Abstract
Structural analysis of MRP1-NBD1 revealed that the Walker A S685 forms hydrogen-bond with the Walker B D792 and interacts with magnesium and the β-phosphate of the bound ATP. We have found that substitution of the D792 with leucine resulted in misfolding of the protein. In this report we tested whether substitution of the S685 with residues that prevent formation of this hydrogen-bond would also cause misfolding. Indeed, substitution of the S685 with residues potentially preventing formation of this hydrogen-bond resulted in misfolding of the protein. In addition, some substitutions that might form hydrogen-bond with D792 also yielded immature protein. All these mutants are temperature-sensitive variants. However, these complex-glycosylated mature mutants prepared from the cells grown at 27 °C still significantly affect ATP binding and ATP-dependent solute transport. In contrast, substitution of the S685 with threonine yielded complex-glycosylated mature protein that is more active than the wild-type MRP1, indicating that the interaction between the hydroxyl group of 685 residue and the carboxyl group of D792 plays a crucial role for the protein folding and the interactions of the hydroxyl group at 685 with magnesium and the β-phosphate of the bound ATP play an important role for ATP-binding and ATP-dependent solute transport.
Keywords: MRP1, Protein folding, Hydrogen-bond formation, Complex-glycosylated mature protein, ATP binding/hydrolysis, ATP-dependent solute transport
1. Introduction
Over-expression of multidrug resistance protein (MRP1) coffers the cancer cells resistant to multiple anticancer drugs (MDR) [1, 2]. Hydrophobicity plot analysis of MRP1 protein [3, 4] reveals that this protein has two nucleotide binding domains (NBD) and two transmembrane domains (TMD), similar to the topological structure of P-glycoprotein (P-gp) [5-8], except that there is an extra N-terminal transmembrane domain and a cytoplasmic linker region. This ATP-binding cassette (ABC) transporter couples ATP binding and hydrolysis to solute, including anti-cancer drugs, transport across biological membrane [9-12]. Due to this ATP-dependent anti-cancer drug transport, the anti-cancer drug concentration inside of the MRP1-over-expressing cancer cell is too low to kill the cell.
Structural analyses of many prokaryotic ABC transporters, including GlcV [13], MalK [14, 15], HisP [16], MJ0796 [17] and HlyB [18], revealed that the residues in Walker A motif, B motif [19], ABC signature sequence, Q-loop, H-loop, D-loop and an aromatic amino acid are involved in ATP binding and hydrolysis. In order to study the relationship between the structure and function of MRP1, we have mutated some of these residues to similar or dissimilar amino acids, such as the amino acids in Walker A motif K684 and K1333 [20, 21] and C682 and A1331 [22]; in Walker B motif D792, D793, D1454 and E1455 [20, 23, 24]; the aromatic amino acid W653 in NBD1 and Y1302 in NBD2 [25]; and H827 and H1486 in H-loop [26]. The results derived from these mutations provide a deep insight of how the protein transports the anticancer drugs out of the cell. However, substitution of the acidic amino acid D792 in Walker B motif with a hydrophobic residue, such as D792L- or D792A-mutated MRP1 [20, 23], caused misfolding of the protein and prevented further analysis of the mutated protein. Although point mutations leading to misfolding or mis-assembly of membrane proteins have been identified in many hereditary diseases [27], little is known regarding the in vivo folding mechanism of these multi-domain proteins [28]. Replacement of F508 residue in NBD1 of cystic fibrosis transmembrane-conductance regulator (CFTR) with other amino acids suggested that hydrophobic side chain interactions of F508 with surrounding residues are required for proper folding of NBD2 and the domain-domain assembly of the protein [29]. However, this mechanism of protein folding may not be applied to the misfolding caused by substitution of the acidic amino acid with a hydrophobic residue, such as D792L- or D792A-mutated MRP1 [20, 23]. Interestingly, structural analysis of human wild-type MRP1-NBD1 in a complex with Mg·ATP revealed that this acidic amino acid D792 in Walker B motif forms a hydrogen-bond with the S685 in Walker A motif [30]. In addition, the corresponding hydrogen-bond was also identified in other ABC transporters, such as rat CFTR-NBD1 [31] and HlyB [18]. We suspected that D792A or D792L mutation abolished the hydrogen-bond formation between D792 and S685 and resulted in misfolding of the mutated MRP1 protein. If that is the case, substitution of S685 with a residue that prevents formation of this hydrogen-bond should also result in misfolding of the protein. In this report we have mutated S685 to T, A, C, H, D or N and found that only S685T mutation formed complex-glycosylated mature protein, implying that the interaction between the hydroxyl group at 685 in Walker A motif and the carboxyl group at 792 in Walker B motif of NBD1 plays a crucial role for MRP1 protein folding. In addition, all these mutants, except S685T, are temperature-sensitive. However, even if they form mature protein at 27 °C, these mature MRP1 proteins bearing S685A, S685C, S685D, S685H or S685N mutations still do not have full ability to transport LTC4, indicating that the hydroxyl group at 685 (in serine or threonine) plays an important role for interacting with Mg·ATP [30].
2. Materials and methods
2.1. Materials
Sodium orthovanadate, EGTA, Triton X-100, bovine serum albumin (BSA) and ATP were purchased from Sigma. [14,15,19,20-3H]-leukotriene C4 (LTC4) was from PerkinElmer Life Sciences. [α-32P]-8-N3ATP and [γ-32P]-8-N3ATP were from Affinity Labeling Technologies. PNGase F and endoglycosidae H were derived from New England BioLabs. Fetal bovine serum was from Gemini Bio-Products. QuikChange site-directed mutagenesis kit was from Stratagene. Anti-mouse Ig conjugated with horseradish peroxidase was from Amersham Biosciences. Chemiluminescent substrates for western blotting were from Pierce.
2.2. Site-directed mutagenesis of MRP1
Wild-type human MRP1 cDNA cloned into pNUT expression vector [32] was used as a template for the in vitro mutagenesis. The serine residue at position of 685 in Walker A motif was mutated to either threonine (T), alanine (A), cysteine (C), histidine (H), asparagine (N) or aspartic acid (D) (Figs. 2A and 4A) by using the forward/reverse primers and the QuikChange site-directed mutagenesis kit from Stratagene [20]. The forward and reverse primers used to introduce these mutations are: S685T/forward, 5’-GTG GGC TGC GGA AAG ACG TCC CTG CTC TCA GCC-3’; S685T/reverse, 5’-GGC TGA GAG CAG GGA CGT CTT TCC GCA GCC CAC-3’; S685A/forward, 5’-GTG GGC TGC GGA AAG GCG TCC CTG CTC TCA GCC-3’; S685A/reverse, 5’-GGC TGA GAG CAG GGA CGC CTT TCC GCA GCC CAC-3’; S685C/forward, 5’-GTG GGC TGC GGA AAG TGC TCC CTG CTC TCA GCC-3’; S685C/reverse, 5’-GGC TGA GAG CAG GGA GCA CTT TCC GCA GCC CAC-3’; S685H/forward, 5’-GTG GGC TGC GGA AAG CAC TCC CTG CTC TCA GCC-3’; S685H/reverse, 5’-GGC TGA GAG CAG GGA GTG CTT TCC GCA GCC CAC-3’; S685N/forward, 5’- GTG GGC TGC GGA AAG AAC TCC CTG CTC TCA GCC-3’; S685N/reverse, 5’-GGC TGA GAG CAG GGA GTT CTT TCC GCA GCC CAC-3’; S685D/forward, 5’-GTG GGC TGC GGA AAG GAT TCC CTG CTC TCA GCC-3’; S685D/reverse, 5’-GGC TGA GAG CAG GGA ATC CTT TCC GCA GCC CAC-3’. The mutation of D792S in Walker B motif (Fig. 4A) was also introduced into the full length MRP1 cDNA by using the same strategy shown above. The forward and reverse primers used to introduce this mutation are: D792S/forward, 5’-GAC ATT TAC CTC TTC AGT GAT CCC CTC TCA GC-3’ and D792S/reverse, 5’-GC TGA GAG GGG ATC ACT GAA GAG GTA AAT GTC-3’. The underlined sequences are codons for mutated residues. The serine residue at position of 685 in Walker A motif in pDual/N-half/C-half [24] was mutated to either threonine (T) or alanine (A) by using corresponding forward/reverse primers and the QuikChange site-directed mutagenesis kit from Stratagene [20]. Recombinant viral DNA preparation and viral particle production were carried out according to the procedures described [24]. Regions containing these mutations were sequenced to confirm that the correct clones were obtained.
Fig. 2. Substitution of S685 in Walker A motif of NBD1 with an amino acid residue that prevents formation of hydrogen-bond with the D792 in Walker B motif significantly affects the protein folding.
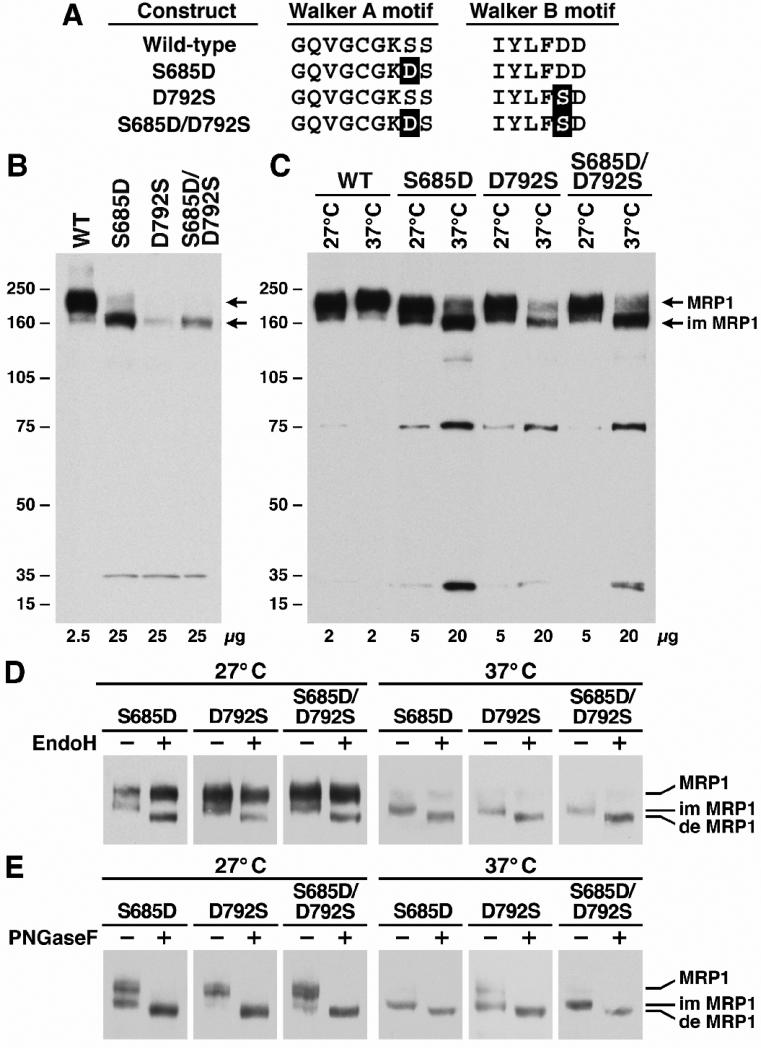
A. Sequence alignment of the Walker A and B motifs in NBD1 of MRP1. The highlighted letters indicate that the serine residue at 685 was substituted with the amino acid residues indicated at that position. The definition of S685A means that the S685 in Walker A motif was substituted with an alanine residue while the Walker B motif was un-mutated. B. Expression of wild-type and variant mutants of human MRP1 proteins in BHK cells at 37 °C. Cells were lysed with 2% SDS and loaded to a 7% polyacrylamide gel. The amounts of proteins loaded in the gel are indicated at the bottom of the gel. Molecular weight markers (kDa) are indicated on the left. MRP1 and im MRP1 on the right indicate the complex-glycosylated mature MRP1 and the core-glycosylated immature MRP1 proteins that were detected in western blot by employing the monoclonal antibody 42.4 against NBD1 [20].
Fig. 4. Switching the two residues in Walker A serine 685 and Walker B aspartic acid 792 did not rescue the misfolding.
A. Sequence alignment of the Walker A and B motifs in NBD1 of MRP1. The highlighted letters indicate that the S685 in Walker A motif was substituted with the D residue (S685D) whereas the D792 in Walker B motif was substituted with the S residue (D792S). B. Expression of wild-type and variant mutants of human MRP1 proteins in BHK cells at 37 °C. The experiments were carried out as described in Figure 2B. C. S685D, D792S and S685D/D792S are temperature-sensitive mutants. The experiments were carried out as described in Figure 3A. D. The complex-glycosylated mature MRP1 protein is endoglycosidase H resistant. The experiments were carried out as described in Figure 3B. E. Both core- and complex-glycosylated MRP1 proteins are sensitive to PNGase F. The experiments were carried out as described in Figure 3C.
2.3. Cell Culture and cell lines expressing MRP1
Baby hamster kidney (BHK-21) cells were cultured in DMEM/F12 medium supplemented with 5% fetal bovine serum at 37 °C in 5% CO2. Subconfluent cells were transfected with pNUT-MRP1/His in the presence of 20 mM HEPES (pH 7.05), 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 6 mM dextrose and 125 mM CaCl2 [32]. Surviving individual colonies in media containing 200 μM methotrexate were picked and amplified. Cells for membrane vesicle preparation were grown in DMEM/F12 media containing 5% fetal bovine serum and 200 μM methotrexate in roller bottles (on a roller machine from BELLCO). Spodoptera frugiperda 21 (Sf21) cells were cultured in Grace's insect cell medium supplemented with 5% heat-inactivated fetal bovine serum at 27 °C. Viral infection was performed according to the Invitrogen's recommendation.
2.4. Identification of MRP1 protein
Western blot was performed according to the method described previously [20]. 42.4 monoclonal antibody was used to identify the NBD1-containing fragments of MRP1, whereas 897.2 or 643.4 (recognize the fragment from 1248 to 1531) monoclonal antibody was used to detect the NBD2-containing fragments [20, 25]. The secondary antibody used was anti-mouse Ig conjugated with horse radish peroxidase. Chemiluminescent film detection was performed according to the manufacturer's recommendations (Pierce).
2.5. Membrane vesicle preparations
Membrane vesicles were prepared according to the procedure described previously [20]. The membrane vesicle pellet was re-suspended in a solution containing 10 mM Tris-HCl (pH 7.5), 250 mM sucrose and 1 X protease inhibitors (2 μg/ml aprotinine, 121 μg/ml benzamidine, 3.5 μg/ml E64, 1 μg/ml leupeptin and 50 μg/ml Pefabloc). After passage through a Liposofast™ vesicle extruder (Avestin, Ottawa, Canada) they were aliquoted and stored in –80 °C.
2.6. Membrane vesicle transport
ATP-dependent LTC4 transport was assayed by a rapid filtration technique [33, 34]. The assays were carried out in a 30 μl solution containing 3 μg of membrane proteins, 50 mM Tris-HCl (pH 7.5), 250 mM sucrose, 10 mM MgCl2, 200 nM LTC4 (17.54 nCi of 3H-labeled LTC4) and 4 mM AMP (used as negative control) or 4 mM ATP (or as indicated in the figure legend). After incubation at 37 °C for indicated time, the samples were brought back to ice and diluted with 1 ml of ice-cold 1 X transport buffer (50 mM Tris-HCl, pH 7.5, 250 mM sucrose and 10 mM MgCl2) and trapped on nitrocellulose membranes (0.2 μm) that had been equilibrated with 1 X transport buffer. The filter was then washed with 10 ml of ice-cold 1 X transport buffer, air-dried and placed in a 10 ml of biodegradable counting scintillant (Amersham Pharmacia Biotech). The radioactivity bound to the nitrocellulose membrane was determined by liquid scintillation counting (Beckman LS 6000SC).
2.7. Endoglycosidase H or PNGase F treatment of proteins
The core-glycosylated oligosaccharides were removed by digestion with endoglycosidase H. Cells were lysed with 2% SDS and sonicated for 20 pulses to break the genomic DNA. 10 μg of the cell lysates were digested overnight at 37 °C with 200 units (New England BioLabs) of endoglycosidase H in 200 μl of solution containing 50 mM sodium acetate buffer (pH5.3), 0.5% NP-40, 1% β-mercaptoethanol, 0.2% SDS (final concentration), 10 μg BSA and 1 X protease inhibitors [23].
The core-glycosylated and the complex-glycosylated oligosaccharides were removed by digestion with PNGase F. 10 μg of the cell lysates were digested overnight at 37 °C with 100 units (New England BioLabs) of PNGase F in 200 μl of solution containing 50 mM sodium phosphate buffer (pH7.5), 0.5% NP-40, 1% β-mercaptoethanol, 0.2% SDS (final concentration), 10 μg BSA and 1 X protease inhibitors [35].
Following digestion, 800 μl of cold ethanol was added to precipitate the proteins. The pellets were collected by centrifugation at 4 °C for 15 minutes. The proteins were resolved by SDS-PAGE (7%), electroblotted to nitrocellulose membrane and probed with monoclonal antibodies against human MRP1 protein [20].
2.8. Photoaffinity labeling of MRP1 protein
Vanadate preparation and photoaffinity labeling of MRP1 protein were performed according to procedures described previously [20]. Briefly, photolabeling experiments were carried out in a 10 μl solution containing 10 μg of membrane proteins, 40 mM Tris-HCl (pH 7.5), 2 mM ouabain, 10 mM MgCl2, 0.1 mM EGTA, 800 μM vanadate and either 10 μM [α-32P]-8-N3ATP or 10 μM [γ-32P]-8-N3ATP. After incubation at 37 °C for 2 minutes, the samples were brought back to ice and UV-irradiated for 2 minutes. The labeled proteins were separated by polyacrylamide gel (7%) electrophoresis and electro-blotted to a nitrocellulose membrane.
2.9. Statistical analysis
The results in Table 1 were presented as means ± SD from three independent triplicate-experiments. The two-tailed P value was calculated based on the unpaired t test from GraphPad Software Quick Calcs. By conventional criteria, if P value is less than 0.05, the difference between two samples is considered to be statistically significant.
Table 1.
Km (Mg·ATP) and Vmax (LTC4) Values of wild-type and mutant MRP1s.
| Vmax (pmol/mg/min)* | Km (μM)* | |
|---|---|---|
| MRP1 | 164.0 ± 7.0 | 59.0 ± 2.2 |
| S685T | 330.7 ± 8.8 | 143.0 ± 8.2 |
| S685D | 65.3 ± 1.2 | 249.3 ± 6.3 |
| D792S | 79.3 ± 2.1 | 245.3 ± 8.2 |
| S685D/D792S | 99.0 ± 2.9 | 151.3 ± 6.8 |
Km (Mg·ATP) and Vmax (LTC4) values for wild-type, S685T, S685D, D792S and S685D/D792S (n = 3) were derived from the corresponding Michaelis-Menten curves shown in Figure 6. The P value for comparison of Km (Mg·ATP) of wild-type versus S685T is 0.0001 and, therefore, the P values for other mutants must be at most 0.0001.
3. Results
3.1. Substitution of S685 in Walker A motif with an amino acid without the hydroxyl group affects the protein folding and processing
Structural analysis of wild-type MRP1-NBD1 revealed that the S685 residue in Walker A motif forms a tight hydrogen-bond with the D792 in Walker B motif [30]. Substitution of the D792 with either leucine or alanine caused misfolding of the mutated MRP1 protein [20, 23]. We suspected that D792A or D792L mutation abolished the hydrogen-bond formation between D792 and S685 and resulted in misfolding of the mutated MRP1 protein. In this report we tested whether substitution of S685 with A, C, H, N or T (Fig. 2A) would also have an effect on the protein folding or not. In order to determine whether or not some of the substitutions, such as T or C, would form hydrogen-bond with D792, the model of NBD1·ATP·ATP·NBD2 sandwich structure was established (Fig. 1). This model indicates that some of the substitutions, such as T, H or C, could potentially form hydrogen-bond with the carboxyl group in D792, whereas the other mutation, such as A, would not.
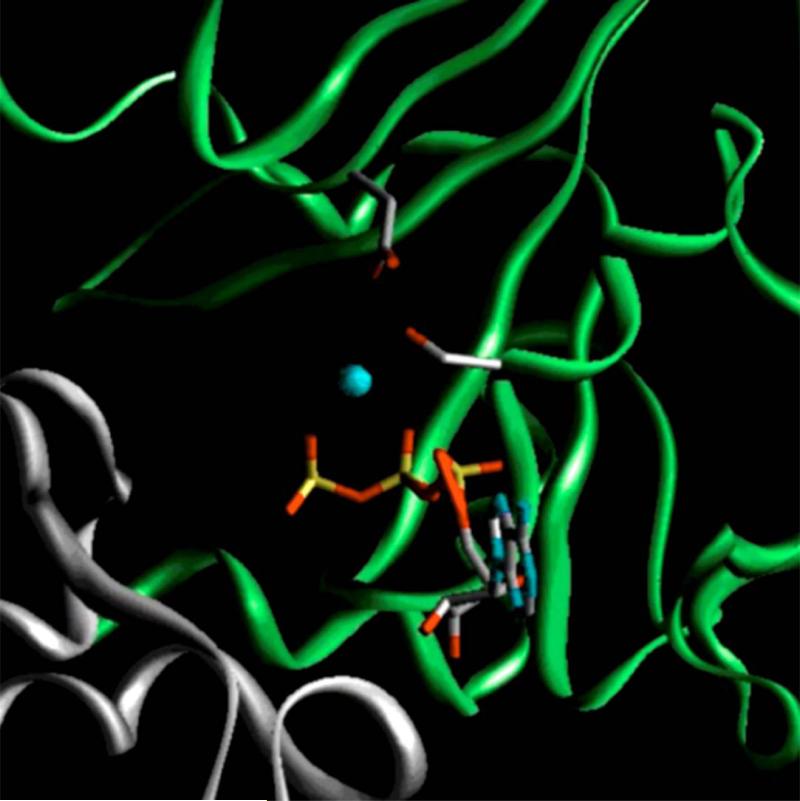
Fig. 1. Model of Mg·ATP-bound form of wild-type nucleotide binding site 1 of human MRP1.
The nucleotide binding domain 1 (NBD1, 643-855) of human MRP1 was modeled based on the crystal structure of Mg·ATP-bound form of wild-type MRP1-NBD1 [30], whereas the NBD2 was modeled using the MJ0796-E171Q homodimer [17] as a template. The gray ribbon indicates the residues from NBD1, whereas the green ribbon represents the residues from NBD2. The gray sticks on the gray ribbon represent S685 in Walker A motif and D792 in Walker B motif. ATP is shown in stick representation. Red indicates oxygen atom; blue, nitrogen; yellow, phosphorus; cyan, Mg++ ion.
Substitution of the S685 with an amino acid that prevents formation of the hydrogen-bond with D792, such as S685A, produced much less MRP1 protein than that of wild-type (Fig. 2B). In addition, majority of S685A is core-glycosylated immature protein at 37 °C, with an apparent molecular weight of ~ 165 kDa (Fig. 2B). Of note, a degradation product (~ 37 kDa) was detected by monoclonal antibody 42.4 [20] against NBD1 in S685A, but not in wild-type MRP1, implying that this mutant MRP1 protein synthesized in BHK cell is not as stable as wild-type. In order to test whether the hydroxyl group in S685 plays such an important role for the protein folding, this serine residue was mutated to threonine (S685T in Fig. 2A). Interestingly, S685T mutant produced similar amount of complex-glycosylated mature protein as wild-type (Fig. 2B), implying that the hydroxyl group in threonine plays a similar role as the one in serine residue. We, then, mutated this serine residue to other amino acids, such as S685C, S685N and S685H (Fig. 2A), that could potentially form hydrogen-bond with the D792 residue in Walker B motif. Although the ratio of mature versus immature MRP1 protein at 37 °C in S685C or S685N is slightly higher than in S685A (Fig. 2B and 3A), substitution of the S685 with either cysteine or asparagine cannot completely rescue the misfolding (Fig. 2B). S685H is an interesting mutant that produces similar amount of immature MRP1 protein and 37 kDa degradation product as S685A, but the amount of complex-glycosylated mature S685H is significantly higher than that of S685A, S685C or S685N (Fig. 2B), implying that the histidine residue at that position might form a weak hydrogen bond or salt bridge with the D792 in Walker B motif.
Fig. 3. S685A, S685C, S685H and S685N are temperature-sensitive mutants.
A. Wild-type and mutant MRP1 expression at either 27 °C or 37 °C. Cells were grown at either 27 °C or 37 °C as indicated on top of the gel, lysed with 2% SDS and loaded to a 7% polyacrylamide gel. B. The complex-glycosylated mature MRP1 protein is endoglycosidase H resistant. The samples were incubated in the absence (-) or in the presence (+) of endoglycosidase H according to the procedure described under “Materials and methods”. C. Both core- and complex-glycosylated MRP1 proteins are sensitive to PNGase F. The samples were incubated in the absence (-) or in the presence (+) of PNGase F according to the procedure described under “Materials and methods”. Molecular weight markers (kDa) are indicated on the left. MRP1, im MRP1 and de MRP1 on the right indicate the complex-glycosylated mature MRP1, the core-glycosylated immature MRP1 and the de-glycosylated MRP1 proteins that were detected in western blots by employing the monoclonal antibodies (mAb 643.4 against NBD2 was used in A, whereas mAb 42.4 against NBD1 and mAb 897.2 against NBD2 were used in B and C) against MRP1.
3.2. S685A, S685C, S685H and S685N are temperature sensitive mutants
Serine residue at 685 may also interact with the metal cofactor [30] and the β-phosphate of the bound ATP [17] and, thus, participates Mg·ATP binding. However, the amounts of mature MRP1 proteins produced by these mutants at 37 °C (Fig. 2B) may not be appropriate for functional analysis. We then tested whether these mutants are temperature sensitive variants or not. As shown in Figure 3A, all these mutants, including S685H, S685N, S685C and S685A, mainly form complex-glycosylated mature MRP1 proteins at 27 °C. In addition, the amount of MRP1 protein accumulated in BHK cells at 27 °C increased at least four fold (comparing to the amount of MRP1 protein accumulated in BHK cells at 37 °C). However, the amounts of MRP1 mutant proteins accumulated in BHK cells at 27 °C is still much less than that of wild-type or S685T (Fig. 3A), probably due to the mutants are not as stable as wild-type, even at 27 °C. This hypothesis is confirmed by the fact that the two degradation products, ~ 75 kDa and ~ 35 kDa, were clearly detected by mAb against NBD2 in the mutants of S685H, S685N, S685C or S685A grown at 27 °C, but not in wild-type or S685T (Fig. 3A).
If these mutants, including S685H, S685N, S685C and S685A, form complex-glycosylated mature MRP1 proteins at 27 °C, they should be distinguished from the core-glycosylated immature protein by digestion with endoglycosidase H [36]. The results in Figure 3B clearly indicate that the 165 kDa immature MRP1s, including S685H, S685N, S685C and S685A, are sensitive to endoglycosidase H digestion whereas the 190 kDa mature MRP1 proteins, regardless whether they are wild-type or mutants, are not (Fig. 3B). In contrast, both 165 and 190 kDa proteins are sensitive to the PNGase F digestion (Fig. 3C).
3.3. Switching the two residues in Walker A and B motifs did not rescue the misfolding
Our results indicate that the interaction between the hydroxyl group of the residue at 685, regardless whether it is a serine or threonine, and the carboxyl group of D792 plays a crucial role for MRP1 protein folding and processing. We, then, tested whether switching these two residues, i.e., substituting the Walker A S685 with aspartic acid and replacing the Walker B D792 with serine (Fig. 4A, S685D/D792S), would promote the proper folding. The results in Figure 4B indicated that substitution of the Walker A serine residue with aspartic acid (S685D) or Walker B mutant D792S resulted in misfolding of the protein. In addition, switching these two residues, S685D/D792S, also did not rescue the misfolding (Fig. 4B). All these three mutants, including S685D, D792S and S685D/D792S, are temperature-sensitive variants (Fig. 4C) that are not as stable as wild-type (Fig. 4C) and mainly degraded by proteasome (Data not shown). The core-glycosylated S685D, D792S and S685D/D792S mutants are sensitive to endoglycosidase H digestion whereas the 190 kDa mature MRP1 proteins are not (Fig. 4D). In contrast, both 165 kDa core-glycosylated and 190 kDa complex-glycosylated MRP1 proteins are sensitive to the PNGase F digestion (Fig. 4E).
3.4. Substitution of Walker A serine or Walker B aspartic acid with a residue that eliminates the hydroxyl group or carboxyl group significantly affects the ATP-dependent LTC4 transport
Since S685 in Walker A motif or D792 in Walker B motif is not only involved in hydrogen-bond formation between them, but also in interacting with the β-phosphate of the bound ATP and the metal co-factor [17, 30], thus, these mutations may not only affect the protein folding, but also the ATP-dependent solute transport. In order to test whether these mutations affect the ATP-dependent solute transport, membrane vesicles were prepared from the cells grown at 27 °C. Although the amounts of MRP1 mutants in the membrane vesicles are much less than that of wild-type or S685T (Fig. 5A), majorities of these mutants in membrane vesicles prepared from the cells grown at 27 °C are complex-glycosylated mature MRP1s (Fig. 5A). In order to compare their relative transport activities, the transport assays were carried out in a total of 3 μg of membrane proteins containing similar amount of MRP1 protein by adding varying amount of membrane vesicles prepared from parental BHK cells (Fig. 5B legend). For un-known reasons, the ATP-dependent LTC4 transport activity catalyzed by S685T is significantly higher than that of wild-type (Fig. 5B). In contrast, substitution of the serine residue with an alanine (S685A) that prevents the interactions with the magnesium co-factor and the β-phosphate of the bound ATP significantly reduced the ATP-dependent LTC4 transport activity (Fig. 5B). However, S685A mutation exerts much higher transport activity than that of the membrane vesicles containing CFTR (Fig. 5B), implying that this mutation did not completely abolish the ATP-dependent LTC4 transport activity. Interestingly, substitution of the Walker A serine residue with a cysteine (S685C), a histidine (S685H), an aspartic acid (S685D) or an asparagine (S685N) that may potentially interact with metal co-factor and the β-phosphate of the bound ATP exerts approximately two fold higher transport activity than that of S685A (Fig. 5B).
Fig. 5. ATP-dependent LTC4 uptake by membrane vesicles prepared from wild-type and mutant MRP1s.
A. Relative amounts of MRP1 proteins in membrane vesicles containing wild-type or mutant MRP1s. The membrane vesicles containing mutant MRP1s were prepared from cells grown at 27 °C so that majorities of the mutant MRP1s are complex-glycosylated mature proteins. The amounts of membrane vesicle proteins loaded in the gel are indicated on the top. MRP1 on the left indicates the 190 kDa complex-glycosylated mature MRP1 proteins that were detected in western blot by employing the monoclonal antibody 42.4 [20]. The intensities of the 190 kDa bands were determined by a scanning densitometer. The mean ratios (n = 3), considering the amount of wild type MRP1 as 1, of the mutant proteins are listed at the bottom of the gel. B. Relative rate of ATP-dependent LTC4 transport activity. The assays were carried out in a 30 μl solution containing 3 μg of membrane vesicles (the amount of MRP1 protein determined in panel A was adjusted to a similar amount by adding varying amount of membrane vesicles prepared from parental BHK cells) and 4 mM AMP (used as a control) or 4 mM ATP at 37 °C for 4 minutes. The amount of LTC4 bound to the membrane vesicles in the presence of 4 mM AMP was subtracted from the corresponding samples in the presence of 4 mM ATP. The data are means ± S.D. of four triplicate-determinations.
Substitution of the Walker B aspartic acid with a serine residue (D792S) that potentially interacts with the magnesium co-factor also exerts approximately two fold higher transport activity than that of S685A (Fig. 5B). In addition, switching these two residues, i.e., S685D/D792S, exerts even higher transport activity than that of S685D or D792S (Fig. 5B), implying that the interactions of these residues with metal co-factor and the β-phosphate of the bound ATP participate the ATP-dependent LTC4 transport.
3.5. The Km (Mg·ATP) values of S685D, D792S and S685D/D792S are significantly higher than that of wild-type MRP1
Six oxygen atoms, including the γ-oxygen of S685 residue in the Walker A motif and the δ-oxygen of D792 in the Walker B motif, interact with the magnesium co-factor to form the octahedral coordination geometry [30]. In addition, the γ-oxygen of S685 residue in the Walker A motif interacts with the β-phosphate of the bound ATP [17]. Therefore, it is reasonable to speculate that substitutions of S685 and D792 with residues that potentially interfere the interactions might decrease the binding force for Mg·ATP and affect ATP-dependent solute transport activity. To test this hypothesis, varying concentrations of ATP were utilized to do the ATP-dependent LTC4 transport in the presence of 10 mM MgCl2 (Fig. 6). As shown in Figure 6 and Table 1, the Vmax (LTC4) value of S685T is significantly higher than that of wild-type, consistent with the results derived from a solution containing 4 mM ATP and 10 mM MgCl2 (Fig. 5B). Although the exact mechanism of why S685T is more active than the wild-type is not clear, the higher Km (Mg·ATP) value of S685T (Table 1) might be a factor. However, the Km (Mg·ATP) values of S685D, D792S and S685D/D792S are even higher than that of S685T (Table 1), whereas the Vmax (LTC4) values of these mutants are much lower than that of wild-type. We simply interpreted these results as that the mutations of S685D, D792S and S685D/D792S not only affected ATP binding but also ATP hydrolysis and ATP-dependent solute transport. These results imply that the Km (Mg·ATP) value of S685A should even be higher than that of S685D or D792S. Unfortunately, the Km (Mg·ATP) value of S685A cannot be accurately determined due to the low transport activity.
Fig. 6. Mg·ATP-dependent LTC4 transport by membrane vesicles containing wild-type and mutant MRP1 proteins.
Membrane vesicles prepared in Figure 5 were used to do the transport experiments. The transport experiments were performed according to the methods described in “Materials and methods”. Briefly, the assays were carried out in a 30 μl solution containing 3 μg of membrane vesicles, 10 mM MgCl2 and varying concentrations of AMP (as a control) or ATP at 37 °C for 1 min. Since the amounts of MRP1 proteins determined in Figure 5A were different, the amounts of membrane vesicles used in these experiments were adjusted to a similar amount of MRP1 by adding varying amounts of membrane vesicles prepared from parental BHK cells: 0.81 μg of wild-type MRP1 + 2.19 μg BHK; 0.82 μg of S685T + 2.18 μg BHK; 3.00 μg of S685D; 2.31 μg of S685D/D792S + 0.69 μg BHK; 1.88 μg of D792S + 1.12 μg BHK. The data are the means ± S.D. of triplicate determinations.
3.6. Walker A serine and Walker B aspartic acid mutations affect ATP binding
The above results [higher Km (Mg·ATP) values] imply that the Walker A serine and Walker B aspartic acid mutations affect Mg·ATP binding. To test this hypothesis, similar amounts of wild-type, S685A-, S685T-, S685D-, D792S- and S685D/D792S-mutated MRP1s were used to do photo-affinity labeling at 37 °C with either [α-32P]-8-N3ATP or [γ-32P]-8-N3ATP in the presence of vanadate. As shown in Figure 7A, MRP1 protein, regardless whether it is wild-type or mutant, is labeled by [γ-32P]-8-N3ATP without UV-irradiation. These results are in harmony with our previous observation that MRP1 protein might be auto-phospharylated [20]. However, the amount of labeling on wild-type MRP1 with [γ-32P]-8-N3ATP, including the phospharylation and UV-cross-linking events, is much less than that of the labeling with [α-32P]-8-N3ATP (Fig. 7A), indicating that most of the bound ATP is hydrolyzed. For S685A-mutated MRP1, the amounts of labeling with either [α-32P]-8-N3ATP or [γ-32P]-8-N3ATP are much less than the corresponding labeling on wild-type MRP1 (Fig. 7A), implying that S685A mutation significantly affects the Mg·ATP binding. However, the amount of labeling on S685A with [α-32P]-8-N3ATP is higher than that of labeling with [γ-32P]-8-N3ATP (Fig. 7A), implying that certain amount of the bound ATP is hydrolyzed. Similar results (as S685A) were obtained from S685D-, D792S- and S685D/D792S-mutated MRP1s (Fig. 7A), implying that all these mutations affect Mg·ATP binding. In contrast, the labeling of S685T with [γ-32P]-8-N3ATP is significantly higher than the corresponding labeling on wild-type MRP1 (Fig. 7A). In addition, the labeling of S685T with [α-32P]-8-N3ATP is also higher than the corresponding labeling on wild-type MRP1 (Fig. 7A). The ratio of [γ-32P]-8-N3ATP labeling, after subtracting the labeling without UV-irradiation, versus [α-32P]-8-N3ATP labeling on wild-type MRP1 is approximately 0.3, whereas this ratio for S685T is approximately 0.6, implying that ATP bound to the S685T-mutated MRP1 may not be efficiently hydrolyzed. However, the ATP-dependent LTC4 transport activity of S685T-mutated MRP1 is much higher than that of wild-type (Fig. 5 and 6). In order to solve this puzzle, S685T was introduced into pDual/N-half/C-half expression vector [24, 25] and expressed in Sf21 cells. Membrane vesicles prepared from Sf21 cells were used to do the photo-affinity labeling at 37 °C with either [α-32P]-8-N3ATP or [γ-32P]-8-N3ATP. The [γ-32P]-8-N3ATP labeling of wild-type NBD1-containing N-half fragment is much higher than the corresponding labeling in NBD2-containing C-half fragment (Fig. 7B), whereas the [α-32P]-8-N3ATP labeling of wild-type NBD2-containing C-half fragment is much higher than the corresponding labeling in wild-type NBD1-containing N-half fragment (Fig. 7B), implying that ATP bound to the wild-type NBD2 is efficiently hydrolyzed. The [γ-32P]-8-N3ATP labeling of S685A-mutated-NBD1-containing N-half fragment is significantly lower than the corresponding labeling in wild-type NBD1-containing N-half (Fig. 7B), implying that S685A mutation affects Mg·ATP binding at the mutated NBD1. However, the [α-32P]-8-N3ATP labeling of the un-mutated NBD2-containing C-half of S685A-mutated MRP1 is much higher than the [γ-32P]-8-N3ATP labeling of the same fragment (Fig. 7B), indicating that ATP bound to this NBD2 is hydrolyzed. In contrast, the [γ-32P]-8-N3ATP or [α-32P]-8-N3ATP labeling of S685T-mutated-NBD1-containing N-half is significantly higher than the corresponding labeling on the wild-type N-half (Fig. 7B), presumably the S685T mutation increased affinity for 8-N3ATP at the mutated NBD1. In addition, the [γ-32P]-8-N3ATP labeling of S685T-mutated-NBD1-containing N-half is much higher than that of the labeling on the un-mutated NBD2-containing C-half (Fig. 7B) and the [α-32P]-8-N3ATP labeling of the un-mutated NBD2-containing C-half of S685T-mutated MRP1 is much higher than the [γ-32P]-8-N3ATP labeling of the same fragment (Fig. 7B), indicating that ATP bound to this NBD2 is efficiently hydrolyzed.
Fig. 7. Photolabeling of wild-type and mutant MRP1 proteins with either [α-32P]-8-N3ATP or [γ-32P]-8-N3ATP.
A. Walker A and B mutants mainly affect nucleotide binding. Membrane vesicles prepared in Figure 5 were used to do the photolabeling experiments. Photolabeling experiments were carried out in a 10 μl of solution containing 10 μg membrane vesicle proteins (the amount of MRP1 protein determined in Figure 5A was adjusted to a similar amount by adding varying amount of membrane vesicles prepared from parental BHK cells), 10 μM [α-32P]-8-N3ATP (α) or 10 μM [γ-32P]-8-N3ATP (γ) and 800 μM vanadate at 37 °C for 2 minutes. The reaction mixture was subjected to UV-irradiation (+) or without UV-irradiation (-) on ice for 2 minutes, separated by SDS-PAGE (7%) and electroblotted to a nitrocellulose membrane. Molecular weight markers (kDa) are indicated on the left. MRP1 on the right of the gel indicates the 32P-nucleotide labeled MRP1 proteins. B. S685A-mutated MRP1 mainly affects ATP binding at the mutated NBD1, but not the ATP hydrolysis at the un-mutated NBD2. S685A or S685T mutation was introduced into the pDual/N-half/C-half and expressed in Sf21 insect cells [24]. The membrane vesicles prepared from the viral particle infected Sf21 cells were used to do photolabeling as described in panel A. NH and CH on the right indicate the 32P-labeled NBD1-containing N-half and NBD2-containing C-half. All the experiments were repeated three times and the counts in each band were determined by Instant Imager [20].
4. Discussion
Structural analyses of human MRP1-NBD1 [30], mouse CFTR-NBD1 [31] and HlyB-NBD [18] revealed that Walker A serine residue forms a strong hydrogen-bond with the Walker B aspartic acid. We have found that substitution of the Walker B D572 of human CFTR-NBD1 with an asparagine [37] or substitution of the Walker B D792 of human MRP1-NBD1 with an alanine or a leucine residue [23] resulted in misfolding of the protein, implying that hydrogen-bond formation between the Walker A serine residue and the Walker B aspartic acid may play a crucial role for the protein folding. In this report we tested this hypothesis by substituting the S685 with a residue that might potentially prevent the hydrogen-bond formation between these two residues. Indeed, substitution of S685 with an alanine residue that eliminates the hydroxyl group in this serine residue resulted in misfolding of the protein at 37 °C (Fig. 2B and 3), indicating that the hydrogen-bond formation between these two residues may play a crucial role for the protein folding. If that is the case, replacement of the hydroxyl group in this serine residue with a thiol group, i.e., S685C mutation [Computer simulation (Fig. 1) of S685C-mutated NBD1 indicates that the distance between the thiol group of C685 and the carboxyl group of D792 is ~ 2.85 Ǻ], should result in a complex-glycosylated mature MRP1 protein. In fact, although the ratio of mature S685C versus immature protein at 37 °C is slightly higher than that of S685A (Fig. 3A), majority of S685C are high-mannose core-glycosylated (endoglycosidase H-sensitive) immature protein at 37 °C (Fig. 2B, 3A and 3B), indicating that the hydrogen-bond formation between these two residues is not the only factor that affects the protein folding. The results derived from S685H, S685N and D792S also support the above conclusion. For example, although majority of the S685H [The five-member imidazole ring in histidine might be partial positively charged under physiological pH and computer simulation (Fig. 1) of S685H-mutated NBD1 indicates that the distance between the nitrogen atom of H685 and the carboxyl group of D792 is ~ 1.93 Ǻ] are core-glycosylated immature protein at 37 °C (Fig. 2B and 3A), the ratio of mature S685H versus immature protein at 37 °C is much higher than that of S685A (Fig. 2B and 3A), implying that the hydrogen-bond formation between these two residues does play a role for the protein folding. Substitution of the hydroxyl group in S685 with a carboxyl group (S685D) will introduce a negatively charged group at that position and may repel each other with the negatively charged D792. As S685A mutant, elimination of the hydrogen-bond formation between D685 (the S685D mutation) and D792 resulted in misfolding of the protein (Fig. 4B and 4C). Interestingly, switching these two residues, i.e., S685D/D792S, might resume the hydrogen-bond formation between them but did not rescue the misfolding of the protein (Fig. 4B and 4C). Therefore, the mechanism by which these substitutions affect the protein folding is not yet clear. Presumably, substitutions of the Walker A serine with an aspartic acid and/or the Walker B aspartic acid with a serine residue might alter the interactions of the hydroxyl group in S685 or the carboxyl group in D792 with surrounding residues and cause overall conformational changes of the full-length molecule that affects the protein folding and trafficking to the plasma membrane. This hypothesis is supported by the fact that even though these mutants form complex-glycosylated mature protein at 27 °C, they are still not as stable as wild-type (Fig. 3A and 4C), implying the conformations of these mutants are different from that of wild-type. Computer simulation of full-length MRP1 molecule using other ABC transporter structure, such as Sav1866 [38], as a template may help to find the conformational changes induced by these mutations. However, computer simulation model may not reflect the real structure of this big membrane-bound glycoprotein. In contrast to other mutations, substitution of the Walker A serine residue with a threonine (S685T) that does not change the position of the hydroxyl group, except introducing an extra methyl group, results in complex-glycosylated mature MRP1 protein (Fig. 2B and 3). Therefore, the precise interactions of the hydroxyl group at 685 or carboxyl group at D792 with the surrounding residues including D792 and/or S685 play a crucial role for the protein folding.
If the above conclusion is a common feature of ABC transporters, substitution of the corresponding residues in other NBDs with amino acid that potentially abolishes the interactions with surrounding residues should also result in misfolding of the protein. Clearly, substitution of D555 in NBD1 or D1200 in NBD2 of P-gp with an asparagine did not cause misfolding of the protein [39]. In addition, substitutions of the Walker B motif D1454 and E1455 in NBD2 of MRP1 with a leucine residue (D1454L/E1455L) also did not cause misfolding of the protein [20]. In order to rule out the possibility that the double mutant D1454L/E1455L might rescue the misfolding caused by D1454L mutation, we have made single mutants including D1454L, D1454N, S1334A, S1334T, S1334C, S1334H, S1334D and S1334N. It turned out that none of these mutations caused misfolding of the protein (manuscript in preparation), implying that the stereo-structure of NBD2 may be different from that of NBD1 and interactions of the hydroxyl group in Walker A serine residue of NBD2 and/or the carboxyl group in Walker B aspartic acid of NBD2 with surrounding residues are not very important for the protein folding.
Structural analyses of human MRP1-NBD1 revealed that the tight hydrogen-bond formation between S685 and D792 stabilizes the protein in a proper conformation and completes the nucleotide binding site [30]. The γ-oxygen of the Walker A serine residue may also interact with the β-phosphate of the bound ATP [17]. In addition, the γ-oxygen of the Walker A serine residue and the δ-oxygen of the Walker B aspartate residue interact, along with other four oxygen atoms from β- and γ-phosphate of the bound ATP and water molecules, with Mg++ ion to form the octahedral coordination geometry [13, 14, 17, 30, 31, 40]. Thus, even though the corresponding mutations form complex-glycosylated mature protein [39], the mutations, such as D555N and D1200N in P-gp, eliminating the carboxyl group in Walker B motif significantly reduced the magnesium-dependent ATP binding [39, 41, 42]. Corresponding mutations in CFTR affect ATP-dependent chloride channel gating [43-45]. It is also true for the corresponding mutations in Walker B motif of NBD2 in MRP1, such as D1454L (manuscript in preparation) and D1454N [46], have a significant effect on the ATP-dependent LTC4 transport. Functional analyses of the Walker A serine mutants in NBD1, including membrane vesicles containing the complex-glycosylated mature and endoglycosidase H-resistant (Fig. 3B and 4D) S685H, S685C, S685A, S685D, S685N, D792S and S685D/D792S prepared from these temperature sensitive variants (Fig. 3 and 4) grown at 27 °C, indicate that these mutations affect ATP binding and ATP-dependent LTC4 transport (Table 1 and Fig. 5, 6 and 7). Interestingly, although the double mutant S685D/D792S did not rescue the misfolding of the protein, the ATP-dependent LTC4 transport activity of this double mutant was significantly higher than the single mutant S685D or D792S (Fig. 5 and Table 1), implying that the hydrogen-bond formation between D685 and S792 may play a role in stabilizing the protein in a proper conformation [30]. In addition, substitution of the Walker A serine residue with a threonine (S685T) without changing the position of the hydroxyl group exerted higher affinity for 8-azido-ATP (Fig. 7) at the mutated NBD1 and significantly increased ATP-dependent LTC4 transport activity (Table 1 and Fig. 5 and 6), emphasizing the importance of the hydroxyl group located at this position.
Acknowledgments
We thank Irene Beauvais for preparation of the manuscript and Marv Ruona for preparation of the graphics.
This work was supported by grant CA89078 (Xiu-bao Chang) from the National Cancer Institute, National Institutes of Health.
The abbreviations used are
- BHK
baby hamster kidney
- Sf21
Spodoptera frugiperda 21
- ABC
ATP binding cassette
- MRP1
multidrug resistance protein 1
- P-gp
P-glycoprotein
- CFTR
cystic fibrosis trans-membrane conductance regulator
- BSA
bovine serum albumin
- NBD
nucleotide binding domain
- LTC4
leukotriene C4
- 8-N3ATP
8-azidoadenosine 5’-triphosphate
- PBS
phosphate-buffered saline
- EGTA
ethylene glycol-bis(β-aminoethyl ether)N,N,N,N-tetraacetic acid
- SDS
sodium dodecyl sulfate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Cole SP, Bhardwaj G, Gerlach JH, Mackie JE, Grant CE, Almquist KC, Stewart AJ, Kurz EU, Duncan AM, Deeley RG. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line [see comments] Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 2.Mirski SE, Gerlach JH, Cole SP. Multidrug resistance in a human small cell lung cancer cell line selected in adriamycin. Cancer Res. 1987;47:2594–2598. [PubMed] [Google Scholar]
- 3.Bakos E, Hegedus T, Hollo Z, Welker E, Tusnady GE, Zaman GJ, Flens MJ, Varadi A, Sarkadi B. Membrane topology and glycosylation of the human multidrug resistance-associated protein. J Biol Chem. 1996;271:12322–12326. doi: 10.1074/jbc.271.21.12322. [DOI] [PubMed] [Google Scholar]
- 4.Hipfner DR, Almquist KC, Leslie EM, Gerlach JH, Grant CE, Deeley RG, Cole SP. Membrane topology of the multidrug resistance protein (MRP). A study of glycosylation-site mutants reveals an extracytosolic NH2 terminus. J Biol Chem. 1997;272:23623–23630. doi: 10.1074/jbc.272.38.23623. [DOI] [PubMed] [Google Scholar]
- 5.Chen CJ, Chin JE, Ueda K, Clark DP, Pastan I, Gottesman MM, Roninson IB. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell. 1986;47:381–389. doi: 10.1016/0092-8674(86)90595-7. [DOI] [PubMed] [Google Scholar]
- 6.Zhang JT, Ling V. Study of membrane orientation and glycosylated extracellular loops of mouse P-glycoprotein by in vitro translation. J Biol Chem. 1991;266:18224–18232. [PubMed] [Google Scholar]
- 7.Gros P, Croop J, Housman D. Mammalian multidrug resistance gene: complete cDNA sequence indicates strong homology to bacterial transport proteins. Cell. 1986;47:371–380. doi: 10.1016/0092-8674(86)90594-5. [DOI] [PubMed] [Google Scholar]
- 8.Gerlach JH, Endicott JA, Juranka PF, Henderson G, Sarangi F, Deuchars KL, Ling V. Homology between P-glycoprotein and a bacterial haemolysin transport protein suggests a model for multidrug resistance. Nature. 1986;324:485–489. doi: 10.1038/324485a0. [DOI] [PubMed] [Google Scholar]
- 9.Leier I, Jedlitschky G, Buchholz U, Cole SP, Deeley RG, Keppler D. The MRP gene encodes an ATP-dependent export pump for leukotriene C4 and structurally related conjugates. J Biol Chem. 1994;269:27807–27810. [PubMed] [Google Scholar]
- 10.Muller M, Meijer C, Zaman GJ, Borst P, Scheper RJ, Mulder NH, de Vries EG, Jansen PL. Overexpression of the gene encoding the multidrug resistance-associated protein results in increased ATP-dependent glutathione S-conjugate transport. Proc Natl Acad Sci U S A. 1994;91:13033–13037. doi: 10.1073/pnas.91.26.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jedlitschky G, Leier I, Buchholz U, Center M, Keppler D. ATP-dependent transport of glutathione S-conjugates by the multidrug resistance-associated protein. Cancer Res. 1994;54:4833–4836. [PubMed] [Google Scholar]
- 12.Loe DW, Almquist KC, Cole SP, Deeley RG. ATP-dependent 17 beta-estradiol 17-(beta-D-glucuronide) transport by multidrug resistance protein (MRP). Inhibition by cholestatic steroids. J Biol Chem. 1996;271:9683–9689. doi: 10.1074/jbc.271.16.9683. [DOI] [PubMed] [Google Scholar]
- 13.Verdon G, Albers SV, Dijkstra BW, Driessen AJ, Thunnissen AM. Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus: nucleotide-free and nucleotide-bound conformations. J Mol Biol. 2003;330:343–358. doi: 10.1016/s0022-2836(03)00575-8. [DOI] [PubMed] [Google Scholar]
- 14.Lu G, Westbrooks JM, Davidson AL, Chen J. ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation. Proc Natl Acad Sci U S A. 2005;102:17969–17974. doi: 10.1073/pnas.0506039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Hung LW, Wang IX, Nikaido K, Liu PQ, Ames GF, Kim SH. Crystal structure of the ATP-binding subunit of an ABC transporter. Nature. 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 17.Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Molecular Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. Embo J. 2005;24:1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. Embo J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou Y, Cui L, Riordan JR, Chang XB. Allosteric interactions between the two non-equivalent nucleotide binding domains of multidrug resistance protein MRP1. J Biol Chem. 2000;275:20280–20287. doi: 10.1074/jbc.M001109200. [DOI] [PubMed] [Google Scholar]
- 21.Buyse F, Hou YX, Vigano C, Zhao Q, Ruysschaert JM, Chang XB. Replacement of the positively charged Walker A lysine residue with a hydrophobic leucine residue and conformational alterations caused by this mutation in MRP1 impair ATP binding and hydrolysis. Biochem J. 2006;397:121–130. doi: 10.1042/BJ20051363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang R, Cui L, Hou Y-X, Riordan JR, Chang XB. ATP Binding to the First Nucleotide Binding Domain of Multidrug Resistance-associated Protein Plays a Regulatory Role at Low Nucleotide Concentration, whereas ATP Hydrolysis at the Second Plays a Dominant Role in ATP-dependent Leukotriene C4 Transport. J. Biol. Chem. 2003;278:30764–30771. doi: 10.1074/jbc.M304118200. [DOI] [PubMed] [Google Scholar]
- 23.Cui L, Hou YX, Riordan JR, Chang XB. Mutations of the Walker B motif in the first nucleotide binding domain of multidrug resistance protein MRP1 prevent conformational maturation. Arch Biochem Biophys. 2001;392:153–161. doi: 10.1006/abbi.2001.2441. [DOI] [PubMed] [Google Scholar]
- 24.Yang R, McBride A, Hou YX, Goldberg A, Chang XB. Nucleotide dissociation from NBD1 promotes solute transport by MRP1. Biochim Biophys Acta. 2005;1668:248–261. doi: 10.1016/j.bbamem.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Q, Chang XB. Mutation of the aromatic amino acid interacting with adenine moiety of ATP to a polar residue alters the properties of multidrug resistance protein 1. J Biol Chem. 2004;279:48505–48512. doi: 10.1074/jbc.M408343200. [DOI] [PubMed] [Google Scholar]
- 26.Yang R, Chang XB. Hydrogen-bond formation of the residue in H-loop of the nucleotide binding domain 2 with the ATP in this site and/or other residues of multidrug resistance protein MRP1 plays a crucial role during ATP-dependent solute transport. Biochim Biophys Acta. 2007;1768:324–335. doi: 10.1016/j.bbamem.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas PJ, Qu BH, Pedersen PL. Defective protein folding as a basis of human disease. Trends Biochem Sci. 1995;20:456–459. doi: 10.1016/s0968-0004(00)89100-8. [DOI] [PubMed] [Google Scholar]
- 28.Sanders CR, Myers JK. Disease-related misassembly of membrane proteins. Annu Rev Biophys Biomol Struct. 2004;33:25–51. doi: 10.1146/annurev.biophys.33.110502.140348. [DOI] [PubMed] [Google Scholar]
- 29.Du K, Sharma M, Lukacs GL. The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat Struct Mol Biol. 2005;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- 30.Ramaen O, Leulliot N, Sizun C, Ulryck N, Pamlard O, Lallemand JY, Tilbeurgh H, Jacquet E. Structure of the human multidrug resistance protein 1 nucleotide binding domain 1 bound to Mg2+/ATP reveals a non-productive catalytic site. J Mol Biol. 2006;359:940–949. doi: 10.1016/j.jmb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, Fowler R, Gao X, Guggino WB, Hendrickson WA, Hunt JF, Kearins MC, Lorimer D, Maloney PC, Post KW, Rajashankar KR, Rutter ME, Sauder JM, Shriver S, Thibodeau PH, Thomas PJ, Zhang M, Zhao X, Emtage S. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. Embo J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang XB, Hou YX, Riordan JR. ATPase activity of purified multidrug resistance-associated protein. J Biol Chem. 1997;272:30962–30968. doi: 10.1074/jbc.272.49.30962. [published erratum appears in J Biol Chem 1998 Mar 27;273(13):7782]
- 33.Leier I, Jedlitschky G, Buchholz U, Keppler D. Characterization of the ATP-dependent leukotriene C4 export carrier in mastocytoma cells. Eur J Biochem. 1994;220:599–606. doi: 10.1111/j.1432-1033.1994.tb18661.x. [DOI] [PubMed] [Google Scholar]
- 34.Loe DW, Almquist KC, Deeley RG, Cole SP. Multidrug resistance protein (MRP)-mediated transport of leukotriene C4 and chemotherapeutic agents in membrane vesicles. Demonstration of glutathione-dependent vincristine transport. J Biol Chem. 1996a;271:9675–9682. doi: 10.1074/jbc.271.16.9675. [DOI] [PubMed] [Google Scholar]
- 35.Almquist KC, Loe DW, Hipfner DR, Mackie JE, Cole SP, Deeley RG. Characterization of the M(r) 190,000 multidrug resistance protein (MRP) in drug-selected and transfected human tumor cell. Cancer Res. 1995;55:102–110. [PubMed] [Google Scholar]
- 36.Tarentino AL, Maley F. Purification and properties of an endo-beta-N-acetylglucosaminidase from Streptomyces griseus. J Biol Chem. 1974;249:811–817. [PubMed] [Google Scholar]
- 37.Gentzsch M, Choudhury A, Chang XB, Pagano RE, Riordan JR. Misassembled mutant {Delta}F508 CFTR in the distal secretory pathway alters cellular lipid trafficking. J Cell Sci. 2007;120:447–455. doi: 10.1242/jcs.03350. [DOI] [PubMed] [Google Scholar]
- 38.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 39.Hrycyna CA, Ramachandra M, Germann UA, Cheng PW, Pastan I, Gottesman MM. Both ATP sites of human P-glycoprotein are essential but not symmetric. Biochemistry. 1999;38:13887–13899. doi: 10.1021/bi991115m. [DOI] [PubMed] [Google Scholar]
- 40.Procko E, Ferrin-O'Connell I, Ng SL, Gaudet R. Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol Cell. 2006;24:51–62. doi: 10.1016/j.molcel.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 41.Sauna ZE, Smith MM, Muller M, Ambudkar SV. Functionally similar vanadate-induced 8-azidoadenosine 5'-[alpha- (32)P]Diphosphate-trapped transition state intermediates of human P- glycoprotin are generated in the absence and presence of ATP hydrolysis. J Biol Chem. 2001;276:21199–21208. doi: 10.1074/jbc.M100886200. [DOI] [PubMed] [Google Scholar]
- 42.Booth CL, Pulaski L, Gottesman MM, Pastan I. Analysis of the properties of the N-terminal nucleotide-binding domain of human P-glycoprotein. Biochemistry. 2000;39:5518–5526. doi: 10.1021/bi992931x. [DOI] [PubMed] [Google Scholar]
- 43.Bompadre SG, Cho JH, Wang X, Zou X, Sohma Y, Li M, Hwang TC. CFTR gating II: Effects of nucleotide binding on the stability of open states. J Gen Physiol. 2005;125:377–394. doi: 10.1085/jgp.200409228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vergani P, Nairn AC, Gadsby DC. On the mechanism of MgATP-dependent gating of CFTR Cl- channels. J Gen Physiol. 2003;121:17–36. doi: 10.1085/jgp.20028673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilkinson DJ, Mansoura MK, Watson PY, Smit LS, Collins FS, Dawson DC. CFTR: the nucleotide binding folds regulate the accessibility and stability of the activated state. J Gen Physiol. 1996;107:103–119. doi: 10.1085/jgp.107.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Payen L, Gao M, Westlake C, Theis A, Cole SP, Deeley RG. Functional Interactions Between Nucleotide Binding Domains and Leukotriene C4 Binding Sites of Multidrug Resistance Protein 1 (ABCC1) Mol Pharmacol. 2005;67:1944–1953. doi: 10.1124/mol.104.007708. [DOI] [PubMed] [Google Scholar]