Summary
Cell migration within a natural context is tightly controlled, often by specific transcription factors. However, the switch from stationary to migratory behavior is poorly understood. Border cells perform a spatially and temporally controlled invasive migration during Drosophila oogenesis. Slbo, a C/EBP family transcriptional activator, is required for them to become migratory. We purified wild-type and slbo mutant border cells as well as nonmigratory follicle cells and performed comparative whole-genome expression profiling, followed by functional tests of the contributions of identified targets to migration. About 300 genes were significantly upregulated in border cells, many dependent on Slbo. Among these, the microtubule regulator Stathmin was strongly upregulated and was required for normal migration. Actin cytoskeleton regulators were also induced, including, surprisingly, a large cluster of “muscle-specific” genes. We conclude that Slbo induces multiple cytoskeletal effectors, and that each contributes to the behavioral changes in border cells.
Introduction
In complex, multicellular animals, cell migration is a very tightly controlled process. Specific cells migrate at specific times during development and, to a lesser extent, in adult animals. While cells of the immune system may be considered professional migratory cells, most other cells are only migratory in a specific phase; their migratory behavior needs to be activated and later inactivated. Examples are neurons, glial cells, neural crest derivatives, germline cells, as well as certain muscle cells. If the migratory cells arise in an epithelium, the cells may undergo an epithelial-to-mesenchymal transition to become migratory. Like other cell fate changes, induction of migratory behavior can be triggered by specific signals and transcription factors and is associated with changes in gene expression (Birchmeier and Brohmann, 2000; Gammill and Bronner-Fraser, 2002; Montell, 2001; Rørth, 2002). The transcription factors are, however, different for different situations. To understand how exactly cells become migratory, it is therefore necessary to understand which genes are regulated downstream of the transcription factors that induce the switch. Once this is understood for specific, well-characterized transitions, the patterns can be compared to determine whether there is a common gene expression cassette that is regulated for cells to become migratory. Alternatively, each cell type may employ a different strategy to release itself from the constraints of a tissue and move away. So far, a complete expression profile reflecting the transcriptional switch has not been obtained from any controlled cell migration process.
While controlled cell migration behavior is useful for animal development, it also has pathological correlates. If tumor cells originating in an epithelium become migratory, this will likely contribute to their ability to metastasize and therefore be dangerous. A large number of studies have analyzed metastasis-associated gene expression. This has been done by comparison of cell lines with different migratory or metastatic potential, which, if done in vitro, may be problematic (Tatenhorst et al., 2005). Another, more physiological approach is to compare gene expression profiles of groups of metastatic and nonmetastatic tumors of a particular type (van 't Veer et al., 2002; Dyrskjot et al., 2003). The latter approach appears to be very useful for collecting predictive markers for metastasis, but, given the complexity of metastasis, it is likely less useful for understanding how cells become migratory. A more refined analysis to identify genes specifically involved in cell migration in this context was done by capturing the cells within a tumor that are most motile and analyzing their expression profiles (Wang et al., 2004). As metastasizing tumor cells may need to reactivate a developmental program normally used by cells to become migratory, studies of normal migratory switches are also likely to contribute to our understanding of metastasis.
To analyze how cells become migratory in response to a normal transcriptional switch, we have analyzed gene expression patterns in border cells of the Drosophila ovary. Border cells are a small group of cells that perform a well-defined and controlled migration in vivo and have become a useful model for studying invasive cell migration (Montell, 2001; Rørth, 2002). Border cells delaminate from the follicular epithelium, invade into the underlying germline tissue, and migrate to the oocyte. One of the key transcription factors inducing migration of border cells is Slbo (Drosophila C/EBP). Expression of Slbo requires a spatial signal (unpaired) from the preexisting anterior polar cells to induce activation of the JAK/STAT pathway in the prospective outer border cells. The anterior polar cells will become the central two cells of the border cell cluster. Together with temporal signals, this leads to the determination of border cells, and thus specific expression of Slbo, at the anterior tip of an egg chamber at a specific time. The effect of Slbo, in turn, appears to be relatively direct, as the border cells migrate a few hours after Slbo is expressed. Slbo expression is essential for border cell migration. Slbo is also expressed in centripetal cells at a slightly later stage, but it is not required for their migration. To understand how border cells become migratory in response to Slbo, we need to know which genes are regulated by Slbo in border cells. In order to determine this, we have carried out genome-wide transcription profiling of border cells purified directly from ovaries. We compared wild-type border cells with their slbo mutant counterparts and also compared border cells with neighboring follicle cells of the same developmental stages. Further functional studies with mutations in these genes identified a number of genes that affect the process, but they have also revealed that the process is fairly robust.
Results and Discussion
Gene Expression Profiling of Border Cells and Validation
Border cells arise from the follicular epithelium at stage 8/9 of oogenesis and then delaminate and migrate. In order to identify genes specifically expressed in this small cluster of invasive cells, we decided to compare the gene expression profile of border cells to that of all other follicle cells from the same stages. This was done to ensure that the cells compared were from the same environment and of the same general maturation stage. For example, all of these cells are postmitotic. Border cells were labeled in vivo by expression of GFP with a specific GAL4 “driver” called c522 that drives expression only in border cells (Figure 1A). The GAL4 driver c323 similarly allowed labeling of the other follicle cells (Figure 1C). To identify genes regulated by Slbo in border cells, we decided to compare the gene expression profile of wild-type border cells to that of slbo mutant border cells, by using the slbo1310 female sterile allele in trans to a null allele (slboe7b). Phenotypically, this combination is very strong, and mutant border cells do not migrate (Figure 1B); thus, the differences in transcriptional output responsible for the migration defect should be revealed. The mutant border cells were labeled as described for the wild-type border cells. To purify the border cells (or follicle cells), stage-9 egg chambers were enriched by timed incubation of the females, and whole ovaries were dissected.
Figure 1. Purification of Border Cells and Follicle Cells, Array Analysis, and Validation.
(A and B) (A) Wild-type and (B) slbo mutant border cells (arrows) labeled with c522-GAL4-driven expression of UAS-actinEGFP (green); late stage-9 egg chambers. Anterior is oriented toward the left; border cells migrate toward the oocyte (right); phalloidin labels F-actin (red). The scale bars = 20 μm.
(C) Wild-type stage-9 egg chamber with follicle cells (arrowhead) labeled by c323-GAL4 + UAS-actinEGFP.
(D) Merged fluorescent and bright-field images of a GFP-labeled border cell cluster in the crude suspension of dissociated cells.
(E) FACS plot used to identify GFP-labeled border cells. GFP-positive events are 0.4% of the total population.
(F) Purified GFP-border cell clusters after FACS.
(G) FACS plot used to purify GFP-labeled follicle cells (~20% of the total population).
(H) Electrophoretic analysis on a few picograms of total RNA extracted from border cells or follicle cells. The ribosomal RNAs are visible as a double band.
(I) Electrophoretic analysis of amplified antisense RNA generated from 30 ng total RNA from each sample after amplification and biotin labeling.
(J–L) scatter plots of gene expression comparisons from different hybridizations. Represented by dots are the 14,010 probe sets present in the Drosophila Genome Array. (J) Technical replicate; two experiments were performed on one RNA collection from wild-type border cells. (K and L) Hybridization profiles of wild-type border cells (WT BCs) versus follicle cells (FCs) and wild-type versus slbo mutant border cells (slbo BCs), respectively.
(M) Validation for selected genes by quantitative real-time RT-PCR, done in triplicate on each of three independent RNA samples (for CG9629 and Hex-C, only on two samples) for each genotype. Data are reported as fold change between wild-type border cells (set to 1) and follicle cells or slbo mutant border cells. “*” indicates a transcript not detected.
A protocol for fluorescence-activated cell sorting (FACS) had been established for isolating GFP-labeled follicle cells (Bryant et al., 1999). We modified the protocol to minimize handling time, but we found in either case that border cells were not recovered from the FACS. Visual inspection of the sample showed that while follicle cells were dissociated to single cells, border cells tended to remain as a cluster (Figure 1D), suggesting that adhesion between border cells is particularly strong. By modifying the FACS sorting such that the pressure on the cells was reduced, we were able to purify these rare border cell clusters to high purity without disrupting them (Figures 1E and 1F). Normal and slbo mutant border cells were purified in this manner, as were the follicle cells for comparison (Figure 1G). Sorted cells from multiple samples were pooled and RNA was extracted (Figure 1H) and subjected to linear mRNA amplification (Figure 1I) and labeling for hybridization to Drosophila Affymetrix arrays. One technical replicate, using the same border cell total RNA pool as starting material, demonstrated the reproducibility of the method (Figure 1J). Multiple biological replicates (independent pools of collected material) allowed us to identify which expression changes could truly be considered cell type and genotype dependent (Figures 1K and 1L; see tables in the Supplemental Data available with this article online). In total, about 5000 genes were reproducibly detected on the array. The remaining 9000 genes represented on the array may not be detectably expressed in this cell type. In addition, some genes may have been missed if the probe set was not optimal.
The array results were externally validated as follows: we selected 20 genes with different levels of expression and different extents of change in the array analysis, and we analyzed their expression by quantitative real-time RT-PCR (QRT-PCR) on RNA not previously amplified. In all cases tested, the changes observed in the arrays were confirmed (Figure 1M). This validates the linear amplification used to generate probe material for the arrays as well as the array hybridization and quantification.
To further confirm that differences in RNA abundance uncovered by the array analysis reflected real differences in gene expression in the tissue, we analyzed expression by in situ methods. We focused on genes upregulated at least 2-fold in border cells relative to follicle cells. Of these, 5 genes were already known to be enriched in border cells, namely, torso-like, singed, eya, fas3, and upd. To analyze more genes in an unbiased manner, we performed RNA in situ analysis and collected available antibodies as well as gene or protein traps, which allow for visualization of endogenous expression by GFP. The results of this survey are presented in Figure S1 and in the panels of the figures contained below and are annotated in Table S1. Previous experiments had indicated that RNA in situ analysis could be difficult for border cells and follicle cells owing to low signal as well as background or obscuring signal from nurse cells. Of 38 probes tested, only 6 gave signal above background in follicle cells, but all 6 verified the predictions from the arrays. All 6 tested antibodies, and 10 of the 11 gene or protein traps verified the expected expression difference. Thus, expression differences were verified in situ for an additional 22 (of 23) “random” genes, giving confidence that the genes identified in this analysis are truly regulated in vivo.
The polar cells are two pairs of specialized cells within the follicular epithelium that serve as signaling centers. Within the border cell cluster, the two central cells are polar cells. These cells do not appear to have a truly migratory phenotype and do not require slbo expression, but they are carried along by the outer border cells. However, polar cell-enriched genes should score as upregulated in border cells by our analysis, as there are only 2 polar cells per 1000 cells in the remaining follicle cell population, compared to 2 out of 8–10 border cells. The in situ analyses showed polar cell-enriched expression for 6 of the 27 analyzed genes (Table S1), indicating that such genes were identified but constitute a minority of the “border cell-enriched” genes.
General Analysis of the Expression Profiles
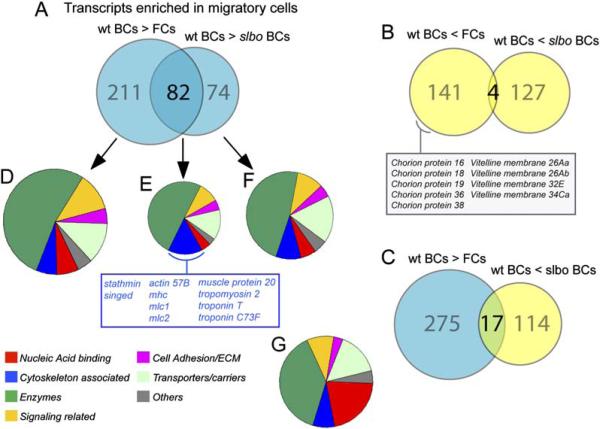
In the overall comparison, we found 293 genes significantly and, on average, at least 2-fold upregulated in border cells relative to follicle cells (Figure 2A and Table S1). When comparing wild-type border cells to slbo mutant border cells, 156 genes were at least 2-fold upregulated. Interestingly, the overlap between these gene sets is very high (82 genes, Figure 2A), with 28% of all border cell-enriched genes positively affected by slbo. The 2-fold cutoff may underestimate the fraction of genes affected by slbo, as the change in signal level from wild-type border cells to slbo mutant tends to be smaller than that to follicle cells (see Table S3). Note that the complimentary overlap between genes reduced in border cells relative to follicle cells and genes reduced in wild-type relative to slbo mutant border cells is low (four genes, Figure 2B, Table S6). Thus, the observed overlap in upregulated genes is significant and does not simply reflect that wild-type border cell samples were used for both comparisons. These results suggest that a large fraction of the genes upregulated in border cells is positively regulated by Slbo. As expected, the polar cell-enriched genes were not regulated by Slbo (Table S1). Gene expression profiling of mutant cells does not, of course, show that affected genes are directly regulated by Slbo. Structure/function analysis of Slbo (DmC/EBP) has previously shown that its essential function is that of a simple transcriptional activator (Rørth, 1994). This was shown for the function of Slbo during development and has subsequently been confirmed for its function in border cell migration (P.R., unpublished data). This would indicate that the key direct target genes of Slbo are positively regulated, in agreement with the present analysis.
Figure 2. Gene Expression Changes Associated with Migratory Border Cells.
(A–C) Diagrams representing genes with significant and at least 2-fold change in RNA level in wild-type border cells (wt BCs) compared to follicle cells (FCs) and slbo mutant border cells (slbo BCs), respectively. (A) Genes upregulated in wild-type border cells. (B) Genes downregulated in wild-type border cells. (C) Overlap of genes expressed at a high level in wild-type border cells (blue) and genes expressed at a higher level in slbo mutant border cells (yellow).
(D–F) Pie charts showing the relative distribution according to seven functional categories of genes found upregulated at least 2-fold in wild-type border cells; the cytoskeleton-associated genes upregulated in both comparisons are listed
(G) Representation of the functional groups in the Drosophila genome. For (D)–(G), only genes with some functional annotation were included: D:216, E:66, F:118, and G:6537 genes.
From this general gene expression analysis, we conclude that the difference between border cells and adjacent follicle cells was larger than the difference between wild-type border cells and those mutant for a single transcription factor (Slbo). This is based on the number of genes that change expression significantly as well as the magnitude of the changes. For example, 91 genes were at least 10-fold more highly expressed in border cells than follicle cells, whereas only 7 genes showed such a difference when comparing wild-type and slbo mutant border cells. This conclusion was not unexpected, as there are multiple differences between border cells and other follicle cells in addition to the level of Slbo. The slbo mutant cells were also not completely devoid of Slbo protein. A complete absence of Slbo in border cells can only be obtained in mosaic clones, as slbo is required for development. This may also contribute to reducing the magnitude of the differences between wild-type and slbo mutant cells. Perhaps a more surprising finding was that a significant number of genes were high in border cells (which contain high levels of Slbo) but even higher in slbo mutant border cells (Figure 2C). One possibility is that they represent compensatory changes in border cells that are not able to migrate due to the slbo mutation.
All genes showing significant changes were categorized by individual inspection as encoding proteins belonging to different functional categories (annotated in Supplemental Tables). In Figures 2D–2F, all genes regulated at least 2-fold and with some functional annotation are represented. This was compared to an annotation of the whole genome (Adams et al., 2000; Figure 2G) to understand which types of genes were affected. For example, one cluster of genes expressed at lower levels in border cells than in the remaining follicle cells encoded chorion and vitelline membrane proteins (Figure 2B), which makes sense given that late-stage follicle cells produce the vitelline membrane and chorion.
For the genes we were most interested in, the 82 genes upregulated in border cells and by Slbo, many functional categories were represented (Figure 2E). Some of these genes should account for the migration phenotype of slbo mutants, although they may also serve other functions in border cells. Transcription factors and other nucleic acid binding proteins were under-represented, possibly reflecting that we are analyzing events at the end of a gene regulatory cascade. Enzymes were somewhat overrepresented but belonged to many different subgroups with no obvious connection. These may be Slbo targets, as many of the known target genes of mammalian C/EBPs encode metabolic enzymes. Another enriched group of genes was those encoding cytoskeleton-associated proteins. These are structural components, motor proteins, or proteins known to directly regulate dynamics of actin filaments or microtubules. Migrating cells make use of their cytoskeleton to change shape and move through a tissue. One obvious idea would be that Slbo controls border cell migration by regulating these genes. To investigate this, we carried out a more detailed functional analysis of genes from this category. They fall into two distinct groups. The first group contains genes, such as stathmin and singed, known from studies in flies or other organisms to function in many cell types. If we include all genes upregulated significantly in border cells, quail, gelsolin, and spire are added. The second group consists of “muscle-specific” genes.
Stathmin, a Regulator of Microtubule Dynamics
Only one of the identified cytoskeletal regulators is known to affect microtubules, namely, Stathmin. Mammalian Stathmin/Op18 protein is well characterized (Belmont and Mitchison, 1996). It binds to microtubules and promotes depolymerization by sequestration of tubulin dimers or direct action at microtubule ends (Ravelli et al., 2004). Interestingly, the activity of Stathmin can be regulated by phosphorylation in response to signaling or cell cycle phases. Drosophila Stathmin appears to have similar biochemical features (Ozon et al., 2002). The availability of an antibody directed against Drosophila Stathmin allowed us to analyze protein levels in situ. As expected, the level of Stathmin was higher in border cells than follicle cells (Figure 3A). When analyzing slbo mutant border cells, we observed a clear difference between the inner polar cells and the outer border cells (Figures 3B–3D). The outer border cells are the migratory cells and require Slbo expression. In these cells, Stathmin expression was undetectable in the absence of Slbo, indicating a very strong dependence on Slbo. In contrast, Stathmin was still expressed in mutant polar cells, explaining why we only saw a moderate reduction of stathmin mRNA levels in whole border cell clusters (Figure 1M).
Figure 3. Stathmin Expression Is Dependent on Slbo and Is Required for Proper Border Cell Migration.
(A) A Stathmin-specific antibody demonstrates high expression in migrating border cells (arrow) compared to follicle cells (arrowhead). (B–D) The Stathmin protein level (red) is severely reduced in slbo mutant border outer border cells (arrows), but it is unaffected by slbo in anterior polar cells (arrowheads). slbo mutant cells are marked by the absence of GFP (green). DAPI (blue): nuclei. In (C), cells are of the slbo mutant; in (D) and (E), some border cells are wild-type for comparison.
(E) The stathmin locus. StathminExC is a viable, fertile deletion of the first two exons of the stathmin C transcript; stathminL27 deletes the entire stathmin gene as well as CG31642, Arc-p20, CG9226, and CG9227.
(F) A stathminL27 mutant border cell clone, marked by the absence of GFP (arrow), does not migrate (n = 3).
(G) Normal migration in the egg chamber from a stathminexC/stathminL27 female, with complete loss of the stathmin C transcript. (H and I) Border cell-specific RNAi of stathmin decreases Stathmin protein levels in a sensitized background, stathminexC/stathminL27; UAS-stathminRNAi/slbo-GAL4 in (I); compare to wild-type (H). In (B)–(D) and (H)–(I), right panels shows anti-stathmin alone, and the left panels show a merged image.
(J and K) Quantification of migration at (J) stage 9 and (K) stage 10 in border cells with reduced Stathmin levels (stathminexC/stathminL27; UAS-stathminRNAi/slbo-GAL4) compared to control (stathminexC/stathminL27; TM3/slbo-GAL4) and “rescue” (UAS-stathmin/+; stathminexC/stathminL27; UAS-stathminRNAi/slbo-GAL4).
(L) Overexpression of stathmin in slbo mutant border cells does not rescue migration. Quantification of slbo1310,slbo-GAL4/slbo1310 and slbo1310,slbo-GAL4/slbo1310;UAS-Stathmin egg chambers at stage 10.
To analyze the function of Stathmin in border cells, we generated stathmin mutants. This was done by imprecise excision of a P element located immediately upstream of the stathmin C transcript (Figure 3E). A mutant deleting only the stathmin C isoform (stathminexC), leaving stathmin A and B intact, was homozygous viable and had no effect on border cell migration. A mutant deleting the complete stathmin locus (stathminL27) and four adjacent genes (including Arc-p20, a component of the Arp2/3 complex) was homozygous lethal, and clones of stathminL27 mutant border cells were unable to migrate (Figure 3F). Both the lethality and the migration block were rescued by reintroducing ubiquitously expressed stathmin and Arc-p20 at the same time. Reintroducing Arc-p20 alone did not rescue border cell migration, indicating that stathmin is essential for this process. To interfere with stathmin upregulation at the time of migration, we expressed a functional stathmin “hairpin”-RNAi construct in the sensitized stathminexC/stathminL27 background (Figure 3G). By using the slbo-GAL4 driver, we could specifically target stathmin RNAi expression to outer border cells right before and during migration. This strongly decreased the amount of Stathmin protein in border cells (compare Figure 3I to Figure 3H) and caused significant delays in migration (Figures 3J and 3K). The delays in migration could be reversed by driving higher levels of stathmin expression from a UAS construct (“rescue” in Figure 3J). These results identify Stathmin as an important regulator downstream of Slbo. To test whether lack of Stathmin was solely responsible for the slbo phenotype, we overexpressed Stathmin in the slbo mutant background (Figure 3L). Migration was not restored, indicating that additional genes downstream of Slbo must also be important.
Singed and Related Regulators of the Actin Cytoskeleton
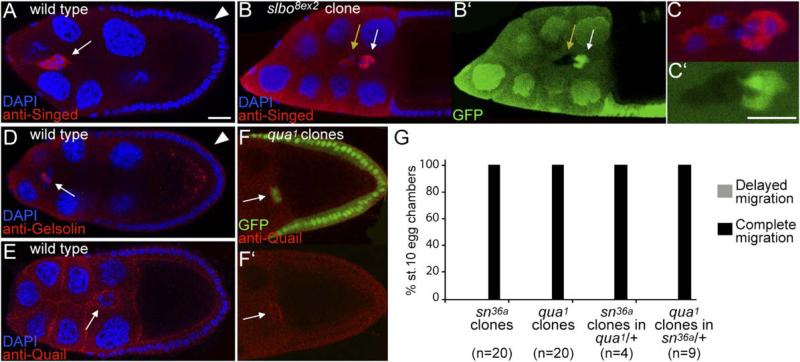
Singed is an actin-bundling protein related to Fascin, highly expressed in border cells (Figures 1M and 4A; Cant et al., 1994). Fascin is important for the formation of cell protrusions and has been implicated in the control of cell migration, also in vivo (reviewed in Adams, 2004). We confirmed by clonal analysis that Singed protein levels were regulated by Slbo (Figures 4B and 4C). Despite the strong and regulated expression, migration was normal in border cells mutant for singed (Figure 4G). The strongest allele of singed available was used, but it retained a low level of protein expression (data not shown). In addition, functional overlap may exist between actin regulators. Quail is an actin binding protein of the villin family (Mahajan-Miklos and Cooley, 1994), and its function in the germline of the ovary genetically overlaps with that of Singed (Cant et al., 1998). quail mRNA was also upregulated in border cells relative to follicle cells (Figure 1M), and Quail protein was detected in border cells (Figures 4E and 4F). Quail is structurally similar to Gelsolin, which was also upregulated in border cells (Figure 1M), as well as the Gelsolin-related FliI, which was not detectably expressed. However, Gelsolin was enriched in polar cells rather than the migratory outer border cells (Figure 4D). As for singed, we observed no migration defects in quail mutant border cells (Figure 4G), nor in cells mutant for quail and only one functional copy of singed or vice versa (Figure 4G). We were not able to recover clones of border cells simultaneously mutant for both singed and quail, which is likely to reflect a functional overlap between the two genes at an earlier stage. The simultaneous upregulation of redundant actin regulators may reflect a genetically robust approach to changing the actin cytoskeleton in border cells.
Figure 4. Singed and Quail Actin Binding Proteins in Border Cells.
(A–C) Egg chambers stained with antibody to Singed. (A) High Singed expression in wild-type stage-9 border cells (arrow) compared to follicle cells (arrowhead). (B and C) Merged and (B′ and C′) single channel images of a mosaic border cell cluster ([C] at high magnification); slbo8ex2 mutant cells are marked by the absence of GFP and by a yellow arrow; wild-type cells are marked by white arrow. The scale bars are 20 μm.
(D) Stage-9 egg chamber stained with anti-Gelsolin. High Gelsolin expression is detected in polar cells within the border cell cluster (arrow) compared to follicle cells (arrowhead).
(E and F) Late stage-9 egg chambers stained with anti-Quail. (E) Wild-type. (F) Egg chamber with a quail germline mutant clone (GFP negative); Quail is detected in border cells (arrow).
(G) Quantification of border cell migration in egg chambers at stage 10, when migration is complete in wild-type.
Activation of a Muscle Gene Expression Program
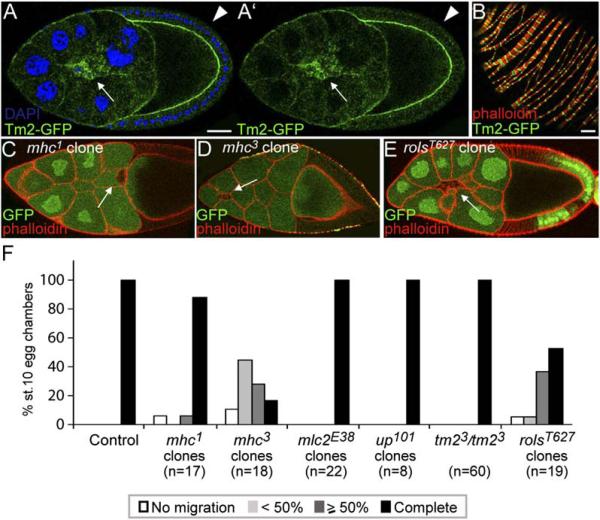
A rather surprising finding of this global expression analysis was that the remaining genes encoding cytoskeleton-associated proteins and upregulated in border cells in a slbo-dependent manner were all “muscle specific” (Figures 2E and 1M). This included a complete palate of structural genes: muscle actin (57B), muscle myosin heavy chain and light chains, tropomyosin 2 (tm2), troponins, and the calponin-related mp20. The muscle-specific expression has been shown for this group of genes in Drosophila embryos as well as mature muscles. For tropomyosin 2, a GFP gene trap allele was available and confirmed expression in border cells (Figure 5A) as well as in the muscle sheath (Figure 5B). The expression profiling indicated that border cells also expressed the corresponding non-muscle forms such as actin42A, zipper (myosin heavy chain), and sqh (myosin light chain), but at the same level as in follicle cells. The nonmuscle proteins are generally required for many cellular processes, including, where tested, migration of border cells (Edwards and Kiehart, 1996). This raised the question of why this large cluster of muscle-specific structural genes would be turned on in border cells as well. To address this, we analyzed migration of border cells mutant for individual muscle genes for which mutants were available (mhc, mlc2, upheld=troponinT and tm2). As mhc and mlc2 are essential genes, this was done by clonal analysis. No defects were seen in border cells mutant for mlc2, upheld, or tm2, but clear migration defects were observed in border cells mutant for mhc (mhc1 or mhc3, Figures 5D and 5F). Thus, while not all of the muscle structural genes are required for border cell migration, at least muscle Mhc expression contributes to effective migration.
Figure 5. Muscle-Specific Genes and Border Cell Migration.
(A and B) (A) Tropomyosin2-GFP (protein trap) in migrating border cells (arrow) compared to follicle cells (arrowhead) and (B) in the muscle sheath that surrounds egg chambers, counterstained for F-actin (phalloidin, red). The scale bars are 20 μm.
(C) Stage-10 egg chamber with an mhc1 mutant border cell clone (marked by the absence of GFP) that shows normal migration.
(D) Stage-10 egg chamber with an mhc3 mutant border cell clone (marked by the absence of GFP) that is delayed (<50%).
(E) Late stage-9 egg chamber with a rolsT627 mutant border cell clone that shows defective migration (50%).
(F) Quantification of border cell migration in egg chambers at stage 10 for mhc1, mhc3, mlc2E38, up101 and rolsT627 mutant border cell clones, or in tm23 homozygous mutant females. Controls for mhc1, mhc3, and rolsT627 mutant clones were: mhc1/+ (n = 46), mhc3/+ (n = 21), and rolsT627/+ (n = 20).
Given that both muscle and nonmuscle forms of the same cytoskeletal proteins have a role in border cell migration, their functions are likely to be different. In agreement with this, we did not observe genetic interactions between mutants affecting muscle and nonmuscle forms of myosin heavy or light chains (data not shown). There is precedence for such nonoverlapping functions. For example, zipper has a unique role in developing muscle cells, which contain plenty of muscle myosin heavy chain (Bloor and Kiehart, 2001). In mammalian cells, different myosin heavy chain isoforms can have distinct subcellular localization (Bresnick, 1999). Also, the actin proteins, despite having few amino acid differences, are functionally distinct in vivo (Fyrberg et al., 1998; Brault et al., 1999).
The muscle gene expression program activated in migratory border cells extended beyond structural genes to regulatory genes. One such gene was bent, encoding a very large titin-like molecule with a myosin light chain kinase domain. Being essential but on the fourth chromosome, bent was not amenable to standard clonal analysis. Genes required for myoblast fusion were also identified, namely, rols/antisocial (Rau et al., 2001; Chen and Olson, 2001) and rost. We had previously shown that mbc, which encodes a DOCK180 family Rac GEF and is required for myoblast fusion, has a role in border cell migration downstream of the PVR guidance receptor (Duchek et al., 2001). Mbc protein interacts physically with the presumed adaptor protein Rols (Chen and Olson, 2001). Clonal analysis with a strong (likely complete loss-of-function) allele of rols showed defects in border cell migration (Figures 5E and 5F), suggesting that Mbc and Rols might act together during migration as well. The defect was milder than for mbc, implying that Mbc activity might not be completely dependent on Rols. For the small transmembrane protein Rost, no useful mutants were available. We also noted that a very closely related and adjacent gene, CG13101, was similarly regulated in border cells and might overlap rost function. Thus, at least mbc and rols function in border cells as well as in muscle (myoblast fusion). Activation of a broad “muscle-specific” gene expression program in border cells may reflect a requirement for a specific subset of the genes within this program.
Analysis of Additional Transcription Factors
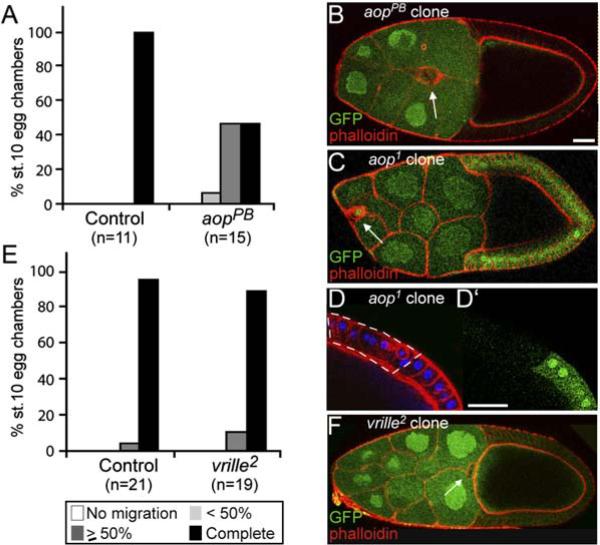
Previous unbiased genetic approaches to identify genes important for border cell migration have largely identified transcription factors or inducing signals. Changes in cell fate can alter cell behavior dramatically without affecting cell survival, thus still allowing analysis of the mutant cells. The transcription factors themselves often show differential expression. In addition to Slbo, the posttranslationally regulated transcription factor STAT, which is important for border cell migration, was also upregulated in border cells (1.6-fold). We tested the transcription factors that were upregulated in border cells and had mutants available for effects on border cell migration. aop/yan transcripts were increased 1.9-fold in border cells. In a PiggyBac transposon-based clonal screen for border cell migration defects, we identified an insertion in aop. Complementation analysis confirmed the gene assignment, and quantification of the phenotype showed a clear effect of aop on border cell migration (Figures 6A and 6B). This requirement for aop has recently been characterized by another group (Schober et al., 2005). We also analyzed a complete loss-of-function allele, aop1. As expected, border cell migration was strongly affected (Figure 6C), but, in addition, clones were rare and morphological abnormalities were seen in other follicle cells (Figure 6D) as well as in germline cells. Thus, aop may affect the behavior of multiple cell types in the ovary. Another transcription factor, vrille, was also upregulated (over 2-fold). vrille has been implicated in signaling, circadian rhythm, and cellular morphogenesis (Szuplewski et al., 2003), but border cells mutant for vrille were largely unaffected and experienced only subtle delays (Figures 6E and 6F).
Figure 6. Function of Aop and Vrille Transcription Factors in Border Cells.
(A–F) Quantification of migration at stage 10 for (A) aop (yan)PB104 and (E) vrille2 mutant border cell clones. Controls are aopPB/+ and vrille2/+. (B) Stage-10 egg chamber with an aopPB104 mutant border cell clone (arrow): delayed migration. (C) Late stage-9 egg chamber with an aop1 mutant border cell clone. Mutant cells are marked by the absence of GFP; one cell is GFP positive. (D and D′)An aop1 mutant follicle cell clone in a stage-10 egg chamber. Phalloidin (red) and DAPI (blue). The scale bar is 20 μm. (F) A vrille2 mutant border cell clone marked by the absence of GFP (arrow).
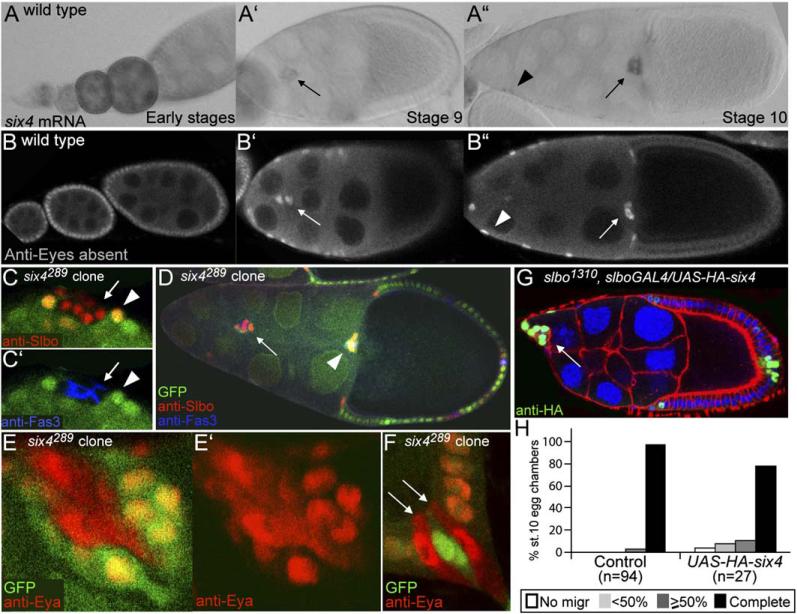
The most border cell-enriched RNA encoding a transcription factor, apart from Slbo, was Six4 (4.5-fold). Six4 expression in border cells was confirmed by in situ analysis (Figure 7A). Drosophila Six4 is the homeo-domain transcription factor most related to mammalian Six4 and Six5 (Kirby et al., 2001). Six family proteins act in complex with proteins of the Eya (Eyes absent) family (Kawakami et al., 2000). eya transcripts were also 2.7-fold enriched in border cells relative to follicle cells of the same stage, and Eya was expressed in a pattern similar to that of six4 (Figure 7B). Both six4 and eya were expressed in earlier-stage follicle cells as well (Figures 7A and 7B), and eya has been shown to function at these stages to repress polar cell fate (Bai and Montell, 2002). Follicle cells mutant for six4 expressed a polar cell marker (Fas3 in Figures 7C′ and 7D) and were functional polar cells, as determined by the ability to induce surrounding anterior follicle cells to become Slbo-positive, migratory border cells (Figures 7C and 7D). This suggested that Six4 cooperates with Eya in repressing polar cell fate. It had been indicated that Six proteins affect nuclear localization of their Eya partner (Ohto et al., 1999). The six4 mutant allowed us to test this in an in vivo context. Although six4 mutant cells were transformed to functional polar cells, Eya protein was not absent as in the endogenous polar cells, showing that Eya accumulation was independently regulated (arrows in Figure 7F). However, Eya protein was partially relocalized to the cytoplasm of six4 mutant cells (Figures 7E and 7F), supporting the hypothesis that Six4 and Eya interact in vivo. As six4 and eya are both upregulated in outer border cells when they migrate, they are likely to act together in this process as well. However, their earlier roles precludes straightforward loss-of-function analyses in border cells, as “border cell clusters” consisting only of six4 or eya mutant cells are not functional simply because polar cells do not migrate on their own. Overexpression of HA-tagged six4 in border cells interfered with migration (Figures 7G and 7H), as found for transcription factors required in border cells slbo (Rørth et al., 2000) and yan (Schober et al., 2005).
Figure 7. six4 and eyes absent in Follicle Cells and Border Cells.
(A) six4 expression detected by RNA in situ hybridization.
(B) Eya expression detected by antibody staining. Early stages are to the left.
(C) Six4289 mutant follicle cell clone (marked by the absence of GFP, arrows) in the anterior of a stage-9 egg chamber stained with anti-Slbo (border cells, red in [C]) and anti-Fas3 (polar cells, blue in [C′]). Wild-type cells directly in contact with the mutant clone express Slbo (arrowheads).
(D) Stage-10 egg chamber with two border cell clusters (arrowhead: wild-type cluster, arrow: ectopically induced cluster).
(E and F) six4289 mutant follicle cell clones stained with anti-Eya (red). (E) Anterior tip and (F) posterior tip of egg chambers.
(G) Delayed migration in a stage 9 slbo1310, slboGAL4/UAS-HA-six4 egg chamber. HA-Six4 is nuclear.
(H) Quantification of migration in stage-10 egg chambers of the genotype slbo1310, slboGAL4/UAS-HA-six4 and control slbo1310, slboGAL4/+.
The expression of Six4 in border cells may contribute to activation of the muscle gene program described above. The conserved muscle transcription factor Mef2, an activator of muscle actin and myosin expression, was not detected in border cells by expression profiling or by antibody staining, nor were Twist and Nautilus/MyoD. Six4 is required for development of muscle and other mesodermal tissues in Drosophila (Kirby et al., 2001). Mutants of Caenorhabditis elegans Unc-39 (Yanowitz et al., 2004), belonging to the Six4/5 family, also affect muscle/mesodermal differentiation as well as directed cell migration. Mammalian Six5, also called myotonic dystrophy-associated homeodomain protein (DMAHP), has been analyzed due to its contribution to DM1 (Groenen and Wieringa, 1998), and Six4/5 affect normal muscle development. Another transcription factor complex that might contribute to the activation of the muscle program is that of MAL-D (or MRTF) and SRF (serum response factor). The MRTF/SRF complex plays an important role in muscle development in mammals and directly regulates muscle (structural) genes (Wang and Olson, 2004). We have previously shown that MAL-D/SRF plays a crucial role in border cell migration and provided evidence that this complex acts to strengthen the cytoskeleton of invasive border cells in response to perceived tension (Somogyi and Rørth, 2004). This mode of regulation makes MAL-D/SRF activity in border cells indirectly dependent on Slbo (Somogyi and Rørth, 2004), which could be responsible for the apparent regulation of the muscle gene cluster by Slbo. We do not, however, want to exclude the possibility that Slbo might affect muscle genes directly, as the mammalian C/EBP transcription factors are known to regulate different differentiation-specific genes in different contexts.
Conclusions and Perspective
In this study, we have analyzed overall gene expression changes resulting from a transcriptional switch that induces invasive migratory behavior in vivo. The major goal of the analysis was to identify transcriptional changes that directly affect cell behavior and make the cells move. Our results indicate that regulation of both the actin cytoskeleton and the microtubule cytoskeleton, likely coordinated regulation, is important for this transition. Identifying Stathmin as an important regulator downstream of Slbo in border cells indicates that microtubule dynamics are critical for border cell migration. Key questions are now how microtubule dynamics affect the process, and whether Stathmin activity is regulated. Two recent findings suggest that Stathmin may be a more general regulator of cell migration: Stathmin-microtubule interactions are spatially regulated in migrating cells in culture (Niethammer et al., 2004), and Stathmin upregulation may promote migration and metastasis of sarcoma cells (Baldassarre et al., 2005). The actin cytoskeleton is clearly crucial for cell migration and is controlled by many regulators. The upregulated modulators identified in this study were different from those identified in a whole-genome study of tumor cells selected, in vivo, to be highly motile (Wang et al., 2004). There are obviously many differences between these studies; for one, a normal transition to migratory behavior may differ from unrestrained, high motility. The activation of a “muscle-specific” program in migratory border cells was unexpected and provides an intriguing connection between these cells that move and the specialized cells that move an animal (muscle). Overall, the analysis of actin regulators indicates that this is a robust system, with many effectors coregulated, even by one transcription factor. Genetically, this is reflected by minor defects in individual “effector” mutants despite absolute dependence on the transcriptional switch. Further analysis in other systems, and subsequent comparisons, will tell us to what extent the gene expression program employed by border cells to become migratory is a general one.
Experimental Procedures
Drosophila Strains and Genetic Analysis
Stathmin mutants were generated by imprecise excision of the GS2209 P element. A total of 640 w− excision alleles were analyzed by PCR, followed by mapping. For lethal alleles, homozygous mutant embryos were recovered from ex/Cyo,twist-GFP stocks. To generate a stathmin RNAi construct, a 500 bp fragment of the stathmin coding region common to all three isoforms was amplified by PCR and cloned in forward and reverse orientation into pGem-WIZ (kind gift of Sujin Bao). For UASt-Stathmin and pCaSpeR-tub-Stathmin, a Not1-Xho1fragment was subcloned from LD04103 (full-length stathmin A). To generate pCaSpeR-tub-Arc-p20, the ORF was obtained by PCR from genomic DNA and cloned with SV40UTR. The complete six4 ORF was cloned by PCR from the cDNA clone GM13131 and three HA tags added at the 5′ end (UASt-HA-six4). The aop/yan allele aopPB104 is a PiggyBac insertion in the first intron of aop-B. The remaining stocks and mutants are described in Flybase: slboe7b, slbory8ex2, slbo1310, sn36a, qua1, mhc1, mhc3, mlc2E38, tm23, up101, rolsT627, aop1, vrille2, six4289, the tm2-GFP protein-trap line, GAL4 drivers GawB-c522 GAL4; GawB-c323a and slboGAL4. Flies, except c323a/UAS-actinEGFP (22°C), were raised on standard medium at 25°C. Crosses for RNAi (and controls) were incubated at 29°C. For clonal analysis, mutants were recombined onto the appropriate FRT chromosomes and checked with deficiencies or other alleles. Clones were induced by heat shocking larvae of the genotype hsFLP/+; FRT, mut/FRT, ubiGFP. For slbory8ex2, stathminL27, aop1, and six4289 clones, heat shock to adults was also used, 3–4 days before analysis.
Purification by Fluorescence-Activated Cell Sorting
Ovaries enriched in stage 9 (females incubated at 25°C for 18 hr on rich food) were dissected (100 pairs/30 min) in ice-cold SFM medium (GIBCO-BRL-Invitrogen). Ovaries were dissociated by incubation at room temperature for 30 min in 0.9 ml Trypsin 0.5%/EDTA (Sigma) + 0.1 ml 67 mg/ml Collagenase in PBS (Sigma). Supernatant was filtered through a 62 μm nylon mesh (Small Parts Incorporated) into tubes containing SFM medium supplemented with 10% fetal calf serum (FCS) to 1 ml final volume, centrifuged at 1000 × g for 7 min, and the pellet was resuspended in 1 ml SFM with FCS and kept on ice until sorting.
Fluorescent-activated cell sorting was performed on a Dako Cytomation MoFlo high-speed flow cytometer, modified in order to recover border cell clusters. A 150 μm nozzle was used at 8 psi sheath pressure. Minimal sample differential pressure was used to create a 1000 events per second acquisition trigger rate or less. The light source used was a Lyt 200-S (iCyt, Urbana IL) 488 nm diode attenuated to 150 mw and shaped to ellipse. GFP expression was captured in FL1 by using a Chroma HQ520/40 reflected from a 500DCSP, reflected from a Chroma Q550LP, and passed through a 605DSP. A scatter plot was used to exclude apex noise and fragments, and a bivariate fluorescence plot using the GFP FL1 on the x axis and the adjacent parameter FL2 on the y axis for dumping background was also used. Sorting gates to satisfy included the scatter gate and positive GFP. Multiple sortings were performed and pooled in order to recover sufficient border cells for microarray experiments.
RNA Extraction, Amplification, and Microarray Hybridization
For RNA extraction, border cell clusters and single follicle cells were sorted directly in lysis buffer (PicoPure RNA isolation kit, Arcturus, Mountain View, CA) and frozen at −80°C until subsequent extraction steps. Total RNA was usually extracted from ~8,000 events of border cell cluster collection and from 20,000 events of follicle cell-sorted collection. RNA quality was assessed by using RNA Pico Lab-Chip together with an Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA) and quantified by using the RiboGreen RNA quantitation kit (Molecular Probes).
From each sample, 30 ng total RNA was linearly amplified by in vitro transcription by using the RiboAmp OA RNA Amplification kit (Arcturus Genomics), followed by the BioArray High Yield transcription kit (ENZO Biochemical, New York, NY) to generate biotinylated cRNA. To verify that amplification was consistent between samples for different RNA lengths and sequences, spike-in controls (Bacillus subtilis gene transcripts detectable by probe sets on the arrays) were included prior to initiation of first-strand synthesis. Equivalent amounts (10–20 μg) of fragmented biotin-labeled cRNA from each sample were hybridized to GeneChip Drosophila Genome Arrays (Affymetrix, Santa Clara, CA). Hybridization and scanning were performed by the GeneChip facility of the Siteman Cancer Center (St. Louis, MO).
Microarray Data Analysis
Raw data have been deposited with ArrayExpress at EBI (E-MEXP-493). Images from scanned chips were processed by using the default settings of GeneChip Operating Software v.1.0 (GCOS) and individually scaled to an average target signal of 1500. Pairwise (border cells versus follicle cells, wild-type versus slbo) comparisons of transcript levels were performed by using GCOS v.1.0, with each experiment treated independently. The final lists include only genes significantly changed in the same direction in all independent experiments (p << 0.05), see tables in the Supplemental Data.
Quantitative Real-Time RT-PCR
Quantitative real-time RT-PCR (QRT-PCR) analysis was performed with SYBR Green PCR Core Reagents (Applied Biosystems, Foster City, CA) and 7700 Sequence Detection Systems (Applied Biosystems). For cDNA synthesis, 10–20 ng total RNA was reverse transcribed in 20 μl containing 1× buffer from Superscript II RNase H- kit (Invitrogen), 100 ng oligo-dT, 10 mM DTT, 0.5 mM dNTPs, 40 U RNasin (Promega), 200 U SS Reverse Transcriptase. A total of 100 pg total RNA was used per QRT-PCR in 25 μl containing 1× SYBR Green PCR Master mix, 0.25 U UDP-N-Glycosidase (Invitrogen), 300 nM each of the gene-specific primer pair. rp49 was used as a reference gene. See Supplemental Data for primer sequences and amplicon sizes.
Tissue In Situ Hybridization
In situ hybridizations to wild-type ovaries were performed by using digoxygenin-labeled antisense RNA probes synthesized with DGC cDNA clones as templates. For six4, a standard protocol was used, but for the rest of the genes, an optimized protocol was used: ovaries were fixed overnight in 4% PFA/PBS Tween 20 (0.1% PBSTw) at 4°C, washed with PBSTw, and kept in methanol for >90 min at −20°C. After rehydration, ovaries were dissociated and treated with 10 μg/ml proteinase K for 8 min, washed twice with PBSTw, and refixed for 20 min with 4% PFA/PBSTw, followed by five quick washes with PBSTw. Egg chambers were prehybridized for 60 min at 65°C in 50% formamide, 5× SSC, 1% Boehringer Block (BB), 1 mg/ml Torula RNA, 0.1 mg/ml heparin, 0.1% Tween 20, 0.1% CHAPS, 5 mM EDTA and hybridized overnight at 65°C with 40–100 ng/ml with DIG-labeled RNA antisense probe. Washes were performed at 65°C: 2× 30 min in 50% fomamide, 5× SSC, 0.1% CHAPS; 15 min in 2× SSC, 0.1% CHAPS; and 2× 30 min in 0.2× SSC, 0.1% CHAPS. Samples were rinsed with MABTw (100 mM maleic acid, 150 mM NaCl, 0.1% Tween 20) and blocked in 5% BB, MABTw for 1 hr. Anti-DIG antibody (Roche) was used at 1:4000 in 5% BB for 2 hr. Samples were washed 6× 20 min in MABTw, incubated 3× 5 min in 100 mM Tris (pH 9.5), 50 mM MgCl2, 100 mM NaCl, 0.1% Tween 20, and stained with BM Purple (Roche).
Antibodies
Standard procedures for staining and detection were used with the following primary antibodies: rat anti-Slbo (1:500); rabbit anti-Stathmin (1:500, Ozon et al., 2002), rabbit anti-Gelsolin (1:1000, Stella et al., 1994), rabbit anti-Wb (1:100), mouse anti-Singed (1:100), rabbit anti-Glutactin (1:500), mouse anti-HA.11 (1:1000, from BAbCO). From the Developmental Studies Hybridoma Bank: anti-Eya (EYA10H6, 1:1000), anti-Quail (6B9, 1:25), anti-Fas3 (7G10, 1:50), anti-Nrt (1:5). Fluorescent secondary antibodies were purchased from (Jackson ImmunoResearch); rhodamine-phalloidin and DAPI were purchased from (Molecular Probes).
Supplementary Material
Acknowledgments
We are very grateful to Silvie Ozon, Stefan Baumgartner, John Fessler, and Maria Leptin for antibodies; Lynn Cooley for both antibody and GFP-trap flies; and Andrew Jarman, Sandy Bernstein, John C. Sparrow, and the Bloomington stock center for mutants. We thank Jeff Gordon's lab and Mark A. Watson (Washington University) and Klaus-Michael Kuerner for technical advice and discussions; Ann Marie Voie for embryo injection; the Cagan and Rørth labs for help and discussions; and Eileen Furlong and Stephen Cohen for comments on the manuscript. G.F. was supported by a fellowship from DAAD. J.M. was supported by a fellowship from HFSP.
Footnotes
Supplemental Data Supplemental Data including confirmation of gene expression patterns by in situ analysis (Figure S1), lists of genes upregulated or downregulated in border cells versus follicle cells or slbo mutant border cells (Tables S1–S6), and a list of primers for QRT-PCR are available at http://www.developmentalcell.com/cgi/content/full/10/4/497/DC1/.
References
- Adams JC. Roles of fascin in cell adhesion and motility. Curr. Opin. Cell Biol. 2004;16:590–596. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Bai J, Montell D. Eyes absent, a key repressor of polar cell fate during Drosophila oogenesis. Development. 2002;129:5377–5388. doi: 10.1242/dev.00115. [DOI] [PubMed] [Google Scholar]
- Baldassarre G, Belletti B, Nicoloso MS, Schiappacassi M, Vecchione A, Spessotto P, Morrione A, Canzonieri V, Colombatti A. p27(Kip1)-stathmin interaction influences sarcoma cell migration and invasion. Cancer Cell. 2005;7:51–63. doi: 10.1016/j.ccr.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- Birchmeier C, Brohmann H. Genes that control the development of migrating muscle precursor cells. Curr. Opin. Cell Biol. 2000;12:725–730. doi: 10.1016/s0955-0674(00)00159-9. [DOI] [PubMed] [Google Scholar]
- Bloor JW, Kiehart DP. zipper Nonmuscle myosin-II functions downstream of PS2 integrin in Drosophila myogenesis and is necessary for myofibril formation. Dev. Biol. 2001;239:215–228. doi: 10.1006/dbio.2001.0452. [DOI] [PubMed] [Google Scholar]
- Brault V, Reedy MC, Sauder U, Kammerer RA, Aebi U, Schoenenberger C. Substitution of flight muscle-specific actin by human (beta)-cytoplasmic actin in the indirect flight muscle of Drosophila. J. Cell Sci. 1999;112:3627–3639. doi: 10.1242/jcs.112.21.3627. [DOI] [PubMed] [Google Scholar]
- Bresnick AR. Molecular mechanisms of nonmuscle myosin-II regulation. Curr. Opin. Cell Biol. 1999;11:26–33. doi: 10.1016/s0955-0674(99)80004-0. [DOI] [PubMed] [Google Scholar]
- Bryant Z, Subrahmanyan L, Tworoger M, LaTray L, Liu CR, Li MJ, van den Engh G, Ruohola-Baker H. Characterization of differentially expressed genes in purified Drosophila follicle cells: toward a general strategy for cell type-specific developmental analysis. Proc. Natl. Acad. Sci. USA. 1999;96:5559–5564. doi: 10.1073/pnas.96.10.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant K, Knowles BA, Mooseker MS, Cooley L. Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. J. Cell Biol. 1994;125:369–380. doi: 10.1083/jcb.125.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant K, Knowles BA, Mahajan-Miklos S, Heintzelman M, Cooley L. Drosophila fascin mutants are rescued by overexpression of the villin-like protein, quail. J. Cell Sci. 1998;111:213–221. doi: 10.1242/jcs.111.2.213. [DOI] [PubMed] [Google Scholar]
- Chen EH, Olson EN. Antisocial, an intracellular adaptor protein, is required for myoblast fusion in Drosophila. Dev. Cell. 2001;1:705–715. doi: 10.1016/s1534-5807(01)00084-3. [DOI] [PubMed] [Google Scholar]
- Duchek P, Somogyi K, Jékely G, Beccari S, Rørth P. Guidance of cell migration by the Drosophila PDGF/VEGF Receptor. Cell. 2001;107:17–26. doi: 10.1016/s0092-8674(01)00502-5. [DOI] [PubMed] [Google Scholar]
- Dyrskjot L, Thykjaer T, Kruhoffer M, Jensen JL, Marcussen N, Hamilton-Dutoit S, Wolf H, Orntoft TF. Identifying distinct classes of bladder carcinoma using microarrays. Nat. Genet. 2003;33:90–96. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- Edwards KA, Kiehart DP. Drosophila nonmuscle myosin II has multiple essential roles in imaginal disc and egg chamber morphogenesis. Development. 1996;122:1499–1511. doi: 10.1242/dev.122.5.1499. [DOI] [PubMed] [Google Scholar]
- Fyrberg EA, Fyrberg CC, Biggs JR, Saville D, Beall CJ, Ketchum A. Functional nonequivalence of Drosophila actin isoforms. Biochem. Genet. 1998;36:271–287. doi: 10.1023/a:1018785127079. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Bronner-Fraser M. Genomic analysis of neural crest induction. Development. 2002;129:5731–5741. doi: 10.1242/dev.00175. [DOI] [PubMed] [Google Scholar]
- Groenen P, Wieringa B. Expanding complexity in myotonic dystrophy. Bioessays. 1998;20:901–912. doi: 10.1002/(SICI)1521-1878(199811)20:11<901::AID-BIES5>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes–structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Kirby RJ, Hamilton GM, Finnegan DJ, Johnson KJ, Jarman AP. Drosophila homolog of the myotonic dystrophy-associated gene, SIX5, is required for muscle and gonad development. Curr. Biol. 2001;11:1044–1049. doi: 10.1016/s0960-9822(01)00319-0. [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Cooley L. The villin-like protein encoded by the Drosophila quail gene is required for actin bundle assembly during oogenesis. Cell. 1994;78:291–301. doi: 10.1016/0092-8674(94)90298-4. [DOI] [PubMed] [Google Scholar]
- Montell DJ. Command and control: regulatory pathways controlling invasive behavior of the border cells. Mech. Dev. 2001;105:19–25. doi: 10.1016/s0925-4773(01)00393-8. [DOI] [PubMed] [Google Scholar]
- Niethammer P, Bastiaens P, Karsenti E. Stathmintubulin interaction gradients in motile and mitotic cells. Science. 2004;303:1862–1866. doi: 10.1126/science.1094108. [DOI] [PubMed] [Google Scholar]
- Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, Kawakami K. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol. Cell. Biol. 1999;19:6815–6824. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozon S, Guichet A, Gavet O, Roth S, Sobel A. Drosophila stathmin: a microtubule-destabilizing factor involved in nervous system formation. Mol. Biol. Cell. 2002;13:698–710. doi: 10.1091/mbc.01-07-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau A, Buttgereit D, Holz A, Fetter R, Doberstein SK, Paululat A, Staudt N, Skeath J, Michelson AM, Renkawitz-Pohl R. rolling pebbles (rols) is required in Drosophila muscle precursors for recruitment of myoblasts for fusion. Development. 2001;128:5061–5073. doi: 10.1242/dev.128.24.5061. [DOI] [PubMed] [Google Scholar]
- Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- Rørth P. Specification of C/EBP function during Drosophila development by the bZIP basic region. Science. 1994;266:1878–1881. doi: 10.1126/science.7997882. [DOI] [PubMed] [Google Scholar]
- Rørth P. Initiating and guiding migration: lessons from border cells. Trends Cell Biol. 2002;12:325–331. doi: 10.1016/s0962-8924(02)02311-5. [DOI] [PubMed] [Google Scholar]
- Rørth P, Szabo K, Texido G. The level of C/EBP protein is critical for cell migration during Drosophila oogenesis and is tightly controlled by regulated degradation. Mol. Cell. 2000;6:23–30. doi: 10.1016/s1097-2765(05)00008-0. [DOI] [PubMed] [Google Scholar]
- Schober M, Rebay I, Perrimon N. Function of the ETS transcription factor Yan in border cell migration. Development. 2005;132:3493–3504. doi: 10.1242/dev.01911. [DOI] [PubMed] [Google Scholar]
- Somogyi K, Rørth P. Evidence for tension-based regulation of Drosophila MAL and SRF during invasive cell migration. Dev. Cell. 2004;7:85–93. doi: 10.1016/j.devcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Stella MC, Schauerte H, Straub KL, Leptin M. Identification of secreted and cytosolic Gelsolin in Drosophila. J. Cell Biol. 1994;125:607–616. doi: 10.1083/jcb.125.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuplewski S, Kottler B, Terracol R. The Drosophila bZIP transcription factor Vrille is involved in hair and cell growth. Development. 2003;130:3651–3662. doi: 10.1242/dev.00588. [DOI] [PubMed] [Google Scholar]
- Tatenhorst L, Puttmann S, Senner V, Paulus W. Genes associated with fast glioma cell migration in vitro and in vivo. Brain Pathol. 2005;15:46–54. doi: 10.1111/j.1750-3639.2005.tb00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Wang DZ, Olson EN. Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr. Opin. Genet. Dev. 2004;14:558–566. doi: 10.1016/j.gde.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, Singer RH, Segall JE, Condeelis JS. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- Yanowitz JL, Shakir MA, Hedgecock E, Hutter H, Fire AZ, Lundquist EA. UNC-39, the C. elegans homolog of the human myotonic dystrophy-associated homeodomain protein Six5, regulates cell motility and differentiation. Dev. Biol. 2004;272:389–402. doi: 10.1016/j.ydbio.2004.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.