Abstract
Paenibacilli are gram-positive, aerobic bacteria that are related to Bacilli but differ in the DNA encoding their 16S rRNA. Until recently, these organisms were not known to cause human disease. There are now several reports of human infection caused by a few members of this genus, most commonly by P. alvei. We report a human infection in a patient with a permacath for chronic hemodialysis who was found to have bacteremia caused by P. thiaminolyticus, which is an environmental bacterium that has never been found to cause human disease. We identified this bacterium by biochemical tests, cloning, sequencing the genomic DNA encoding its 16S rRNA, growth characteristics, and electron microscopic studies. This constitutes the first report of a human infection caused by this organism.
Keywords: Paenibacillus thiaminolyticus infection, hemodialysis, bacteremia, 16S rRNA
Introduction
Paenibacillus is a genus of aerobic, gram-positive bacilli that were originally included in the genus Bacillus. Based on the finding that these organisms share a highly conserved genome encoding their 16S rRNA, which differs from that of the Bacilli, they were reclassified under the genus Paenibacillus. The Paenibacilli have rarely been found to cause disease in humans [1]. Recently, human infections by a number of species in this group have been described, the most common being P. alvei, which occurs in honeybees. We now report a case of bacteremia in a patient with renal failure caused by P. thiaminolyticus, which has hitherto not been found to cause human disease. This report constitutes the first documented case of human infection caused by this organism.
Case Report
The patient is an 80-yr-old man with multiple medical problems that included end-stage renal disease on hemodialysis, hypertension, congestive heart failure, and Parkinson's disease. The patient also has a subtotal colectomy with ileostomy for colon cancer. A left subclavian permacath (Bard Accos Systems, Salt Lake City, UT) was placed by the interventional radiology service for hemodialysis in mid-May, 2007. In an outpatient visit, about one mo later, the patient complained of itching and irritation at the catheter site. His leukocyte (WBC) count was 12.6 × 103/μl, which was compatible with his usual WBC count in the 11-13 × 103/μl range. Topical hydrocortisone was prescribed.
About one mo later, the patient presented to the Emergency Department complaining of pain in the left anterior chest wall at the catheter site. The patient was found to be afebrile (37.3°C). Physical examination showed a 5×5 cm area over the left clavicle that was swollen and tender to palpation. The permacath site and tunnel under the fossa appeared uninvolved and minimally tender to palpation. The WBC count was 20.5 × 103/μl with a high granulocyte percentage (84%) and a left-shift. The patient was admitted to the hospital. Blood cultures from both arms, distant from the permacath site, were obtained upon admission.
The patient received regular hemodialysis treatments and was given single doses of vancomycin and gentamicin after each treatment. On the second hospital day, the blood cultures taken on the day of admission yielded pure growth of gram-variable rods from both aerobic and anaerobic bottles. Consequently, the patient was placed on a regimen of iv Zosyn (piperacillin and tazobactam) and amikacin. The blood cultures were subsequently identified biochemically as Bacillus species that was sensitive to all of the antibiotics used in sensitivity testing (ie, ampicillin, cefazolin, cefotaxime, cefuroxime, levofloxacin, erythromycin, gentamicin, imipenem, linezolid, penicillin, rifampin, synercid, teracyclin, trimeth/sulfa, and vancomycin), except clindamycin. Blood cultures repeated on the second hospital day confirmed these results. Subsequent biochemical and genetic testing, described below, identified the organism as Paenibacillus thiaminolyticus. Blood cultures taken on the third hospital day were found to be negative.
On third hospital day, the WBC count decreased to 17.5 × 103/μl, and continued to decrease to 16.8 and 11.6 × 103/μl on the fourth and sixth hospital days, respectively. A CT scan with contrast of the neck and thorax on the fourth hospital day suggested the presence of osteomyelitis, abscess, and subclavian vein thrombus, but all signs and symtoms resolved after the access catheter was removed on the same day and was replaced with a non-tunneled temporary venous catheter inserted in the right femoral vein. Culture of the permacath revealed two colonies of Staphylococcus epidermidis. Vancomycin, at dosages for hemodialysis, was continued. On the sixth hospital day, iv amikacin and zosyn were discontinued and were replaced by po Levofloxacin. The patient experienced no subsequent pain or swelling in the left clavicular area, the site of the original catheter, and his WBC count remained in his baseline range. He was discharged from the hospital after 10 days. He was subsequently seen in the hemodialysis clinic and post-dialysis vancomycin was continued for 3 more weeks. Another permacath was subsequently placed in the right subclavian vein for hemodialysis.
Biochemical Studies
We performed biochemical testing on cultures using the API 50 CHB kit (bioMerieux, Inc., Durham, NC). The results are shown in Table 1 for the patient's cultures and for type strains of P. thiaminolyticus (ATCC11377) and P. alvei (DSM29).
Table 1.
| Test | Carbohydrate | P. thiaminolyticus | BKBLC 2156 | P. alvei |
|---|---|---|---|---|
| Control (NG = no growth) | NG | NG | NG | |
| GLY | glycerol | + | + | + |
| ERY | erythritol | - | - | - |
| DARA | D-arabinose | - | - | - |
| LARA | L-arabinose | - | - | - |
| RIB | D-ribose | + | + | + |
| DXYL | D-xylose | - | - | - |
| LXYL | L-xylose | - | - | - |
| ADO | D-adonitol | - | - | + |
| MDX | methyl-βD-xylopyranoside | - | - | - |
| GAL | D-galactose | + | - | + |
| GLU | D-glucose | + | + | + |
| FRU | D-fructose | + | + | - |
| MNE | D-mannose | + | + | + |
| SBE | L-sorbose | - | - | - |
| RHA | L-rhamnose | - | - | - |
| DUL | dulcitol | - | - | - |
| INO | inositol | - | - | - |
| MAN | D-manitol | - | - | - |
| SOR | D-sorbitol | - | - | - |
| MDM | methyl αD- annopyranoside | - | - | - |
| MDG | methyl αD- glucopyranoside | - | - | - |
| NAG | N-acetylglucosamine | + | + | + |
| AMY | amygdaline | + | + | + |
| ARB | arbutine | + | + | + |
| ESC | esculin | + | + | + |
| SAL | salicin | + | + | + |
| CEL | D-cellobiose | - | - | - |
| MAL | D-maltose | + | + | + |
| LAC | D-lactose (origin bovine) | - | - | - |
| MEL | D-melobiose | + | + | - |
| SAC | D-saccharose | + | + | - |
| TRE | D-trehalose | + | + | - |
| INU | inulin | - | - | - |
| MLZ | D-melezitose | - | - | - |
| RAF | D-rafinose | + | + | - |
| AMD | amidon | + | + | + |
| GLYG | glycogene | + | + | - |
| XLT | xylitol | - | ± | - |
| GEN | gentiobiose | + | + | - |
| TUR | D-turanose | + | + | - |
| LYX | D-lyxose | - | - | - |
| TAG | D-tagatose | - | - | - |
| DFUC | D-fucose | - | - | - |
| LFUC | L-fucose | - | - | - |
| DARL | D-arabitol | - | - | - |
| LARL | L-arabitol | - | - | - |
| GNT | potassium gluconate | + | + | - |
| 2KG | potassium 2-cetogluconate | - | - | - |
| 5KG | potassium 5-cetogluconate | - | - | - |
From type strain ATCC11377.
From type strain DSM29.
As shown in Table 2, the patient's cultures were identified with 99.9% probability as P. thiaminolyticus. The only other organism that could be considered was P. alvei at low probability (0.1%). As controls, we tested cultures of the type strains for P. thiaminolyticus and P. alvei using the same method. As shown in Table 2, each type strain tested positive at the 99.9 percent level.
Table 2.
Results of biochemical testing of patient cultures and type strains of Paenibacillus thiaminolyticus and Paenibacillus alvei using the API 50 CHB format.
| Culture | Organism Identity | % Confidence |
|---|---|---|
| BKBLC 2156 (patient) | Paenibacillus thiaminolyticus | 99.9 |
| Paenibacillus alvei | 0.1 | |
| Paenibacillus thiaminolyticus type strain1 | Paenibacillus thiaminolyticus | 99.9 |
| Paenibacillus glucanolyticus | 0.1 | |
| Paenibacillus alvei type strain2 | Paenibacillus alvei | 99.9 |
| Paenibacillus coagulans | 0.1 |
From type strain ATCC11377.
From type strain DSM29.
Genetic Identification
The Paenibacillus genus was established based on phenotypic characteristics and comparative 16S rRNA sequences of gram-positive organisms formerly classified as Bacilli [1]. Therefore, to confirm our identification of the organism as P. thiaminolyticus, we performed the sequencing of the 16S rRNA gene. Briefly, the clinical isolate, called BKBLC2156, was cultured on a blood agar plate and colonies were harvested in 0.5 ml PBS. Bacterial genomic DNA was prepared by using DNeasy tissue kit (Qiagen). 16S rRNA genes were amplified by PCR. For each PCR, 5 μl of the extracted DNA was added to 45 μl of PCR mixture containing 5 μl of 10× PCR buffer (Qiagen), 1.5 mM MgCl2, 200 μM each dNTP, 50 pmol of each primer, and 5 units of TaqDNA polymerase. Reactions were run at 94°C for 2 min, followed by 30 cycles of amplification at 94°C for 30 sec, 52°C for 30 sec, and 72°C for 90 sec, and extension for 20 min at 72°C. Universal bacterial 16S rRNA gene primer pair (forward, 8F AGAGTTTGATYMTGGCTCAG; and reverse, 1492R, TACGGYTACCTTGTTACGACTT) produced PCR products of ≈1,500 bp, spanning positions 8–1513 of the E. coli 16S rRNA gene [2]. The PCR products were separated from free PCR primers by using a PCR purification kit (Qiagen), ligated with the pGEM T Easy (Promega) cloning vector, and used to transform E. coli DH5α competent cells. The cloned inserts underwent sequence analysis using both forward (8F) and reverse (1510R) primers. In total, 11 clones from BKBLC2156 were sequenced. As references, 12 clones from P. thiaminolyticus (ATCC 11377), 10 clones from P. alvei (ATCC 14040), and 12 clones from P. popilliae (ATCC 14706) were sequenced.
For taxonomic identification, the sequences were searched using SEQMATCH against the type strain sequences in the 16S rRNA gene database at RDP II (http://rdp.cme.msu.edu/). Nine of the 11 clones from BKBLC2156 best matched with a 16S rRNA gene (AB073197) from a type strain of P. thiaminolyticus (DSM7262) (Table 3). Degree of similarity between them (similarity scores 0.970-0.980) is comparable to that (0.963-0.993) between the known P. thiaminolyticus strains (ATCC11377 and DSM7262 (Table 4). As shown in Table 1, P. alvei was an unlikely candidate based on the API biochemical results. We therefore compared our results with known sequences of P. alvei. As shown in Table 3, the similarity score between BKBLC2156 and the two available 16S rRNA gene sequences from a P. alvei type strain (DSM29) is substantially below the similarity threshold (0.875, equivalent to 97% sequence identity) [2] for being considered as the same species.
Table 3.
Similarity between 16S rRNA genes of BKBLC 2156 from the patient's culture and Paenibacillus type strains.
| BKBLC2156 clones | Similarity to type strain sequences in 16S rRNA gene database | |||||
|---|---|---|---|---|---|---|
| P. thiaminolyticusa | P. popilliaeb | P. alveic | ||||
| AJ320490d | AB073197 | AB073198 | AF071859 | AJ320491 | AB073200 | |
| 1 | 0.963 | 0.975 | 0.974 | 0.956 | <0.770 | <0.770 |
| 2 | 0.958 | 0.974 | 0.968 | 0.950 | <0.770 | <0.770 |
| 3 | 0.962 | 0.977 | 0.972 | 0.955 | <0.770 | <0.770 |
| 5 | 0.964 | 0.977 | 0.970 | 0.955 | <0.770 | <0.770 |
| 6 | 0.960 | 0.973 | 0.973 | 0.957 | <0.770 | <0.770 |
| 7 | 0.960 | 0.973 | 0.973 | 0.956 | <0.770 | <0.770 |
| 8 | 0.957 | 0.972 | 0.965 | 0.955 | <0.770 | <0.770 |
| 9 | 0.960 | 0.976 | 0.968 | 0.950 | <0.770 | <0.770 |
| 10 | 0.962 | 0.977 | 0.970 | 0.952 | <0.770 | <0.770 |
| 11 | 0.964 | 0.980 | 0.973 | 0.954 | <0.770 | <0.770 |
| 12 | 0.955 | 0.970 | 0.961 | 0.954 | <0.770 | <0.770 |
Table 4.
Similarity between 16S rRNA genes of Paenibacillus test and type strains.
| Strain number | Number of clones | P. thiaminolyticusa | P. popilliaeb | P. alveic | |||
|---|---|---|---|---|---|---|---|
| AJ320490d | AB073197d | AB073198d | AF071859d | AJ320491d | AB073200d | ||
| BKBLC 2156 | 11 | 0.955-0.964 | 0.970-0.980 | 0.961-0.974 | 0.950-0.957 | <0.770 | <0.770 |
| P. thiaminolyticuse | 12 | 0.953-0.979 | 0.963-0.993 | 0.947-0.971 | 0.929-0.949 | <0.770 | <0.770 |
| P. popilliaef | 12 | 0.926-0.942 | <0.931-0.957 | 0.981-1.000 | 0.962-0.981 | <0.770 | <0.770 |
| P. alveig | 10 | <0.791 | 0.795-0.813 | 0.791-0.809 | 0.779-0.798 | 0.930-0.945 | 0.940-0.955 |
DSM7262
ATCC14706
DSM29
GenBank accession number
ATCC11377
ATCC14706
ATCC14040
Highest similarity is highlighted in boldface type
As shown in Table 3, BKBLC2156 also closely resembles P. popilliae. Two of the 11 clones from BKBLC 2156 (clones 6 and 7) are as similar to the type strain of P. thiaminolyticus as to the type stain of P. popilliae. To resolve the ambiguity we performed a phylogenetic analysis. To construct a phylogenetic tree, the sequences were aligned with NAST at Greengenes (http://greengenes.lbl.gov/cgi-bin/nph-index.cgi) [3]. Misalignments were manually curated in Molecular Evolutionary Genetics Analysis 3.1 (MEGA 3.1) [4]. As shown in Fig. 1, a phylogenetic tree was generated by using MEGA 3.1. Evolutionary distances were calculated with the Jukes–Cantor algorithm [4,5]. The statistical strength of the neighbor-joining method was assessed by bootstrap resampling (500 replicates) [6]. The phylogenetic tree has two main branches, corresponding to P. thiaminolyticus and P. popilliae type strain sequences, respectively, and all BKBLC2156 clones are located on the P. thiaminolyticus branch. This result indicates that, although clones 6 and 7 cannot distinguish between P. thiaminolyticus and P. popilliae by sequence similarity, they are evolutionarily more closely related to P. thiaminolyticus. Overall, the 16S rRNA genes analyses support assignment of BKBLC 2156 to P. thiaminolyticus and indicate that BKBLC 2156 is highly unlikely to belong to P. alvei.
Fig. 1.
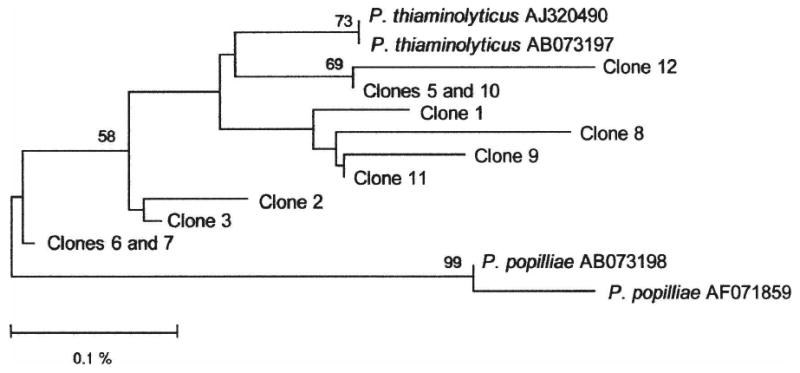
Identification of BKBLC2156 by 16S rRNA gene-based phylogenetic analysis. Bootstrap values (based on 500 replicates) are represented at each node when >50%, and the branch length index is represented below the phylogram. Included in the analyses are clones of 16S rRNA genes from BKBLC 2156 as well as 16S rRNA gene sequences of P. thiaminolyticus (DSM 7262) and P. popilliae (ATCC 14706) type strains. Their Genbank accession numbers are shown after the species name.
Although there was a high degree of similarity between the sequences for the cultured organism and P. popilliae, the latter organism can be ruled out by the absence of positive testing on the API biochemical test results (Table 1) and our finding that P. popilliae shows slow growth on blood agar and does not swarm, whereas BKBLC2156 was found to grow rapidly on blood agar and swarmed, a characteristic of type strains of both P. thiaminolyticus and P. alvei.
Finally, a well-known feature of P. popilliae is the presence of para-spore crystals [7] that can be best visualized by electron microscopy (EM). We therefore performed EM with a JEM 1010 (JEOL) electron microscope on 3% phosphate-buffered glutaraldehyde-fixed samples of the type strains of P. popilliae, alvei, thiaminolyticus, and a sample of BKBLC 2156. After fixation, these samples were post-fixed in 1% osmium tetroxide (1 hr), dehydrated, and then placed in a 1:1 mixture of propylene oxide and embed-812 (50/50 mixture, EM Sciences, Hatfield, PA), and rotated overnight. The samples were then embedded in Beem capsules at 60°C. The epon blocks were cut at 70 nm; sections were placed on copper grids and stained with uranyl acetate and lead citrate. We found that many of the bacilli in the type strain of P. popilliae contained paraspore crystals. However, neither BKBL C2156 nor the type strain of P. thiaminolyticus contained any such structures. This result further confirmed that P. popilliae was not the causative organism.
Discussion
From the patient's clinical course, it seems clear that his fever and elevated white cell count on admission were caused by blood-borne P. thiaminolyticus. Not only were two successive blood cultures positive for this organism, but therapy with iv antibiotics to which the organism was found to be sensitive (pipericillin and an aminoglycoside) resulted in a significant reduction in the WBC count and elimination of the fever. The decreased WBC count correlated with the disappearance of the organism from the blood cultures, ie, on the third hospital day when the blood cultures were negative, the WBC count had decreased from the admission value of 20,500 to 17,500/μl and then decreased to the baseline level in a few days.
Our identification of the causative organism as P. thiaminolyticus is based on biochemical API test results and on the sequencing of the DNA encoding its 16S rRNA. While there was little ambiguity in the API results, there was a small possibility (0.1%) of the organism's being P. alvei. However, this possibility was eliminated in our sequencing results that showed that the DNA sequence encoding the 16S rRNA had a high level of identity to that of P. thiaminolyticus but much less similarity to that of P. alvei; the identity level was below the matching threshold. Interestingly, the 16S rRNA sequence of the organism also was closely related to that of P. popilliae. The latter, however, could be ruled out on the basis of the phylogenetic analysis, biochemical results, its growth properties, and the EM finding that the causative organism did not contain para-spore crystals as seen the bacilli of P. popilliae [7].
P. thiaminolyticus, like the other members of the Paenibacilli, was originally classified as a Bacillus but then reclassified as Paenibacillus genus [1] because of unique phenotypic characteristics and comparative 16S rRNA sequence [8-10]. It was later thought to be related to Bacillus (and then Paenibacillus) alvei, and lost standing in bacteriological nomenclature [9]. However, it was subsequently found that this group is highly homogeneous for 49 phenotypic characteristics and distinguishable from P. alvei [9].
P. thiaminolyticus has been found to cause disease in ruminants, in particular, cerebrocortical necrosis (also known as polioencephalomalacia) in these animals [11-14]. This disease is postulated to be induced at least in part by a thiaminase that results in the inhibition of one or more thiamin-requiring reactions necessary for energy metabolism in the central nervous systems of ruminants [15]. However no human disease has hitherto been found related to P. thiaminolyticus. This is, therefore, the first report of P. thiaminolyticus infection in humans (Table 5).
Table 5.
Summary of human infections caused by Paenibacilli.
| Patient | Condition | Species | Source | Identification method | Reference | |
|---|---|---|---|---|---|---|
| Age | Sex | |||||
| n/a* | n/a* | Meningitis | P. alvei | Cerebrospinal fluid | Biochemical tests | 19 |
| 20 da | F | Meningitis | P. alvei | Cerebrospinal fluid | Biochemical tests | 21 |
| 26 yr | F | Sickle cell disease; hip prosthesis infection | P. alvei | Blood (hip prothesis) | Biochemical tests | 20 |
| 22 yr | M | Traumatic injury; endophthalmitis | P. alvei | Foreign body from eye | Biochemical tests | 16 |
| 62 yr | M | Pneumonitis; pleuritis | P. alvei | Pleural fluid | Biochemical tests | 17 |
| 62 yr | M | Leg cellulitis | P. alvei | Culture from site | Biochemical tests | 18 |
| 93 yr | F | Cerebral infarction | P. polymyxa | Blood | Biochemical tests | 23 |
| 18 yr | F | Munchausen syndrome |
P. polymyxa Bacillus licheniformis Bacillus pumilus |
Blood from self-inflicted compound | Biochemical tests | 22 |
| 52 yr | M | Periorbital trauma | P. macerans | Brain abscess | Biochemical tests | 24 |
| 9 yr | M | Neutropenic fever | P. hongkongensis | Pseudobacteremia | 16S rDNA | 26 |
| 57 yr | M | Endocarditis | P. popilliae | Blood (heart valve) | Biochemical tests | 27 |
| 49 yr | M | Carcinoma of oropharynx | P. sanguinis | Blood | Biochemical tests; 16S rRNA | 28 |
| 13 yr | M | Lymphoblastic leukemia | P. massiliensis | Blood | Biochemical tests; 16S rRNA | 28 |
| 75 yr | F | Nephropathy, hemodialysis | P. timonensis | Blood | Biochemical tests; 16S rRNA | 28 |
| 35 yr | F | Drug abuse, night sweats, pulmonary nodules | P. provencensis | Cerebrospinal fluid | Biochemical tests; 16S rRNA | 29 |
| 54 yr | M | Cerebellar syndrome; Whipple's disease | P. urinalis | Urine | Biochemical tests; 16S rRNA | 29 |
| 80 yr | M | Renal failure, hemodialysis, colon cancer | P. thiaminolyticus | Blood (possible permacath) | Biochemical tests; 16S rRNA | current case |
Neonate, age and sex information not available (n/a).
As summarized in Table 5, even though more than one hundred species have been identified in the genus Paenobacillus, relatively few have been found to cause human disease. The most common Paenobacillus species in human infections is P. alvei [16-21]. Isolated single cases of infection in man by P. polymyxa [22,23], P. macerans [24,25], and P. hongkongensis (possibly a contaminant) [26] have also been described. One patient was reported as having endocarditis caused by P. popilliae [27]. Recently, five new species of Paenibacillus, isolated from human blood, cerebrospinal fluid, and urine, have been described [28,29]. Of the 17 reported cases of Paenibacillus infection in humans, including the present study, 8 were found to have positive blood cultures, making blood the most common source for isolation of the bacteria from this genus.
The sources of infection by P. thiaminolyticus are unknown. Paenibacillus species are mainly environmental organisms [30]. They are not detectable on human skin [31]. Of the 8 reported cases of bacteremia (including this one), 2 identified medical devices as the source, ie, a hip prosthesis and a heart valve (Table 5). Since the indwelling subclavian catheter in our patient was the cause of pain that resulted in his coming to the emergency room, one may speculate that this device was the source of the bacteremia. However, culture of the indwelling subclavian catheter was negative for P. thiaminolyticus. On the other hand, this negative culture was obtained after antimicrobial therapy was instituted and one day after the third blood culture, which was negative for this organism. Thus this finding may have been due to antimicrobial therapy, especially since the organism was susceptible to all antimicrobial agents tested, except clindamycin. Furthermore, no other organisms were recovered from the catheter (except S. epidermidis as a skin contaminant) or the blood cultures.
In summary, this is the first reported case of human infection, ie, bacteremia, by P. thiaminolyticus. There seems to be a predilection of the Paenibacilli to cause bacteremia. Since most of these organisms are susceptible to a wide range of anti-microbial agents, early identification will enable rapid, effective treatment.
Acknowledgments
This work was supported in part by National Institutes of Health Grant R01AI063477. The sequence reported in this paper has been deposited in the GenBank database (accession number EU420075).
References
- 1.Ash C, Priest FG, Collins MD. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek. 1993;64:253–260. doi: 10.1007/BF00873085. [DOI] [PubMed] [Google Scholar]
- 2.Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. Bacterial biota in the human distal esophagus. PNAS USA. 2004;101:4250–4255. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 5.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis, version 1.02. Pennsylvania State University; University Park, PA: 1997. [Google Scholar]
- 6.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Schairer HU, Schnetter W, Lereclus D, Agaisse H. Bacillus popilliae cry18Aa operon is transcribed by sigmaE and sigmaK forms of RNA polymerase from a single initiation site. Nucleic Acids Res. 1998;26:1288–1293. doi: 10.1093/nar/26.5.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heyndrickx M, Vandemeulebroecke K, Scheldeman P, Kersters K, de Vos P, Logan NA, Aziz AM, Ali N, Berkeley RC. A polyphasic reassessment of the genus Paenibacillus, reclassification of Bacillus lautus (Nakamura 1984) as Paenibacillus lautus comb. nov. and of Bacillus peoriae (Montefusco et al. 1993) as Paenibacillus peoriae comb. nov., and emended descriptions of P. lautus and of P. peoriae. Int J Syst Bacteriol. 1996;46:988–1003. doi: 10.1099/00207713-46-4-988. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura LK. Bacillus thiaminolyticus sp. nov., nom. rev. Int J Syst Bacteriol. 1990;40:242–246. doi: 10.1099/00207713-40-3-242. [DOI] [PubMed] [Google Scholar]
- 10.Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K. Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int J Syst Bacteriol. 1997;47:289–298. doi: 10.1099/00207713-47-2-289. [DOI] [PubMed] [Google Scholar]
- 11.Ramos JJ, Marca C, Ferrer LM, Loste A, Cebrian LM. Faecal thiaminase, plasma lactate and pyruvate concentrations and erythrocyte transketolase activity changes in apparently normal replacement ewes after the initiation to the pasture. Res Vet Sci. 2006;80:11–16. doi: 10.1016/j.rvsc.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Thomas KW. The effect of thiaminase-induced subclinical thiamine deficiency on growth of weaner sheep. Vet Res Commun. 1986;10:125–141. doi: 10.1007/BF02213975. [DOI] [PubMed] [Google Scholar]
- 13.Thomas KW, Griffiths FR. Natural establishment of thiaminase activity in the alimentary tract of newborn lambs and effects on thiamine status and growth rates. Aust Vet J. 1987;64:207–210. doi: 10.1111/j.1751-0813.1987.tb15183.x. [DOI] [PubMed] [Google Scholar]
- 14.Tirrell IM, Wall JL, Daley CJ, Denial SJ, Tennis FG, Galens KG, O'Handley SF. YZGD from Paenibacillus thiaminolyticus, a pyridoxal phosphatase of the HAD (haloacid dehalogenase) superfamily and a versatile member of the Nudix (nucleoside diphosphate x) hydrolase superfamily. Biochem J. 2006;394:665–674. doi: 10.1042/BJ20051172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brent BE, Bartley E. Thiamin and niacin in the rumen. J Anim Sci. 1984;59:813–822. doi: 10.2527/jas1984.593813x. [DOI] [PubMed] [Google Scholar]
- 16.Antonello A, Weinstein GW. Successful treatment of Bacillus alvei endophthalmitis. Am J Ophthalmol. 1989;108:454–455. doi: 10.1016/s0002-9394(14)73321-5. [DOI] [PubMed] [Google Scholar]
- 17.Coudron PE, Payne JM, Markowitz SM. Pneumonia and empyema infection associated with a Bacillus species that resembles B. alvei. J Clin Microbiol. 1991;29:1777–1779. doi: 10.1128/jcm.29.9.1777-1779.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin JH, Sung SK, Shim JD, Kim HR, Park SW, Lee JN. A case of Paenibacillus alvei cellulitis in immuno-competent patient. Korean J Lab Med. 2005;25:53–55. [Google Scholar]
- 19.Park SJ, Cong W, Lee SY. Bacterial and fungal species from cerebrospinal fluid in the past five years. Korean J Pathol. 1976;10:137–142. [Google Scholar]
- 20.Reboli AC, Bryan CS, Farrar WE. Bacteremia and infection of a hip prosthesis caused by Bacillus alvei. J Clin Microbiol. 1989;27:1395–1396. doi: 10.1128/jcm.27.6.1395-1396.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiedermann BL. Non-anthrax Bacillus infections in children. Pediatr Infect Dis J. 1987;6:218–220. [PubMed] [Google Scholar]
- 22.Galanos J, Perera S, Smith H, O'Neal D, Sheorey H, Waters MJ. Bacteremia due to three Bacillus species in a case of Munchausen's syndrome. J Clin Microbiol. 2003;41:2247–2248. doi: 10.1128/JCM.41.5.2247-2248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nasu Y, Nosaka Y, Otsuka Y, Tsuruga T, Nakajima M, Watanabe Y, Jin M. A case of Paenibacillus polymyxa bacteremia in a patient with cerebral infarction. Kansenshogaku Zasshi. 2003;77:844–848. doi: 10.11150/kansenshogakuzasshi1970.77.844. [DOI] [PubMed] [Google Scholar]
- 24.Bert F, Ouahes O, Lambert-Zechovsky N. Brain abscess due to Bacillus macerans following a penetrating periorbital injury. J Clin Microbiol. 1995;33:1950–1953. doi: 10.1128/jcm.33.7.1950-1953.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrero F, Galan F, Marin P, Garcia-Martos P, Capote FJ. Catheter-associated infection by Bacillus macerans in a patient with acute leukemia. Enferm Infect Microbiol Clin. 1996;14:628–629. [PubMed] [Google Scholar]
- 26.Teng JL, Woo PC, Leung KW, Lau SK, Wong MK, Yuen KY. Pseudobacteraemia in a patient with neutropenic fever caused by a novel Paenibacillus species: Paenibacillus hongkongensis sp. nov. Mol Pathol. 2003;56:29–35. doi: 10.1136/mp.56.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu YJ, Hong TC, Hou CJ, Chou YS, Tsai CH, Yang DI. Bacillus popilliae endocarditis with prolonged complete heart block. Am J Med Sci. 1999;317:263–265. doi: 10.1097/00000441-199904000-00010. [DOI] [PubMed] [Google Scholar]
- 28.Roux V, Raoult D. Paenibacillus massiliensis sp. nov., Paenibacillus sanguinis sp. nov. and Paenibacillus timonensis sp. nov., isolated from blood cultures. Int J Syst Evol Microbiol. 2004;54:1049–1054. doi: 10.1099/ijs.0.02954-0. [DOI] [PubMed] [Google Scholar]
- 29.Roux V, Fenner L, Raoult D. Paenibacillus provencensis sp. nov., isolated from human cerebrospinal fluid, and Paenibacillus urinalis sp. nov., isolated from human urine. Int J Syst Evol Microbiol. 2008;58:682–687. doi: 10.1099/ijs.0.65228-0. [DOI] [PubMed] [Google Scholar]
- 30.McSpadden-Gardener B. Ecology of Bacillus and Paenibacillus spp. in agricultural systems. Phytopathology. 2004;94:1252–1258. doi: 10.1094/PHYTO.2004.94.11.1252. [DOI] [PubMed] [Google Scholar]
- 31.Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. PNAS USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]


