Abstract
The LAH4 family of histidine-rich peptides exhibits potent antimicrobial and DNA transfection activities, both of which require interactions with cellular membranes. The bilayer association of the peptides has been shown to be strongly pH-dependent, with in-planar alignments under acidic conditions and transmembrane orientations when the histidines are discharged. Therefore, we investigated the pH- and temperature-dependent conformations of LAH4 in DPC micellar solutions and in a TFE/PBS solvent mixture. In the presence of detergent and at pH 4.1, LAH4 adopts helical conformations between residues 9 and 24 concomitantly with a high hydrophobic moment. At pH 6.1, a helix-loop-helix structure forms with a hinge encompassing residues His10–Ala13. The data suggest that the high density of histidine residues and the resulting electrostatic repulsion lead to both a decrease in the pK values of the histidines and a less stable α-helical conformation of this region. The hinged structure at pH 6.1 facilitates membrane anchoring and insertion. At pH 7.8, the histidines are uncharged and an extended helical conformation including residues 4–21 is again obtained. LAH4 thus exhibits a high degree of conformational plasticity. The structures provide a stroboscopic view of the conformational changes that occur during membrane insertion, and are discussed in the context of antimicrobial activity and DNA transfection.
Abbreviations: CSI, chemical shift index; DPC, dodecyl phosphocholine; DQF-COSY, double quantum filtered correlation spectroscopy; Fmoc, 9-fluorenylmethyloxycarbonyl; HPLC, high-performance liquid chromatography; MALDI, matrix-assisted laser desorption ionization; NMR, nuclear magnetic resonance; NOESY, nuclear Overhauser effect spectroscopy; PBS, phosphate-buffered saline; RMSD, root mean-square deviation; ROESY, rotating frame Overhauser effect spectroscopy; TFA, trifluoroacetic acid; TFE, trifluoroethanol; TOCSY, total correlation spectroscopy
Introduction
Amphipathic peptide sequences are produced, stored, and secreted by many organisms, including fungi, plants, insects, amphibia, and humans (1–4). They serve many important functions in life, such as cell-to-cell signaling and antimicrobial protection, and are used in biotechnological applications such as cell penetration and transfection—activities that are linked to the transport of large cargo molecules into the cell interior (5–8). For example, to fight antimicrobial infections, the innate immune system of many species employs cationic peptides (1,2), and many of these or related sequences also exhibit cell-penetrating activities (4,9). Therefore, intricate interactions with cellular membranes are a key prerequisite for many of the above-mentioned biological functions. To develop their biological activities, the peptides have to interact with the lipid bilayer, which directly constitutes their site of action and/or has to be passed before the peptides can enter the cell interior (1,10).
Studies using biophysical methods to examine the membrane interactions of amphipathic peptides have revealed a complex behavior with lipid-, temperature-, and pH-dependent morphologies of the supramolecular complexes and a wide variety of peptide topologies and aggregation states in the membrane (11,12). Furthermore, after they are released, many amphipathic peptides are soluble in water, where they occur as monomers or aggregates and subsequently target the membranes of other species (12). Several models have been proposed to explain the pore-forming properties of these peptides, including the barrel-stave, wormhole, and carpet models (13–15). These models have been included in phase diagrams similar to those established for lipid or detergent-lipid mixtures, as such complex representations are needed for a complete description of all the possible membrane-peptide morphologies (11).
To investigate membrane insertion and the interactions that control the membrane topology of amphipathic peptides, researchers designed a series of sequences in which the core regions are composed predominantly of leucines, alanines, and four histidines, resulting in a family of peptides known as LAH4 (16,17). Due to the presence and arrangement of the histidines, one can not only change the membrane topology in a pH-dependent manner (16,18), but also test the energetic contributions associated with the transition of the peptide orientation from parallel to the surface to a transmembrane alignment (19–21). The transition pH reflects a fine balance between the ionization state of the histidines, as well as hydrophobic and other interactions. The parent LAH4 polypeptide consists of 26 amino acids, is soluble in aqueous solution (where it is monomeric under acidic conditions), and forms small aggregates at neutral pH (22). Of note, the pronounced antibiotic activity of LAH4 at pH 5.5 indicates that antibiotic action is caused by amphipathic helices aligned along the surface of the membrane, rather than by the formation of a specific transmembrane pore structure (17,23,24). More recently, it was shown that LAH4 and several of its derivatives exhibit potent DNA and siRNA transfection activities while at the same time the transfection complex maintains its antibacterial activity (25–28). During transfection, the peptide/DNA complexes use an endosomal pathway that involves a shift to acidic environments and thus correlates with the pH-induced topological transitions of the membrane-associated peptide (25,26).
So far, the molecular details of membrane insertion of amphipathic peptides remain largely unknown (29,30). In this work, we sought to characterize the conformations of membrane-associated LAH4 using two-dimensional (2D) solution NMR spectroscopy in micellar solutions as a function of temperature and pH, as well as in a membrane-mimetic solvent mixture. We were able to obtain a stroboscopic view of the conformational and topological changes that occur during membrane insertion. The structural data are discussed in the context of the peptides' membrane insertion and the concomitant antibiotic and DNA transfection activities.
Materials and Methods
Peptide synthesis
The peptide LAH4 (MW: 2777; sequence: KKALLALALHHLAHLALHL ALALKKA-NH2) was prepared by solid-phase peptide synthesis on Millipore 9050 or ABI433 automatic peptide synthesizers using Fmoc chemistry. A fourfold excess of Fmoc-protected amino acids (Bachem, Heidelberg, Germany; Applied Biosystems, Weiterstadt, Germany) was used during chain elongation. After TFA cleavage was achieved, the peptide was purified by preparative HPLC using an acetonitrile/water gradient in the presence of 0.1% TFA and a 214 nm detection wavelength. The identity and high purity of the product was confirmed by MALDI mass spectrometry and analytical HPLC. After lyophilization, the TFA counterions were exchanged in 5% acetate (v/v).
NMR experiments
To investigate the structure of LAH4, 6 mg of the lyophilized peptide were dissolved in a mixture of 50% (v/v) deuterated TFE (TFE-d3) in PBS (115 mM NaCl, 8 mM KH2PO4, 20 mM Na2HPO4, pH 7.0) or added to a DPC-d38 micellar solution in PBS. The TFE-d3 and perdeuterated DPC (DPC-d38) were purchased from Promochem (Wesel, Germany). When different peptide/detergent ratios were screened (31), the best resonance linewidths were obtained from samples consisting of 200 mM DPC, 2 mM peptide, 0.02% sodium azide, and 10% of D2O. Without further titration, the addition of peptide decreased the pH to ∼6. The pH values for the samples were adjusted to 4.1 or 7.8 by the addition of 1 M stock solutions of H3PO4 or Na3PO4. Before NMR measurements were performed, the micellar solutions were vortexed for 2 h followed by 5 min of centrifugation at 1000 g to remove impurities.
One-dimensional (1D) and 2D NMR experiments were performed at 300 K or 317 K on a DRX500 spectrometer (Bruker Biospin, Rheinstetten, Germany) equipped for pulsed field gradient spectroscopy. For 1H assignments, 2D TOCSY (32), NOESY (33), ROESY (34), and DQF-COSY (35) spectra were recorded. Water suppression in TOCSY was performed by means of a jump-return pulse sequence (36), and a MLEV17 spin-lock (37) of either 25 ms or 70 ms was used. In addition, NOESY and ROESY experiments with 100 ms mix times and a WATERGATE pulse sequence (38) were recorded. When considered useful, additional NOESY experiments with 200 ms mix times were acquired. For the DQF-COSY experiment, presaturation of the water resonance during the relaxation delay was performed. All phase-sensitive 2D experiments were recorded using the time-proportional phase incrementation method (39). For these experiments, 96–160 transients for 600–650 t1 increments with 2048 complex data points were collected. The relaxation delay between successive transients was 1.2–1.5 s. The spectral width was set to 7002 Hz in both dimensions. Before Fourier transformation was performed, the data along the t1 dimension were zero-filled to 1024, and a sine square apodization function in t1 and a Gaussian window function with −10 Hz line-broadening in the t2 dimension were applied. All of the data were processed with XWINNMR software (Bruker Biospin, Rheinstetten, Germany). Residues were assigned by using the in-house-written software ccnmr and glxcc (40). Proton chemical shifts are reported with respect to the H2O signal (4.75 ppm relative to tetramethylsilane).
The chemical shift assignments for LAH4 in 50% TFE or DPC micelles at different pH values were obtained using the standard method as previously described (41). Additionally, to improve the assignment of ambiguous sequential resonances, spin diffusion was considered.
Structure calculations
Distance constraints were extracted from the NOESY and ROESY spectra with a 100 ms mix time, and, when available, NOE cross-peaks that became visible after 200 ms of mix time were taken into consideration to confirm the presence of otherwise weak intensities. The cross-peaks were classified according to their intensities as weak, medium, or strong, with upper-limit distances of 5.0, 3.4, and 2.8 Å, respectively. Only the interresidual NOE-derived restraints were used during the calculation procedure, resulting in a total of 178 and 157 distance restraints for the structures at pH 4.1 and 6.1, respectively. Hydrogen-bonding restraints were also used for the determination of structures (14 and 11 hydrogen bonds for the structures at pH 4.1 and 6.1, respectively). Calculations were performed using the Xplor-NIH v2.17 program with only a few changes (42). The calculations started from extended conformations using the torsion angle dynamics simulated annealing protocol written by Stein et al. (43). During the high-temperature dynamics, the first cooling period was set to 10,000 steps per cycle. The second cooling involved 6000 steps, and the final Powell minimization was increased to 100 steps per cycle with kNOE = 50 kcal/Å2. This calculation was followed by molecular-dynamics refinement in explicit water (44). For each condition, a total of 200 structures with no distance restraint violation higher than 0.5 Å were calculated, and the 20 most stable conformations were extracted to represent the peptide structure. The quality of the resulting ensembles was assessed by application of the PROCHECK-NMR and AQUA alghorithms (45). The program MOLMOL 2K.2 was used to calculate the pairwise RMSDs for both sets of calculated structures and to generate the structural models shown (46).
As another calculation methodology, we also employed the program CNS, which takes into account intraresidual restraints as well as restraint upper limits that are derived from internuclear distances between histidine ring hydrogens. This procedure is described in more detail in the Supporting Material. The mean structures over the corresponding ensemble and the best structures at pH 6.1 and pH 4.1 in the presence of DPC are accessible through the Protein Data Base (2KJO and 2KJN).
Chemical shift information is available through the BMRB Database (accession numbers 16332 and 16332 for pH 4.1 and 6.1 (317 K), respectively). Additional chemical shift data in 50% TFE in PBS (6827) or in the presence of DPC at pH 6.1/300 K and pH 7.8/317 K (6886) are also available.
Results
Secondary structure in 50% TFE-PBS
The graphic representation shown in Fig. 1 of NOEs obtained in 50% TFE in PBS, a solvent that mimics the hydrophobicity of membranes, reveals a number of dαN (i, i+3) and dαβ (i, i+3) correlations. At the same time, the dαN (i, i+4) and dNN (i, i+1) NOE cross-peaks are weak or absent even at long NOE mixing intervals. Furthermore, the CSIs of the Cα protons lack a clear high local density of upfield shifts (Fig. 1 A), which would be indicative of stable α-helical structures (47). This ensemble of data suggests that in TFE solution, the peptide exhibits a modest propensity for helical structures in the L4–L23 region.
Figure 1.
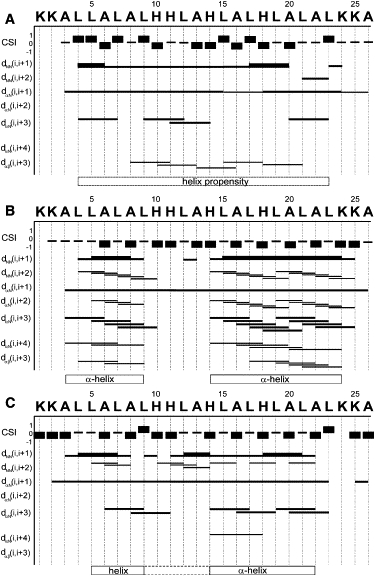
Graphic summary of NOEs observed for LAH4 in (A) 50% TFE-PBS, (B) DPC micellar solutions at pH 6.1 and 317 K and (300 K. The thickness of the lines represents strong, medium, and weak NOE intensities. The helical domains indicated correspond to the statistical analysis of simulated annealing calculations (panel B) or to regions where at least two of the following are observed: dαN (i, i+3), dαN (i, i+4), dαβ (i, i+3), dNN (i, i+2), strong dNN (i, i+1), and high density of helical CSI (47).
Under these conditions, the first two lysines, the NH resonance of A3, and the side chains of K24 and K25 remain undetectable. The C-terminal amide exhibits resonances at 6.88 and 7.31 ppm, and these chemical shifts are summarized in Table S1.
Secondary structure in DPC micelles
Structure of LAH4 at different temperatures and pH 6.1
Although isotropic TFE/water mixtures are advantageous for solution-state NMR spectroscopy due to the peptides' fast reorientational correlation times in this environment, a much better representation of the interfaces of phosphatidylcholine bilayers can be obtained from DPC micelles. Therefore, NOESY and TOCSY spectra of the LAH4 peptide at pH 6.1 were recorded in DPC micellar solution at 300 K and 317 K. Because the spectra at 317 K were better resolved, assignment of the peptide was initially performed at this temperature and thereafter compared with the spectra obtained at 300 K. The NH-Hα region of the TOCSY spectra of LAH4 at 317 K and pH 6.1 is shown in Fig. 2 A. The corresponding NOE correlations in the NH-NH region are summarized in Fig. 1 B. At this temperature, a continuous pattern of dαN (i, i+3), dαN (i, i+4), a number of dαβ (i, i+3), and several strong dNN (i, i+1) connectivities are indicative of α-helical structures encompassing residues A3–L9 and H14–K24.
Figure 2.
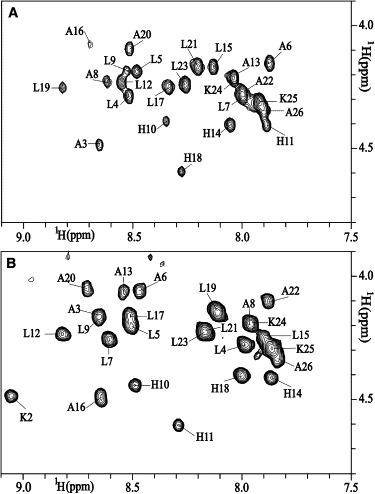
The NH-Hα region of TOCSY spectra of LAH4 in DPC micellar solutions at 317 K is shown. (A) pH = 6.1. (B) pH = 4.1.
At 300 K, the interresidual contacts in the region encompassing residues L5–A22 are suggestive of helical structures (Fig. 1 C). Between L9 and H14, contacts such as NH-NH or dαN (i, i+3) or dαN (i, i+4), which are typical for the definition of helical structures, are less abundant or missing, suggesting a less stable helical conformation in the central region of the peptide (hatched box in Fig. 1 C), and reflecting to some extent the situation observed at the higher temperature (Fig. 1 B).
Structures of LAH4 at 317 K and different pH values
Because proton activity plays such an important role in the topology and biological activities of LAH4, we investigated the structure of membrane-associated LAH4 as a function of pH (16,17,20,23). Significant changes in the number, magnitude, and position of the cross-peaks were observed upon alterations in pH, and required independent assignments of the resonances at each pH condition (Fig. 2). Although an overall high degree of α-helicity was observed in the presence of DPC under all conditions investigated, changes in the chemical shifts as well as the NOE patterns (compare Fig. 1 B and Fig. 4) indicated the occurrence of pH-dependent conformational differences and/or changes in the interaction with the micellar environment, in agreement with histidine pK values of ∼6 (16,18).
Figure 4.
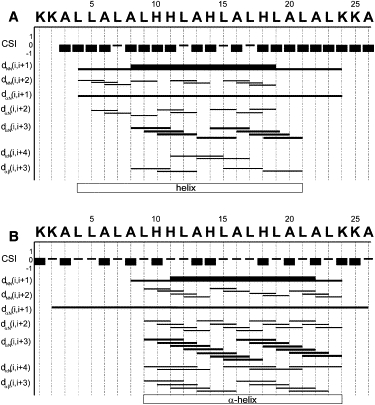
Graphic representation of NOEs observed for LAH4 in DPC micellar solutions at 317 K and pH values of (A) 7.8 and (B) 4.1. The thickness of the lines represents strong, medium, and weak NOE intensities. The helical domains have been identified by the rules specified in the legend to Fig. 1.
At pH 7.8 the pattern of NOE intensities indicates an α-helical structure between L4 and L21 (see Fig. 4 A). The helical structure at pH 7.8 is confirmed by the CSIs of the Cα protons, which show a strong tendency toward negative values throughout the sequence. The small number of NOE cross-peaks, the missing NH resonances, and the weak NH-NH contacts suggest that the termini, involving K1–A3 and A22–A26, are unstructured. At pH 7.8 only the HD2 atoms of the histidines are visible in the spectra, whereas at pH 6.1 and pH 4.1 the HE1 protons are also detectable (Fig. 3).
Figure 3.
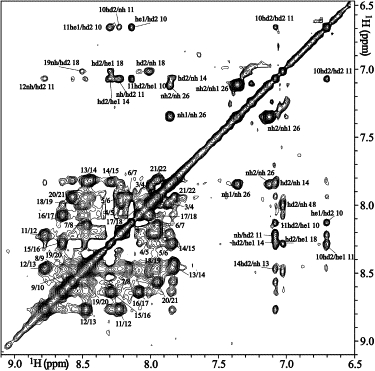
The NH-NH region of the 2D NOESY spectrum in DPC recorded at pH 6.1 and 300 K, with the assignment of cross-peaks shown.
A graphic representation of unambiguously assigned NOE intensities at pH 4.1 is shown in Fig. 4 B. The pattern and intensities of the interresidual contacts are consistent with α-helical conformations in the L9–K24 region. Of note, the number of medium-range NOEs observed for residues A9–H14 is greater under acidic conditions (Fig. 4 B) than at pH 6.1 (Fig. 1 B), suggesting that LAH4 adopts a better-defined α-helical structure. Although the CαH chemical shifts, as represented by their CSIs (Fig. 1 B and Fig. 4 C), are less negative than those at pH 7.8, on average they are considerably below those observed for random coil conformations. Furthermore, the secondary shifts of CαH and NH show an oscillating pattern with a 3–4 residue repeat as observed for other amphipathic helices (48). Although the N-terminal part seems unstructured at all pH values, its overall character appears to be different at acidic pH, where the NH of K2 is visible (Fig. 4).
Detailed structural folds of micelle-associated LAH4 at pH 6.1 and pH 4.1
Starting from extended conformations and using the experimental NOE intensities obtained at 317 K, we performed simulated annealing calculations for LAH4 in the presence of DPC at pH 6.1 and pH 4.1. Fig. 5 shows the backbone superposition of the 20 lowest-energy structures that were generated using Xplor-NIH. Fig. 5 A represents the set of structures for LAH4 at pH 4.1, and panels B and C show that obtained for pH 6.1. Whereas at pH 4.1 an uninterrupted α-helical region encompassing residues L9–K24 is evident, the structures at pH 6.1 are characterized by two helical domains (A3–L9 and H14–K24) separated by a mostly unstructured region comprising residues H10–A13, where only a few experimental restraints are available (Fig. 1 B).
Figure 5.
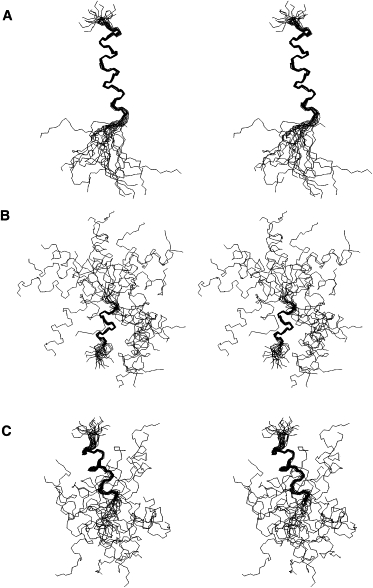
Stereo view of the backbone superposition of the 20 lowest-energy structures at pH 4.1, where region 9–24 (A) is considered for superposition of the coordinates, and at pH 6.1, where the regions 3–9 (B) and 14–24 (C) are considered separately for superposition of the structures.
When the Ramachandran angular distribution is analyzed, 75.2% and 73.3% of the residues occur in the most favored regions for the structural models obtained at pH 4.1 and 6.1, respectively (Table 1). When only the residues belonging to the well-structured regions are taken into account, these fractions rise to 91.2% at pH 4.1 and to 89.7% at pH 6.1. This shows that most of the residues that fall into the least-favored regions of the Ramachandran plot belong to regions in which only a few distance restraints are measured. The assessment of the RMSD values for each ensemble showed a marked convergence of the conformation of the well-defined segments in the polypeptide chain. For the calculated models at pH 4.1, the RMSD is 0.43 Å for the backbone of the α-helix comprising L9–K24. For the data set obtained at pH 6.1, the backbone RMSDs of the helical regions A3–L9 and H14–K24 are 0.48 Å and 0.53 Å, respectively.
Table 1.
Statistics of the NMR structural evaluation of LAH4 in DPC micellar solution at pH 4.1 and 6.1 using Xplor-NIH
| Structural restraints from NMR spectroscopy | pH 4.1 | pH 6.1 | |
|---|---|---|---|
| Total NOE | 262 | 238 | |
| Intraresidue | 84 | 81 | |
| Interresidue | 178 | 157 | |
| Hydrogen bonds | 14 | 11 | |
| Ramachandran Analysis | |||
| Residues in most favored regions | 75.2% | 73.3% | |
| Residues in additionally allowed regions | 7.2% | 17.3% | |
| Residues in generously allowed regions | 1.9% | 5.0% | |
| Residues in disallowed regions | 6.1% | 4.4% | |
| Coordinate precision for selection (residue numbers) | 9–24 | 3–9 | 14–24 |
| Pairwise RMSD of the backbone (Å) | 0.43 ± 0.14 | 0.48 ± 0.20 | 0.53 ± 0.18 |
| Pairwise RMSD for all heavy atoms (Å) | 1.11 ± 0.24 | 1.31 ± 0.26 | 1.44 ± 0.27 |
Discussion
The cationic linear amino acid sequence LAH4 is a representative of a large family of antimicrobial peptides (3,23), and in addition has been found to be a potent vector for carrying nucleic acids into the eukaryotic cell interior (6,28). To perform these biological activities, the peptide has to interact with and/or cross cellular membranes (1,10,26). When amphipathic peptides transfer from the water to the membrane phase, they are thought to first interact with the membrane interface, where they adopt an (helical) amphipathic 3D structure (30,49) (Fig. 6, C, D, and G). Although this initial membrane-interacting state is characterized by alignments of the peptide along the membrane surface (Fig. 6 G), transmembrane orientations of some of the more hydrophobic sequences (Fig. 6 I) are subsequently observed in a second step (12,50). These transitions have been well investigated and characterized in a quantitative manner for LAH4 in lipid bilayers, where pH-dependent in-plane and transmembrane alignments have indeed been observed (16,19,20).
Figure 6.
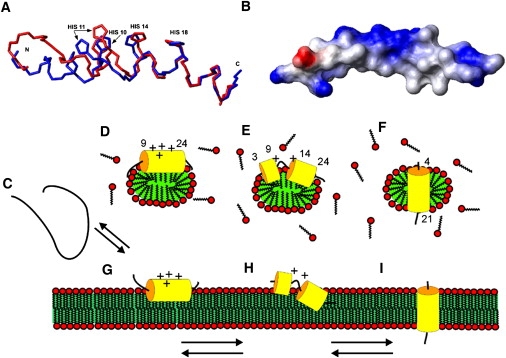
(A) Backbone traces of the mean structures of LAH4 showing the histidine heavy atoms mostly affected by pH changes. The mean structure of the peptide at pH 6.1 is displayed in blue, and the red trace represents the mean structure at pH 4.1. The superposition of both structures is based on the helical region encompassing the region between H14 and L23. (B) Space-filling model of the structure obtained at pH 6.1 and 317 K, representing the positive surface charges of the polypeptide in blue and the negative charges in red. The C-terminus is shown to the left. (C) Model representations of the LAH4 peptide in solution (random coil and monomeric at low pH (22,23)), of the structures in micellar environments (D–F) and in lipid bilayers (G–I). The surface-oriented topology at acidic pH (D and G), the transition state (E and H), and the transmembrane alignments at pH > 7 are shown (F and I) as measured in oriented phospholipid bilayers (G–I) (16) or by analogy (D–F). Panels D–F summarize the structural features observed in this work, whereas panels G–I combine these findings with the membrane topology obtained from oriented phospholipid bilayers (16,18).
Here, we studied the structural details that are associated with such topological transitions by using multidimensional solution-state NMR spectroscopy in micellar and isotropic membrane-mimetic environments. A first inspection of the chemical shift and NOE information indicates that, although helical secondary structures are predominant, the peptide undergoes a number of structural transitions upon changes in solvent, temperature, or pH (Fig. 1, Fig. 4, and Fig. 5). When added at concentrations > 0.1 mM to PBS buffer, pH 7.0, without cosolvent, LAH4 exhibits considerable line-broadening and little chemical shift dispersion. This is in agreement with previous studies in which, under related conditions, the peptide exhibited a predominantly random coil character (23) and formed aggregates of ∼40 nm hydrodynamic diameter (22). However, upon addition of 50% TFE, the NMR spectra suggest that LAH4 occurs as a monomer, while at the same time the NOEs indicate a modest propensity of the peptide to form helical conformations between residues 4 and 23 (Fig. 1 A). Of interest, the helical conformations are more stable in DPC micellar environments than in TFE, although this solvent has been suggested to promote the formation of helical secondary structures (51), which underlines the importance of choosing the appropriate membrane model system. A comparison of the LAH4 spectra obtained in TFE (Fig. 1 A) with those recorded in the presence of DPC at 300 K or 317 K (Fig. 1, B and C, and Fig. 4) suggests that in TFE/PBS 50/50 the structural features represent an intermediate state, and the full helical character is induced only upon association with membrane interfaces.
Therefore, we focused our attention on the structural analysis of LAH4 in the presence of DPC micelles, which better mimic the membrane and at the same time can be characterized by rotational correlation times sufficiently fast for solution-state NMR structural investigations (52). Although in the presence of detergent LAH4 adopts predominantly α-helical conformations under all conditions tested, the delineations of these helices vary in a pH-dependent fashion (Fig. 1, B and C, Fig. 4, and Fig. 5). A long, well-defined amphipathic helix spanning residues 9–24 is present at pH 4.1, and at the same time the most N- and C-terminal residues remain largely disordered (Fig. 5 A). At this pH value, the histidines are charged (16) and all locate at the polar face of an amphipathic helical structure (Fig. 6 A). Therefore, under acidic conditions the peptide aligns parallel to the surface of lipid bilayers (16,18), as illustrated in Fig. 6 G.
When the pH is increased to 6.1 (317 K), subtle conformational transitions occur and two helical domains are formed encompassing residues 3–9 and 14–24 (Fig. 1 B and Fig. 5, B and C). The pH is within the range in which LAH4 changes from in-plane to transmembrane topologies (16,18), and therefore this structure is of particular interest. The flexible loop region connecting the two helices probably enables the membrane insertion of one helix at a time, thus facilitating the realignment process in a manner comparable to models proposed for the voltage-gating mechanism of alamethicin (53).
Finally, when the pH is changed from 6.1 to 7.8, LAH4 reverts to a long α-helix encompassing residues 4–21 (Fig. 4 A). At this pH the peptide adopts transmembrane alignments in oriented phosphatidylcholine membranes (Fig. 6 I) (16,18). Correlation times faster than the micellar complex, concomitant with isotropic averaging, suggest that the termini are unstructured and exposed to the water phase. The chemical shifts for the histidine side-chain resonances indicate that all four of them are deprotonated at pH 7.8. In addition, the transmembrane alignment of the peptide in oriented bilayers (Fig. 6 I) and paramagnetic shift measurements in DPC micellar environments (P. Bertani and B. Bechinger, unpublished results) suggest that they are deeply inserted into the hydrophobic core of the micelle (Fig. 6 F) (16,18).
Therefore, when changing from acidic to neutral conditions, the helical domain shifts from a C-terminal to a more N-terminal localization, and at the same time the histidine side chains lose their charges and pronounced alterations in membrane topology occur. The concomitant environmental changes, as well as the favorable interactions of the protonated histidine side chains (at acidic pH) with the negative end of the helix dipole (54), probably contribute to these modulations of the secondary structure.
It should be noted that the NOE-based calculations yield a family of structures (Fig. 5) that include conformations with helix delineations somewhat different from the consensus regions indicated throughout this work, reflecting the large conformational space and the dynamics of these peptides. Not only do they exhibit conformational freedom when associated with the membranes, they can also exchange between the water and the micellar environment. Indeed, membrane insertion and helix formation have been shown to be tightly connected processes (55), and the observed temperature dependence of the LAH4 spectra at pH 6.1 (Fig. 1, B and C) suggests that entropic contributions related to hydrophobic interactions have an effect on the penetration of the helices into the micelle and the conformational preferences of the peptide. Furthermore, temperature-dependent changes in the shape and phase behavior of the detergent/peptide mixtures may play a role in determining LAH4's interactions with the micellar interface (56).
By assigning resonances at three different pH values, we were able to correlate the previously determined titration curves with specific histidine residues (16). The H10, H11, H14, and H18 side chains exhibit pK values of 5.8, 5.4, 5.7, and 6.0, respectively, and on average are significantly reduced when compared to histidine protonation in aqueous solution. In an α-helical conformation, these sites are in close proximity, and repulsion between like charges potentially contributes to the decrease in pK and a more unstable helix conformation in the center of the peptide (Fig. 5 A and Fig. 6 A). Of note, the H11 ring system exhibits the highest degree of conformational freedom (Fig. 6 A) and is well positioned to interact with the DPC interface (16). The early deprotonation of the histidines probably correlates with a deeper penetration of these residues into the membrane and ultimately a transmembrane alignment of the peptide.
Circular dichroism and magic-angle spinning solid-state NMR results indicate that LAH4 also adopts predominantly α-helical conformations in the transfection complex with DNA (26). Of interest, LAH4 has the required length of 26 amino acids and at pH 6.1 forms a helix-turn-helix motif similar to other DNA-binding motifs (57). Therefore, it is possible that a related conformation is also present in the transfection complex, although at the present time the detailed structure of the peptide in this environment remains a matter of speculation. However, a thermodynamic analysis of the LAH4-DNA association indicates a high coverage of DNA by the peptide (1 peptide/2 bp at pH 7.5), which is reduced by about twofold when the pH is decreased (26). The peptide-DNA interactions are predominantly electrostatic at neutral pH; however, van der Waals, hydrophobic, and other energies contribute under acidic conditions (26). Such changes in the thermodynamic signature indicate considerable pH-dependent structural rearrangements within this supramolecular complex. It is noteworthy that the transfection process occurs via an endosomal pathway, and that acidification of this organelle results in the release of a large number of peptides and consequently lysis of the endosomal membranes (26) by a mechanism that is probably related to the lysis of bacterial membranes.
When Vogt and Bechinger (23) investigated the antimicrobial activity of LAH4 in a pH-dependent manner, they found that the peptide killed bacteria at neutral and acidic pH, although it was more active at low pH when oriented along the membrane surface. Here, we present the structure of LAH4 as obtained under acidic conditions in membrane environments (Fig. 5 A and Fig. 6 A) where the peptide forms an amphipathic α-helix that is inserted into the membrane interface and is oriented parallel to the membrane surface (Fig. 6 G) in agreement with its high hydrophobic moment (16). Under these conditions, the peptide imposes a high curvature strain on the membrane and severely disrupts the bilayer packing, which can explain the breakdown of electric resistance, pore formation, and cell killing (11,58). When the pH is increased, the peptide transiently adopts a wedge-like conformation. Bent helical structures have also been observed for the highly lytic peptide melittin, for the channel-former alamethicin, and for antimicrobial peptides of the magainin class (e.g., pardaxin and LL-37) (53,59–62). Of note, our work shows that the conformation of the polypeptide in DPC micelles exhibits a high degree of plasticity, being temperature-, pH-, and solvent-dependent, and therefore can adjust to a variety of environmental parameters, with several of these configurations being involved in its interesting biological activities.
Acknowledgments
We thank Roland Graff and the team of Luis Moroder for their help and advice regarding the acquisition of NMR spectra in TFE and during peptide synthesis, respectively. We also thank the Institute of Supramolecular Chemistry, University of Strasbourg, for hosting our laboratory.
This study was supported by grants from the Deutsche Forschungsgemeinschaft (SFB266), the Max Planck Society, the Agence National de la Recherche (TRANSPEP), and Vaincre la Mucoviscidose. J.G. received a postdoctoral grant from Louis Pasteur University (from 2009 on, the University of Strasbourg), and V.H.O.M. received an exchange fellowship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil.
Supporting Material
References
- 1.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Boman H.G. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 2003;254:197–215. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]
- 3.Bechinger B., Lohner K. Detergent-like action of linear cationic membrane-active antibiotic peptides. Biochim. Biophys. Acta. 2006;1758:1529–1539. doi: 10.1016/j.bbamem.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Sugawara M., Resende J.M., Bechinger B. Membrane structure and interactions of human catestatin by multidimensional solution and solid-state NMR spectroscopy. FASEB J. 2010;24:1737–1746. doi: 10.1096/fj.09-142554. [DOI] [PubMed] [Google Scholar]
- 5.Ziegler A. Thermodynamic studies and binding mechanisms of cell-penetrating peptides with lipids and glycosaminoglycans. Adv. Drug Deliv. Rev. 2008;60:580–597. doi: 10.1016/j.addr.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Kichler A., Mason A.J., Bechinger B. Cationic amphipathic histidine-rich peptides for gene delivery. Biochim. Biophys. Acta. 2006;1758:301–307. doi: 10.1016/j.bbamem.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Heitz F., Morris M.C., Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br. J. Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mäe M., Langel U. Cell-penetrating peptides as vectors for peptide, protein and oligonucleotide delivery. Curr. Opin. Pharmacol. 2006;6:509–514. doi: 10.1016/j.coph.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Henriques S.T., Melo M.N., Castanho M.A. Cell-penetrating peptides and antimicrobial peptides: how different are they? Biochem. J. 2006;399:1–7. doi: 10.1042/BJ20061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brogden K.A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 11.Bechinger B. Rationalizing the membrane interactions of cationic amphipathic antimicrobial peptides by their molecular shape. Curr. Opin. Colloid Interface Sci. 2009;14:349–355. [Google Scholar]
- 12.Bechinger B. A dynamic view of peptides and proteins in membranes. Cell. Mol. Life Sci. 2008;65:3028–3039. doi: 10.1007/s00018-008-8125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers. 2002;66:236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 14.Hancock R.E., Rozek A. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol. Lett. 2002;206:143–149. doi: 10.1111/j.1574-6968.2002.tb11000.x. [DOI] [PubMed] [Google Scholar]
- 15.Ludtke S.J., He K., Huang H.W. Membrane pores induced by magainin. Biochemistry. 1996;35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- 16.Bechinger B. Towards membrane protein design: pH-sensitive topology of histidine-containing polypeptides. J. Mol. Biol. 1996;263:768–775. doi: 10.1006/jmbi.1996.0614. [DOI] [PubMed] [Google Scholar]
- 17.Mason A.J., Gasnier C., Bechinger B. Enhanced membrane disruption and antibiotic action against pathogenic bacteria by designed histidine-rich peptides at acidic pH. Antimicrob. Agents Chemother. 2006;50:3305–3311. doi: 10.1128/AAC.00490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bechinger B., Ruysschaert J.M., Goormaghtigh E. Membrane helix orientation from linear dichroism of infrared attenuated total reflection spectra. Biophys. J. 1999;76:552–563. doi: 10.1016/S0006-3495(99)77223-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aisenbrey C., Goormaghtigh E., Bechinger B. Translocation of amino acyl residues from the membrane interface to the hydrophobic core: thermodynamic model and experimental analysis using ATR-FTIR spectroscopy. Mol. Membr. Biol. 2006;23:363–374. doi: 10.1080/09687860600738742. [DOI] [PubMed] [Google Scholar]
- 20.Aisenbrey C., Kinder R., Bechinger B. Interactions involved in the realignment of membrane-associated helices: an investigation using oriented solid-state NMR and attenuated total reflection Fourier transform infrared spectroscopies. J. Biol. Chem. 2006;281:7708–7716. doi: 10.1074/jbc.M513151200. [DOI] [PubMed] [Google Scholar]
- 21.Aisenbrey C., Bechinger B., Gröbner G. Macromolecular crowding at membrane interfaces: adsorption and alignment of membrane peptides. J. Mol. Biol. 2008;375:376–385. doi: 10.1016/j.jmb.2007.10.053. [DOI] [PubMed] [Google Scholar]
- 22.Marquette A., Mason A.J., Bechinger B. Aggregation and membrane permeabilizing properties of designed histidine-containing cationic linear peptide antibiotics. J. Pept. Sci. 2008;14:488–495. doi: 10.1002/psc.966. [DOI] [PubMed] [Google Scholar]
- 23.Vogt T.C.B., Bechinger B. The interactions of histidine-containing amphipathic helical peptide antibiotics with lipid bilayers. The effects of charges and pH. J. Biol. Chem. 1999;274:29115–29121. doi: 10.1074/jbc.274.41.29115. [DOI] [PubMed] [Google Scholar]
- 24.Mason A.J., Moussaoui W., Bechinger B. Structural determinants of antimicrobial and antiplasmodial activity and selectivity in histidine-rich amphipathic cationic peptides. J. Biol. Chem. 2009;284:119–133. doi: 10.1074/jbc.M806201200. [DOI] [PubMed] [Google Scholar]
- 25.Kichler A., Leborgne C., Bechinger B. Histidine-rich amphipathic peptide antibiotics promote efficient delivery of DNA into mammalian cells. Proc. Natl. Acad. Sci. USA. 2003;100:1564–1568. doi: 10.1073/pnas.0337677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prongidi-Fix L., Sugawara M., Bechinger B. Self-promoted cellular uptake of peptide/DNA transfection complexes. Biochemistry. 2007;46:11253–11262. doi: 10.1021/bi700766j. [DOI] [PubMed] [Google Scholar]
- 27.Mason A.J., Leborgne C., Kichler A. Optimising histidine rich peptides for efficient DNA delivery in the presence of serum. J. Control. Release. 2007;118:95–104. doi: 10.1016/j.jconrel.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Langlet-Bertin B., Leborgne C., Kichler A. Design and evaluation of histidine-rich amphipathic peptides for siRNA delivery. Pharm. Res. 2010;27:1426–1436. doi: 10.1007/s11095-010-0138-2. [DOI] [PubMed] [Google Scholar]
- 29.Leontiadou H., Mark A.E., Marrink S.J. Antimicrobial peptides in action. J. Am. Chem. Soc. 2006;128:12156–12161. doi: 10.1021/ja062927q. [DOI] [PubMed] [Google Scholar]
- 30.White S.H., Wimley W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 31.Verly R.M., Moraes C.M., Bechinger B. Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy. Biophys. J. 2009;96:2194–2202. doi: 10.1016/j.bpj.2008.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braunschweiler L., Ernst R.R. Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 1983;53:521–528. [Google Scholar]
- 33.Jeener J., Meier B., Ernst R.R. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 1979;71:4546–4553. [Google Scholar]
- 34.Bothner-By A.A., Stephens R., Jeanloz R. Structure determination of a tetrasaccharide: transient nuclear Overhauser effect in the rotating frame. J. Am. Chem. Soc. 1984;106:811–813. [Google Scholar]
- 35.Piantini U., Sorensen O.W., Ernst R.R. Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc. 1982;104:6800–6801. [Google Scholar]
- 36.Plateau P., Gueron M. Exchangeable proton NMR without base-line distortion using new strong pulse sequences. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 37.Bax A., Davis D.G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–360. [Google Scholar]
- 38.Sklenar V., Piotto M., Saudek V. Gradient-tailored water suppression for 1H–15N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson. A. 1993;102:241–245. [Google Scholar]
- 39.Derome A.E., Williamson M.P. Rapid-pulsing artifacts in double-quantum filtered COSY. J. Magn. Reson. 1990;88:177–185. [Google Scholar]
- 40.Cieslar C., Ross A., Holak T.A. Efficiency in multidimensional NMR by optimized recording of time point phase pairs in evolution periods and their selective linear transformation. J. Magn. Reson. B. 1993;101:97–101. [Google Scholar]
- 41.Wüthrich K. John Wiley & Sons; New York: 1986. NMR of Proteins and Nucleic Acids. [Google Scholar]
- 42.Schwieters C.D., Kuszewski J.J., Clore G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 43.Stein E.G., Rice L.M., Brünger A.T. Torsion-angle molecular dynamics as a new efficient tool for NMR structure calculation. J. Magn. Reson. 1997;124:154–164. doi: 10.1006/jmre.1996.1027. [DOI] [PubMed] [Google Scholar]
- 44.Linge J.P., Williams M.A., Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski R.A., Rullmannn J.A., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 46.Koradi R., Billeter M., Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. 29–32. [DOI] [PubMed] [Google Scholar]
- 47.Wishart D.S., Sykes B.D. Chemical shifts as a tool for structure determination. Methods Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 48.Kuntz I., Kosen P., Craig E. Amide chemical shifts in many helices in peptides and proteins are periodic. J. Am. Chem. Soc. 1991;113:1406–1408. [Google Scholar]
- 49.Wieprecht T., Apostolov O., Seelig J. Membrane binding and pore formation of the antibacterial peptide PGLa: thermodynamic and mechanistic aspects. Biochemistry. 2000;39:442–452. doi: 10.1021/bi992146k. [DOI] [PubMed] [Google Scholar]
- 50.Huang H.W. Action of antimicrobial peptides: two-state model. Biochemistry. 2000;39:8347–8352. doi: 10.1021/bi000946l. [DOI] [PubMed] [Google Scholar]
- 51.Sönnichsen F.D., Van Eyk J.E., Sykes B.D. Effect of trifluoroethanol on protein secondary structure: an NMR and CD study using a synthetic actin peptide. Biochemistry. 1992;31:8790–8798. doi: 10.1021/bi00152a015. [DOI] [PubMed] [Google Scholar]
- 52.Damberg P., Jarvet J., Gräslund A. Micellar systems as solvents in peptide and protein structure determination. Methods Enzymol. 2001;339:271–285. doi: 10.1016/s0076-6879(01)39318-7. [DOI] [PubMed] [Google Scholar]
- 53.Sansom M.S.P. The biophysics of peptide models of ion channels. Prog. Biophys. Mol. Biol. 1991;55:139–235. doi: 10.1016/0079-6107(91)90004-c. [DOI] [PubMed] [Google Scholar]
- 54.Resende J.M., Moraes C.M., Bechinger B. Solution NMR structures of the antimicrobial peptides phylloseptin-1, -2, and -3 and biological activity: the role of charges and hydrogen bonding interactions in stabilizing helix conformations. Peptides. 2008;29:1633–1644. doi: 10.1016/j.peptides.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 55.Liu L.P., Li S.C., Deber C.M. Threshold hydrophobicity dictates helical conformations of peptides in membrane environments. Biopolymers. 1996;39:465–470. doi: 10.1002/(sici)1097-0282(199609)39:3<465::aid-bip17>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 56.Mattila K., Kinder R., Bechinger B. The alignment of a voltage-sensing peptide in dodecylphosphocholine micelles and in oriented lipid bilayers by nuclear magnetic resonance and molecular modeling. Biophys. J. 1999;77:2102–2113. doi: 10.1016/S0006-3495(99)77051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pellegrini-Calace M., Thornton J.M. Detecting DNA-binding helix-turn-helix structural motifs using sequence and structure information. Nucleic Acids Res. 2005;33:2129–2140. doi: 10.1093/nar/gki349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salnikov E.S., Mason A.J., Bechinger B. Membrane order perturbation in the presence of antimicrobial peptides by 2H solid-state NMR spectroscopy. Biochimie. 2009;91:743. doi: 10.1016/j.biochi.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 59.Okada A., Wakamatsu K., Higashijima T. Vesicle-bound conformation of melittin: transferred nuclear Overhauser enhancement analysis in the presence of perdeuterated phosphatidylcholine vesicles. Biochemistry. 1994;33:9438–9446. doi: 10.1021/bi00198a009. [DOI] [PubMed] [Google Scholar]
- 60.Porcelli F., Verardi R., Veglia G. NMR structure of the cathelicidin-derived human antimicrobial peptide LL-37 in dodecylphosphocholine micelles. Biochemistry. 2008;47:5565–5572. doi: 10.1021/bi702036s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhunia A., Domadia P.N., Bhattacharjya S. NMR structure of pardaxin, a pore-forming antimicrobial peptide, in lipopolysaccharide micelles: mechanism of outer membrane permeabilization. J. Biol. Chem. 2010;285:3883–3895. doi: 10.1074/jbc.M109.065672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhunia A., Ramamoorthy A., Bhattacharjya S. Helical hairpin structure of a potent antimicrobial peptide MSI-594 in lipopolysaccharide micelles by NMR spectroscopy. Chemistry. 2009;15:2036–2040. doi: 10.1002/chem.200802635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


