype identity and motor circuit formation. Int J Dev Neurosci. 2001;19:175–182. doi: 10.1016/s0736-5748(00)00090-3. [DOI] [PubMed] [Google Scholar]
- Li X, McGinnis W. Activity regulation of Hox proteins, a mechanism for altering functional specificity in development and evolution. Proc Natl Acad Sci U S A. 1999;96:6802–6807. doi: 10.1073/pnas.96.12.6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JP, Laufer E, Jessell TM. Assigning the positional identity of spinal motor neurons: rostrocaudal patterning of Hox-c expression by FGFs, Gdf11, and retinoids. Neuron. 2001;32:997–1012. doi: 10.1016/s0896-6273(01)00544-x. [DOI] [PubMed] [Google Scholar]
- Livet J, Sigrist M, Stroebel S, De Paola V, Price SR, Henderson CE, Jessell TM, Arber S. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron. 2002;35:877–892. doi: 10.1016/s0896-6273(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Maconochie M, Nonchev S, Morrison A, Krumlauf R. Paralogous Hox genes: function and regulation. Annu Rev Genet. 1996;30:529–556. doi: 10.1146/annurev.genet.30.1.529. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- McIntyre DC, Rakshit S, Yallowitz AR, Loken L, Jeannotte L, Capecchi MR, Wellik DM. Hox patterning of the vertebrate rib cage. Development. 2007;134:2981–2989. doi: 10.1242/dev.007567. [DOI] [PubMed] [Google Scholar]
- Nordstrom U, Maier E, Jessell TM, Edlund T. An early role for WNT signaling in specifying neural patterns of Cdx and Hox gene expression and motor neuron subtype identity. PLoS Biol. 2006;4:e252. doi: 10.1371/journal.pbio.0040252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Hollyday M. Development and migration of avian sympathetic preganglionic neurons. J Comp Neurol. 1991;307:237–258. doi: 10.1002/cne.903070207. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronshaugen M, Biemar F, Piel J, Levine M, Lai EC. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005;19:2947–2952. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousso DL, Gaber ZB, Wellik D, Morrisey EE, Novitch BG. Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons. Neuron. 2008;59:226–240. doi: 10.1016/j.neuron.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- Shah V, Drill E, Lance-Jones C. Ectopic expression of Hoxd10 in thoracic spinal segments induces motoneurons with a lumbosacral molecular profile and axon projections to the limb. Dev Dyn. 2004;231:43–56. doi: 10.1002/dvdy.20103. [DOI] [PubMed] [Google Scholar]
- Shen WF, Rozenfeld S, Lawrence HJ, Largman C. The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. J Biol Chem. 1997;272:8198–8206. doi: 10.1074/jbc.272.13.8198. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Smith CL, Hollyday M. The development and postnatal organization of motor nuclei in the rat thoracic spinal cord. J Comp Neurol. 1983;220:16–28. doi: 10.1002/cne.902200104. [DOI] [PubMed] [Google Scholar]
- Sockanathan S, Jessell TM. Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons. Cell. 1998;94:503–514. doi: 10.1016/s0092-8674(00)81591-3. [DOI] [PubMed] [Google Scholar]
- Soshnikova N, Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science. 2009;324:1320–1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
- Tarchini B, Huynh TH, Cox GA, Duboule D. HoxD cluster scanning deletions identify multiple defects leading to paralysis in the mouse mutant Ironside. Genes Dev. 2005;19:2862–2876. doi: 10.1101/gad.351105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA, Krumlauf R. Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat Rev Neurosci. 2000;1:116–124. doi: 10.1038/35039056. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Tumpel S, Wiedemann LM, Krumlauf R. Hox genes and segmentation of the vertebrate hindbrain. Curr Top Dev Biol. 2009;88:103–137. doi: 10.1016/S0070-2153(09)88004-6. [DOI] [PubMed] [Google Scholar]
- van der Lugt NM, Alkema M, Berns A, Deschamps J. The Polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mech Dev. 1996;58:153–164. doi: 10.1016/s0925-4773(96)00570-9. [DOI] [PubMed] [Google Scholar]
- Vermot J, Schuhbaur B, Le Mouellic H, McCaffery P, Garnier JM, Hentsch D, Brulet P, Niederreither K, Chambon P, Dolle P, et al. Retinaldehyde dehydrogenase 2 and Hoxc8 are required in the murine brachial spinal cord for the specification of Lim1+ motoneurons and the correct distribution of Islet1+ motoneurons. Development. 2005;132:1611–1621. doi: 10.1242/dev.01718. [DOI] [PubMed] [Google Scholar]
- Wellik DM. Hox genes and vertebrate axial pattern. Curr Top Dev Biol. 2009;88:257–278. doi: 10.1016/S0070-2153(09)88009-5. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Wu Y, Wang G, Scott SA, Capecchi MR. Hoxc10 and Hoxd10 regulate mouse columnar, divisional and motor pool identity of lumbar motoneurons. Development. 2008;135:171–182. doi: 10.1242/dev.009225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.