Abstract
Molecular modelling and docking studies along with three-dimensional quantitative structure relationships (3D-QSAR) studies have been used to determine the correct binding mode of glycogen synthase kinase 3β (GSK-3β) inhibitors. The approaches of Comparative Molecular Field Analysis (CoMFA) and Comparative Molecular Similarity Index Analysis (CoMSIA) are used for the 3D-QSAR of 51 substituted 3-benzofuranyl-4-indolyl-maleimides as GSK-3β inhibitors. Two binding modes of the inhibitors to the binding site of GSK-3β are investigated. The binding mode 1 yielded better 3D-QSAR correlations using both CoMFA and CoMSIA methodologies. The three-component CoMFA model from the steric and electrostatic fields for the experimentally determined pIC50 values has the following statistics: R2(cv) = 0.386 and SE(cv) = 0.854 for the cross-validation, and R2 = 0.811 and SE = 0.474 for the fitted correlation. F (3,47) = 67.034, and Probability.of R2 = 0 (3,47) = 0.000. The binding mode suggested by the results of this study is consistent with the preliminary results of X-ray crystal structures of inhibitor-bound GSK-3β. The 3D-QSAR models were used for the estimation of the inhibitory potency of two additional compounds.
Introduction
Originally identified as a modulator of glycogen metabolism about 20 years ago, glycogen synthase kinase 3β (GSK-3β) is now found to be a Ser/Thr protein kinase with key roles in transduction of regulatory role in a variety of pathways. These include the initiation of protein synthesis, cell proliferation, cell differentiation, and apoptosis. This kinase is also essential for embryonic development.1–4 In humans, two genes are present that encode the related GSK-3 isoforms GSK-3α and GSK-3β, which exhibit approximately 98% sequence identity within their catalytic domains.
Many different kinds of GSK-3 inhibitors have been studied by various researchers.4–27 Our attention was directed to the discovery of inhibitors of the GSK-3β to be used possibly in the treatment of a number of CNS disorders including Alzheimer’s disease, Parkinson’s disease, bipolar disorders, and traumatic brain injury. Our work in this area was influenced by the maleimide-bearing natural product staurosporine.19, 24
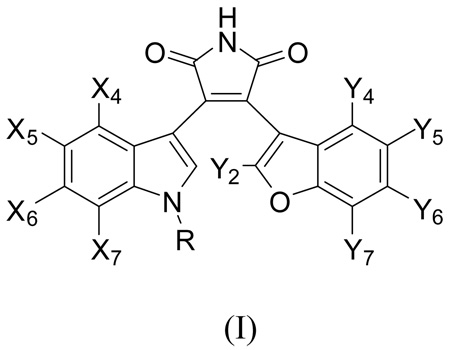
In our previous paper, we reported on the chemical synthesis and the biological activities of a number of substituted maleimides as inhibitors of GSK-3β and additionally examined their selectivity for inhibition of CDK2/cyclinE.28 In this paper, we report on our study of the molecular modelling and docking of the inhibitors into the binding site of GSK-3β, together with 3D-quantitative structure-activity relationships (3D-QSAR) using the comparative molecular field analysis (CoMFA)29–31 and the comparative molecular similarity indices analysis (CoMSIA).32 A specific aim of this study is to identify the correct binding mode of the substituted maleimide compounds included in this study using the computer-aided molecular modelling techniques. Fifty-one 3-benzofuranyl-4-indolyl-maleimide-based GSK-3β inhibitors of structural type I are included in the present work. Two possible binding modes are examined to determine the correct interaction mode of these compounds with the enzyme. Superpositions of the two alignments are obtained by docking the inhibitors to the known X-ray crystal structure of GSK-3β (1R0E), where a similar ligand to our inhibitors is bound.
Results and Discussion
Studies on the Binding Mode of the Inhibitors
In order to study the binding mode of the inhibitors, we chose to utilize 3D-QSAR methodologies. For such 3D-QSAR studies employing both the CoMFA or CoMSIA methodologies, all compounds need to be superimposed under the assumption that they bind in a similar manner to the same binding site. Different methods have been used in the literature for the superposition of the compounds of interest. We decided to dock the inhibitors to the binding site of GSK-3β protein and use the docked conformation of the inhibitors in our CoMFA and CoMSIA studies. In previous publications from this laboratory we assumed that the binding mode of the substituted maleimides, either 3-indolyl-4-indazolylmaleimides or 3-benzofuranyl-4-indolylmaleimides, is similar to that found for staurosporine in its X-ray co-crystal structure with GSK-3β (1Q3D).33
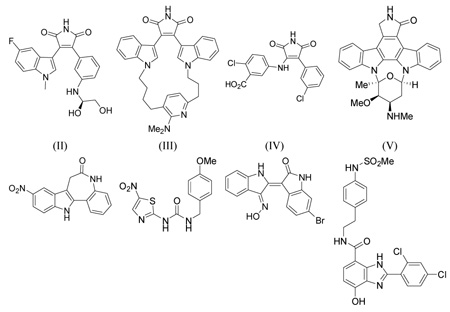
In this study, we reinvestigated the possible binding mode of the 3-(benzofuran-3-yl)-4-(indol-3-yl)maleimides (I) to GSK-3β in an effort to develop a potent and selective GSK-3β inhibitor. In order to find relevant information about the binding mode and conformation of the inhibitors, we first examined the known X-ray crystal structures of GSK-3β currently available in the RCSB PDB Protein Data Bank.34 Table 1 lists the X-ray structures of the GSK-3β complexes that were examined. Four of the eight ligands in Table 1 are similar to our GSK-3β inhibitors.
Table 1.
Known GSK-3β X-ray Structures.
 | ||||||
|---|---|---|---|---|---|---|
| (VI) | (VII) | (VIII) | (IX) | |||
| Group | PDB | Resolution | R-value | Bound Ligand | Ref | |
| A. | 1R0E-like | 1R0E | 2.25 | 0.225 | (II) | 47 |
| 2OW3 | 2.80 | 0.248 | (III) | 48 | ||
| 1GNG | 2.60 | 0.196 | 49 | |||
| 1O9U | 2.40 | 0.233 | 50 | |||
| B. | 1Q4L-like | 1Q4L | 2.77 | 0.212 | (IV) | 51 |
| 1H8F | 2.80 | 0.220 | 52 | |||
| 1I09 | 2.70 | 0.242 | 53 | |||
| 1J1B | 1.80 | 0.216 | 54 | |||
| 1J1C | 2.10 | 0.218 | 54 | |||
| 1Q3D | 2.20 | 0.230 | (V: Staurosporine) | 51 | ||
| 1Q3W | 2.30 | 0.225 | (VI) | 51 | ||
| 1Q5K | 1.94 | 0.222 | (VII) | 55 | ||
| 1UV5 | 2.80 | 0.193 | (VIII) | 6 | ||
| 1PYX | 2.40 | 0.206 | 51 | |||
| 1Q41 | 2.10 | 0.229 | 51 | |||
| 2O5K | 3.20 | 0.240 | (IX) | 23 | ||
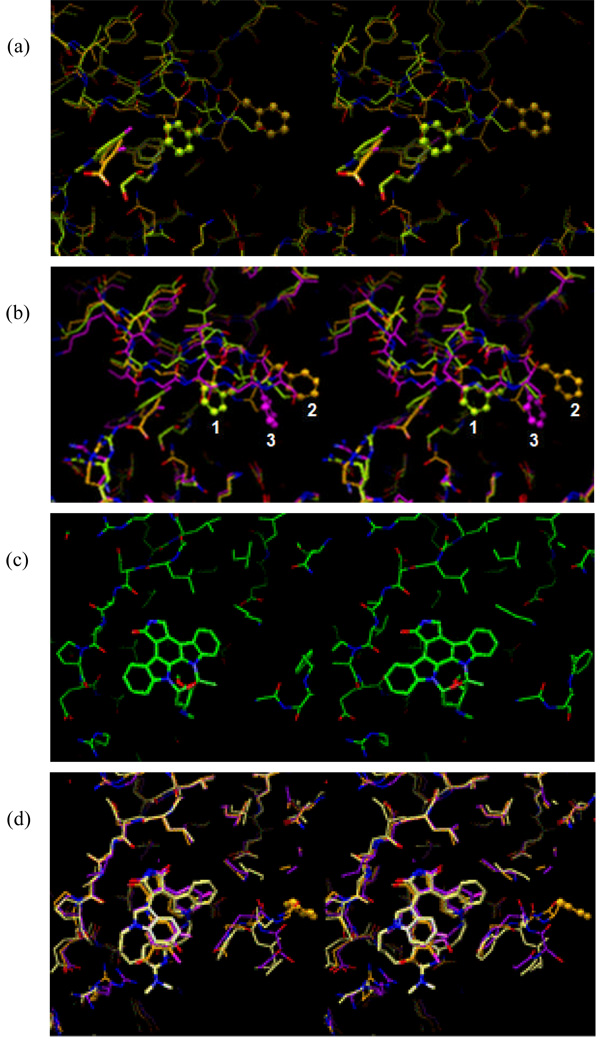
Examination of the X-ray crystal structures of GSK-3β in Table 1 revealed that there are roughly two types of GSK-3β structures with respect to Phe67: one is 1R0E-like (in yellow), and the other is 1Q4L-like (in orange) (Figure 1a). Between these two extreme structures, there are intermediate ones like that represented by the 1Q41 structure (Figure 1b, in pink). The changes in position of the Phe67 residue are due to the differences in the conformation of the Gly-rich loop observed in essentially all Ser/Thr and Tyr protein kinase structures.35–37 Another major change in the conformations observed among these structures is the movement of the Arg141 side chain (see the discussion below). The side chain movements of Arg141 in the GSK-3β structures can be seen at the lower left corner of Figure 1b.
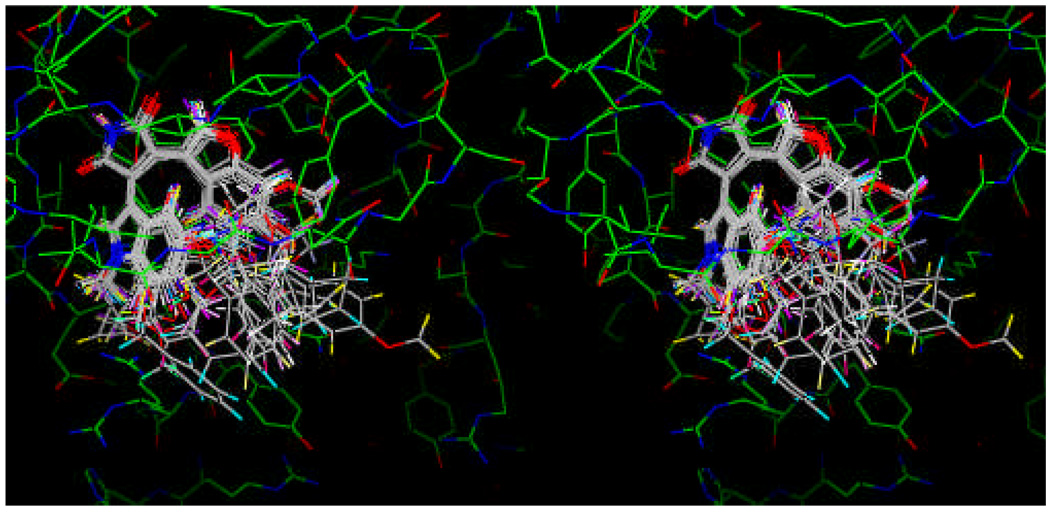
Figure 1.
X-ray crystal structures of ligand-bound GSK-3β structures listed in Table 1. (a) Approximately two groups of GSK-3β structures are shown with respect to the residue Phe67: one is 1R0E-like (yellow), and the other is 1Q4L-like (orange). The two groups of Phe67 positions of GSK-3β structures are illustrated by the two different positions of the phenyl ring shown in the ball-and-stick model in Figure 1a (labeled 1 and 2 in Figure 1b). (b) Between the two extreme structures shown in (a), there are intermediate ones represented by 1Q41 in pink (labeled 3 in Figure 1b). The phenyl ring of this intermediate ones of GSK-3b is illustrated by the phenyl ring (ball-and-stick model in pink) shown between the two different positions of the phenyl rings shown in Figure 1a. Multiple conformations of Arg141 can be seen at the lower left corner. (c) Binding mode of staurosporine (1Q3D). (d) Binding modes of II (1R0E), III (2OW3), and IV (1Q4L) from the superimposed GSK-3β structures. They show similar binding conformation of the three compounds. (e) Binding modes of II and IV showing similar binding conformation of these two compounds.
The binding mode of staurosporine, the compound that the design of our inhibitors was initially influenced by, is shown in Figure 1c. The X-ray crystal structure shows that the binding pose of the staurosporine is guided by the two adjacent intermolecular hydrogen bonds of the pyrrolidin-2-one moiety. The NH group of the pyrrolidin-2-one ring of staurosporine forms a hydrogen bond to the backbone carbonyl oxygen of Asp133, and the carbonyl oxygen of the same ring forms a hydrogen bond to the backbone NH of Val135. Hydrophobic side chains of Leu132 and Lys85 are present around the methylene group (carbon 7 indicated in the staurosporine structure) of staurosporine. In addition, there are two water molecules connecting the carbonyl oxygen of the pyrrolidin-2-one moiety and Glu97 nearby. The two indole rings of staurosporine are fixed by a phenyl group connecting the two rings, and so do their binding positions.
Figure 1d shows the binding modes of II, III, and IV in Table 1. Compound III has two indole rings as does staurosporine, whereas II has only one indole ring, and IV has an aminophenyl and a phenyl ring instead of the two indole rings. The two indole rings in III are partially rigidified through macrocycle formation encompassing the two indole rings. Even though the two indole rings are semi-rigid, the binding conformations of the two indole rings of III are significantly different from that of staurosporine, and similar to those of II and IV.
Figure 1e shows the binding modes of II and IV. The indole ring conformation of II is very similar to that of III as well as that of the aminophenyl ring of IV. The conformation of the phenyl ring of II and IV is similar to one another.
Based on the X-ray structures of II, III, and IV, we proposed that the binding mode of 3-(benzofuran-3-yl)-4-(indol-3-yl)maleimides (I) in Table 6 would be similar to those of II, III, and IV. This is contrary to the binding mode suggested in the earlier publications for 3-indolyl-4-indazolylmaleimides or benzofuran-3-yl-(indol-3-yl)maleimides.19, 24 Since the indole ring of II is on the left side and the substituted phenyl ring of II or IV is on the right side in Figure 1d, it was thought that the indole ring of the maleimides in Table 6 is on the left and the benzofuran ring is on the right in this view. Therefore, all 51 maleimides in Table 6 were docked into the binding site of GSK-3β (1R0E) in this postulated manner. We chose the GSK-3β structure of 1R0E for our docking study, because its bound ligand is the most similar to our inhibitors. The binding conformation of the benzofuran ring could be similar to that of the indole ring of III if the substituent on the benzofuran ring is not large. However, if the substituent on the benzofuran ring is of sufficient bulk, then there is not enough space for the conformation observed for III. In such a case, the benzofuran ring would be rotated by 180°. For the purpose of the CoMFA and CoMSIA studies, the conformation of the rotated benzofuran ring was selected, because some of our inhibitors have a larger substituent than can be accommodated by the pocket present in the unrotated-binding conformation. When a large substituent is present at the X5 position of the indole ring of the benzofuran-3-yl-(indol-3-yl)maleimide, the substituent would clash sterically with Phe67, which would result in the movement of the glycine-rich loop. The consequence of this movement would result in the conformational change of Phe67 from the position of 1R0E-like structure to the position of 1Q4L-like structure (see Figure 1a). For consistency, all the compounds listed in Table 6 were docked in this conformation to the GSK-3β binding site. (See further discussion below.) The starting conformation of the initial conformation of each compound was manually superimposed over the pyrrolidin-2-one ring of the ligand-bound GSK-3β structure (1R0E) because of the reason discussed above.
Table 6.
Inhibitory potency of 51 GSK-3β inhibitors included in this study.
 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| pIC50 | |||||||||
| IC50 | CoMFA (BM1) |
CoMSIA (BM1) |
|||||||
| X | Y | R | (nM) | Obs | Cala | Deva | Calb | Devb | |
| 1 | 7-CH2OMe | H | CH3 | 0.12 | 9.921 | 9.076 | 0.845 | 8.663 | 1.258 |
| 2 | 5-F | 6-CH2OH | CH3 | 0.35 | 9.456 | 9.114 | 0.342 | 8.960 | 0.496 |
| 3 | 5-Br | 6-CH2OH | CH3 | 0.51 | 9.292 | 9.196 | 0.096 | 8.958 | 0.334 |
| 4 | 5-F, 6-Cl | 6-CH2OH | CH3 | 0.95 | 9.022 | 8.409 | 0.613 | 8.588 | 0.434 |
| 5 | 5-Br | H | (CH2)3OH | 1.60 | 8.796 | 8.305 | 0.491 | 8.221 | 0.575 |
| 6 | 5-F | 6-OH | CH3 | 3.50 | 8.456 | 8.598 | −0.142 | 7.740 | 0.716 |
| 7 | 7-CH2OH | 6-CH2OH | CH3 | 5.10 | 8.292 | 9.305 | −1.013 | 9.475 | −1.183 |
| 8 | 7-CH2OH | H | CH3 | 5.40 | 8.268 | 8.371 | −0.103 | 8.442 | −0.174 |
| 9 | 5-Br | H | CH3 | 7.00 | 8.155 | 7.578 | 0.577 | 7.602 | 0.553 |
| 10 | 5-Br | 7-OCH3 | CH3 | 7.50 | 8.125 | 7.182 | 0.943 | 7.086 | 1.039 |
| 11 | 5-CN | 6-CH2OH | CH3 | 13.20 | 7.879 | 8.495 | −0.616 | 8.188 | −0.309 |
| 12 | 6-OH | 5-F | CH3 | 14.00 | 7.854 | 6.916 | 0.938 | 6.739 | 1.115 |
| 13 | 6-OH | H | CH3 | 15.00 | 7.824 | 7.601 | 0.223 | 7.581 | 0.243 |
| 14 | 5-F | CH3 | 22.60 | 7.646 | 6.970 | 0.676 | 6.733 | 0.913 | |
| 15 | 5-F | 6-CH2OCH3 | CH3 | 23.80 | 7.623 | 7.848 | −0.225 | 8.190 | −0.567 |
| 16 | 5-Br | CH3 | 25.30 | 7.597 | 7.913 | −0.316 | 7.647 | −0.050 | |
| 17 | 5-F | H | CH3 | 26.00 | 7.585 | 7.209 | 0.376 | 7.527 | 0.058 |
| 18 | 5-I | H | CH3 | 34.50 | 7.462 | 7.410 | 0.052 | 7.658 | −0.196 |
| 19 | H | H | CH3 | 35.00 | 7.456 | 6.911 | 0.545 | 7.038 | 0.418 |
| 20 | 5-Cl | 5-F | CH3 | 42.00 | 7.377 | 7.099 | 0.278 | 7.069 | 0.308 |
| 21 | 5-Br | CH3 | 48.30 | 7.316 | 7.350 | −0.034 | 7.469 | −0.153 | |
| 22 | 7-OH | H | CH3 | 55.00 | 7.260 | 7.326 | −0.066 | 7.764 | −0.504 |
| 23 | 5-,7-di-Br | 7-OCH3 | CH3 | 88.70 | 7.052 | 7.226 | −0.174 | 7.205 | −0.153 |
| 24 | 5-F, 6-Cl | 7-OCH3 | CH3 | 114.00 | 6.943 | 6.424 | 0.519 | 6.706 | 0.237 |
| 25 | 5-OCH3 | H | CH3 | 125.00 | 6.903 | 7.148 | −0.245 | 6.862 | 0.041 |
| 26 | 5-CN | 5,6-di-F | 131.00 | 6.883 | 7.196 | −0.313 | 6.825 | 0.058 | |
| 27 | 6-OBn | 5-F | CH3 | 160.00 | 6.796 | 6.880 | −0.084 | 6.026 | 0.770 |
| 28 | H | 7-OCH3 | CH3 | 180.00 | 6.745 | 6.381 | 0.364 | 6.696 | 0.049 |
| 29 | 5-I | 5-F | CH3 | 180.00 | 6.745 | 7.505 | −0.760 | 7.290 | −0.545 |
| 30 | 5-F, 6-Cl | H | CH3 | 184.00 | 6.735 | 6.661 | 0.074 | 7.244 | −0.509 |
| 31 | 5-OBn | H | (CH2)3OH | 220.00 | 6.658 | 7.175 | −0.517 | 7.113 | −0.455 |
| 32 | 7-OBn | H | CH3 | 220.00 | 6.658 | 6.918 | −0.260 | 7.071 | −0.413 |
| 33 | 5-OCH3, 6-I | H | CH3 | 223.00 | 6.652 | 6.703 | −0.051 | 6.948 | −0.296 |
| 34 | 5-cyclopropyl | H | CH3 | 235.00 | 6.629 | 6.495 | 0.134 | 6.719 | −0.090 |
| 35 | 5-F, 6-I | 7-OCH3 | CH3 | 247.00 | 6.607 | 6.533 | 0.074 | 6.736 | −0.129 |
| 36 | benzo[g] | 5-,6-di-F | CH3 | 314.00 | 6.503 | 6.134 | 0.369 | 6.354 | 0.149 |
| 37 | 5-Br | 6-O-(p-CH3O)-Bn | CH3 | 335.00 | 6.475 | 6.160 | 0.315 | 5.952 | 0.523 |
| 38 | 5-F | H | H | 360.00 | 6.444 | 6.570 | −0.126 | 6.873 | −0.429 |
| 39 | 5-OCH3, 6-Cl | H | CH3 | 440.00 | 6.357 | 6.536 | −0.179 | 6.513 | −0.156 |
| 40 | H | 5-Br | CH3 | 550.00 | 6.260 | 6.811 | −0.551 | 7.020 | −0.760 |
| 41 | H | 5-F | H | 670.00 | 6.174 | 7.120 | −0.946 | 7.187 | −1.013 |
| 42 | 5-OH | H | CH3 | 690.00 | 6.161 | 7.112 | −0.951 | 7.085 | −0.924 |
| 43 | 5,6-Methylenedioxy | 5-F | CH3 | 708.00 | 6.150 | 6.359 | −0.209 | 6.228 | −0.078 |
| 44 | 6-CF3 | 7-OCH3 | CH3 | 831.00 | 6.080 | 5.933 | 0.147 | 6.486 | −0.406 |
| 45 | 5-F, 6-Cl | 6-OCH3 | CH3 | 866.00 | 6.062 | 6.304 | −0.242 | 6.384 | −0.322 |
| 46 | 6-OBn | H | CH3 | 900.00 | 6.046 | 6.195 | −0.149 | 5.968 | 0.078 |
| 47 | 5-F, 6-Cl- | CH3 | 1040.00 | 5.983 | 6.011 | −0.028 | 6.034 | −0.051 | |
| 48 | 5-Morpholine-4-yl | H | CH3 | 1304.00 | 5.885 | 5.670 | 0.215 | 5.771 | 0.114 |
| 49 | 5-OBn | H | H | 1650.00 | 5.783 | 6.074 | −0.291 | 5.878 | −0.095 |
| 50 | 5-F, 6-Cl- | CH3 | 4092.00 | 5.388 | 5.800 | −0.412 | 5.366 | 0.022 | |
| 51 | 5-F, 6-p-Cl-Ph | 7-OCH3 | CH3 | 7000.00 | 5.155 | 5.359 | −0.204 | 5.694 | −0.539 |
Calculated using the three-component CoMFA model (eq 1) derived from 51 compounds using the binding mode 1.
Calculated using the three-component CoMSIA model (eq 3) derived from 51 compounds using binding mode 1.
Even though the binding mode of the compounds in Table 6 is likely to be the one (binding mode 1) described above, we decided to investigate other possible binding modes, especially in light of the fact that a different binding conformation was previously proposed.19,24,56 First, we examined the relative conformational energy of different possible binding conformations of an unsubstituted 3-(benzofuran-3-yl)-4-(indol-3-yl)maleimide to the GSK-3β binding pocket. Figure 2 shows eight different binding modes with four different conformations that are possible in the GSK-3β binding site. In 2a and 2b in Figure 2, the positions of the indole and the benzofuran rings are switched. With respect to the indole and the benzofuran ring positions, the conformations 2a and 2b in Figure 2 are the same when only the ligand is considered, but they would be different in the binding site. The conformational energies of these conformations are summarized in Table 2.
Figure 2.
Four representative conformations (1 – 4) of benzofuran-3-yl-(indol-3-yl)maleimides in binding mode 1 (a) and 2 (b).
Table 2.
Relative conformational energy of four representative conformations of 3-(benzofuran-3-yl)-4-(indol-3-yl)maleimides shown in Figure 2.
| Binding Mode 1 |
Binding Mode 2 |
ΔEnergy (kcal/mole) |
|---|---|---|
| 1a | 1b | 1.6 |
| 2a | 2b | 0.0 |
| 3a | 3b | 1.4 |
| 4a | 4b | 2.1 |
The conformational energy differences among these four conformers are very small, thus suggesting that any of these conformers may represent their actual binding conformation to the GSK-3β. Nonetheless, it is interesting to note that conformation 2a (and 2b), which is the one believed to represent the likely binding mode of the compounds in Table 6, has the lowest conformational energy. Among the eight possible binding modes shown in Figure 2, binding modes 2a and 2b are the two most likely binding modes based upon an analysis of the known ligand-bound GSK-3β X-ray crystal structures as discussed above. Therefore, we chose to investigate 3D-QSAR based on these two binding modes in order to determine the binding mode of these compounds.
Comparative Molecular Field Analysis
The 51 compounds included in this study are listed in Table 6 along with the IC50 values toward GSK-3β. The IC50 values were determined for their potency to inhibit GSK-3β. Commercially available human GSK-3β, was assayed for its ability to phosphorylate the primed peptide substrate (RRRPASVPPSPSLSRHSS(P)HQRR; 10 µM) in the presence of 0–10 µM of the maleimides.57 The inhibitory potency expressed as pIC50 values is the negative logarithm of IC50 value. Therefore, the larger the pIC50 value is, the more potent the compound is as an inhibitor of the kinase. The two binding modes (2a and 2b in Figure 2) of these compounds were obtained by docking each molecule into the binding site of GSK-3β starting from two different initial binding conformations as described above.
I. CoMFA for Binding Model 1
Figure 3 shows all the compounds superimposed of the docked conformation in binding mode 1. Binding mode 1 corresponds to the conformation 2a shown in Figure 2. The best CoMFA model of the 51 substituted maleimides obtained is a three-component model from the steric and electrostatic fields with the following statistics (see Table 3): R2(cv) = 0.386 and SE(cv) = 0.854 for the cross-validation, and R2 = 0.811 and SE = 0.475 for the fitted. F (3,47) = 67.034, and Prob. of R2 = 0 (3,47) = 0.000. The steric component of these maleimide analogs on the inhibitory potency described by this model is 48%, whereas the electrostatic portion is 52%. The first component explains 51% of the variance in the pIC50, and the second and the third components account for additional 22% and 8% of the variance, respectively. An essentially identical CoMFA model was obtained when the steric and the electrostatic fields were considered separately.
Figure 3.
Superposition of 51 substituted maleimides as GSK-3β inhibitors obtained from docking into the binding site of GSK-3β structure (1R0E) in binding mode 1.
Table 3.
CoMFA Models for 51 Substituted Maleimides from the Binding Modes 1 and 2.
| Cross-validation | Fitted | Contribution | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Eq | Fields | Na | Lb | SE(cv) | R2(cv) | SE | R2 | Steric | Electrostatic |
| Binding Mode 1 | |||||||||
| 1 | Steric+Electrostatic | 51 | 3 | 0.854 | 0.386 | 0.475 | 0.811 | 48% | 52% |
| 51 | 2 | 0.857 | 0.370 | 0.565 | 0.726 | 52% | 48% | ||
| 51 | 1 | 0.901 | 0.288 | 0.745 | 0.513 | 47% | 53% | ||
| Binding Mode 2 | |||||||||
| 2 | Steric+Electrostatic | 51 | 3 | 0.915 | 0.296 | 0.507 | 0.784 | 49% | 51% |
| 51 | 2 | 0.956 | 0.216 | 0.622 | 0.668 | 50% | 50% | ||
| 51 | 1 | 1.026 | 0.078 | 0.746 | 0.512 | 50% | 50% | ||
Number of compounds used in the model.
Number of components in the model.
In our previous unpublished study, the classical QSAR result shown below was obtained.28 It is interesting that the statistical quality (R2 and SE) of the classical QSAR and the 3D-QSAR method using the CoMFA methodology is similar. One compound (Compound 1 in Table 6) was treated as an outlier in the classical QSAR, but this compound was included in the CoMFA study.
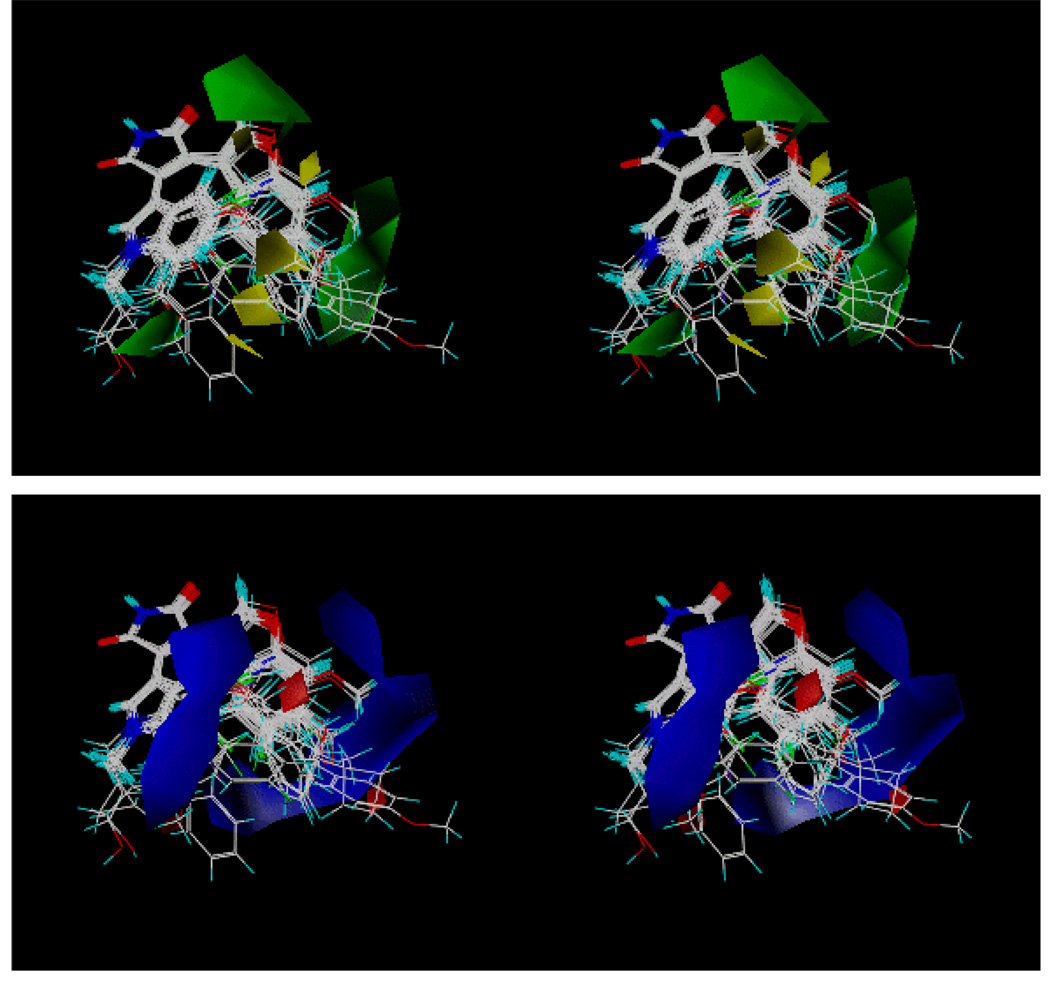
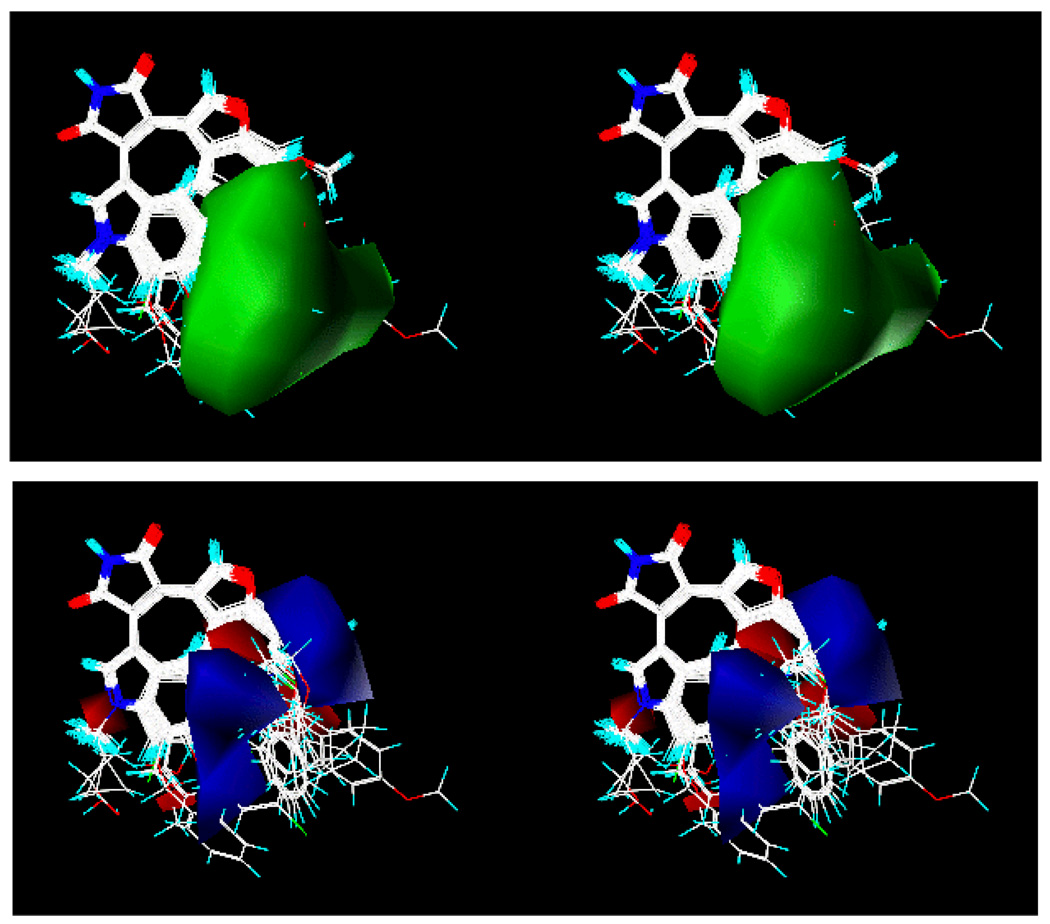
Figure 4 is the coefficient contour map of the three-component model derived from all 51 compounds. In this contour map, the sterically favored regions are shown in green, and the sterically disfavored regions are shown in yellow. The positive electrostatic contours are shown in blue, and the negative electrostatic contours are shown in red. Table 6 shows the observed and the calculated pIC50 values from this three-component CoMFA model along with their observed values. It is interesting to note that there is a sterically favored region in the steric contour map (colored in green in Figure 4a) near the Y2 position of the benzofuran-3-yl-(indol-3-yl)maleimide. The hydrophobic residue Leu132 and the four methylene groups of Lys85 are located near the X2 group and the adjacent carbonyl group of the pyrrolidine-2-one ring. This indicates that a hydrophobic group at this position may improve the inhibitory potency of the compound. As is the case for the binding orientation of staurosporine observed in Figure 1c (1Q3D), the binding orientation of the GSK-3β inhibitors in Table 6 is fixed by the two hydrogen bonds involving the pyrrolidin-2-one rings of the inhibitors. One of the carbonyl oxygens (left one in Figure 3) of the pyrrolidin-2-one ring forms a hydrogen bond with the backbone NH of Val135, and the NH group of the pyrrolidin-2-one ring interacts with the backbone carbonyl group of Asp133 residue. Both residues are in the hinge region of GSK-3β.
Figure 4.
(a) Steric contour of the three-component CoMFA model from the binding mode 1. The regions in green represent sterically favored, whereas the regions in yellow represent disfavored. (b) Electrostatic contour of the three-component CoMFA model from the binding mode 1. The regions in blue represent electrostatically favored, whereas the regions in red represent disfavored.
Four compounds (1 – 4) in Table 6 have IC50 values in sub-nanomolar inhibitory potency. Compounds 2, 3, and 4 have a 6-CH2OH at the Y6 position, and Compound 1 has a 7-CH2OMe at the X7 position. There are other compounds with 6-CH2OH at the Y6 position among the compounds in Table 6. These compounds are in general potent inhibitors. Two compounds (7 and 8) have a similar substituent 7-CH2OH at X7 position to the 7-CH2OMe of Compound 1.
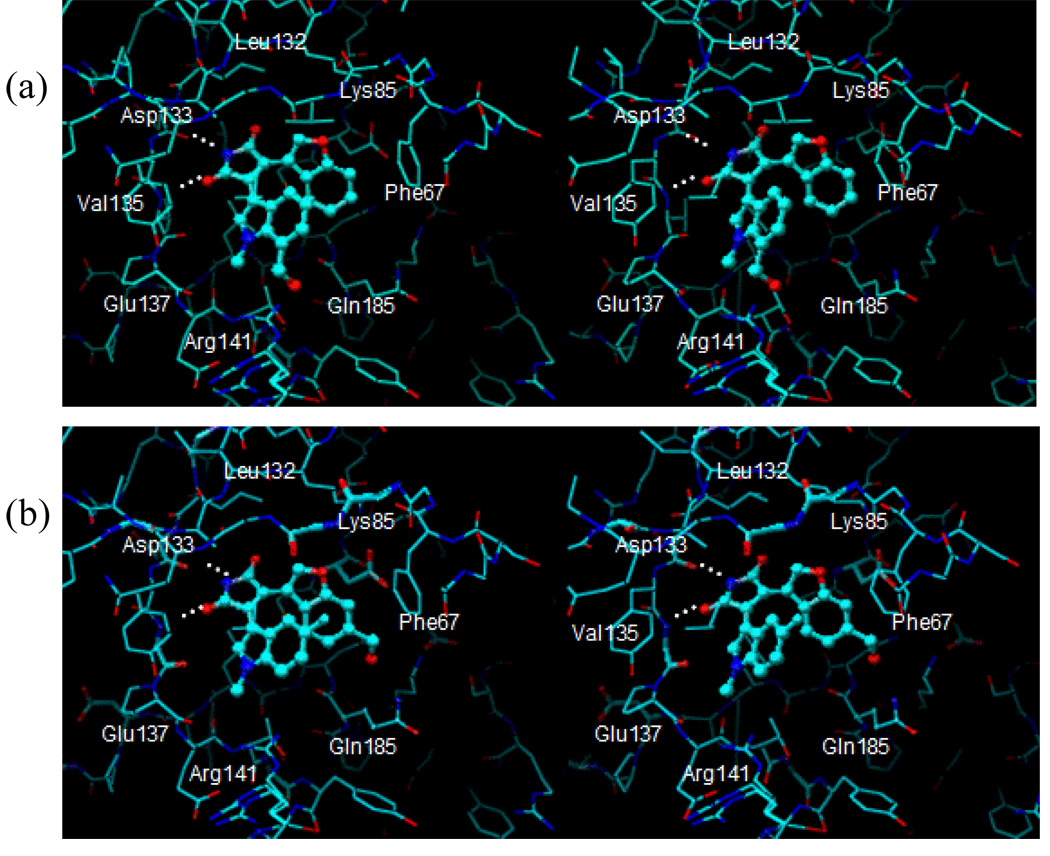
Figure 5 shows the binding site amino acid residues around the 7-CH2OH group at X7 position and the 6-CH2OH at Y6 position of Compound 2 or 8 as a representative case, respectively. The 7-CH2OH group of Compound 8 is in the hydrogen bonding distance with the side chain of Arg141, whereas the 6-CH2OH group of Compound 2 is in the hydrogen bonding distance with the side chain of Gln185.
Figure 5.
Docked binding mode of Compounds 2 (a) or Compound 8 (b) in the binding site of GSK-3β structure in binding mode 1.
Whereas the 7-CH2OH group can act as a hydrogen-bond donor as well as a hydrogen-bond acceptor, the 7-CH2OMe group can only act as a hydrogen-bond acceptor. Because the 7-CH2OMe or 7-CH2OH group would interact with Arg141, and should act as a hydrogen-bond acceptor, the 7-CH2OMe group would be preferred to 7-CH2OH at this position if the hydrogen bonding interactions are the major factor. In addition, if the 7-CH2OH or 7-CH2OMe group interacts with the hydrophobic methylene groups of Arg141, the 7-CH2OMe group would be preferred to 7-CH2OH. Comparison of componds 1 and 8 shows this is indeed the case.
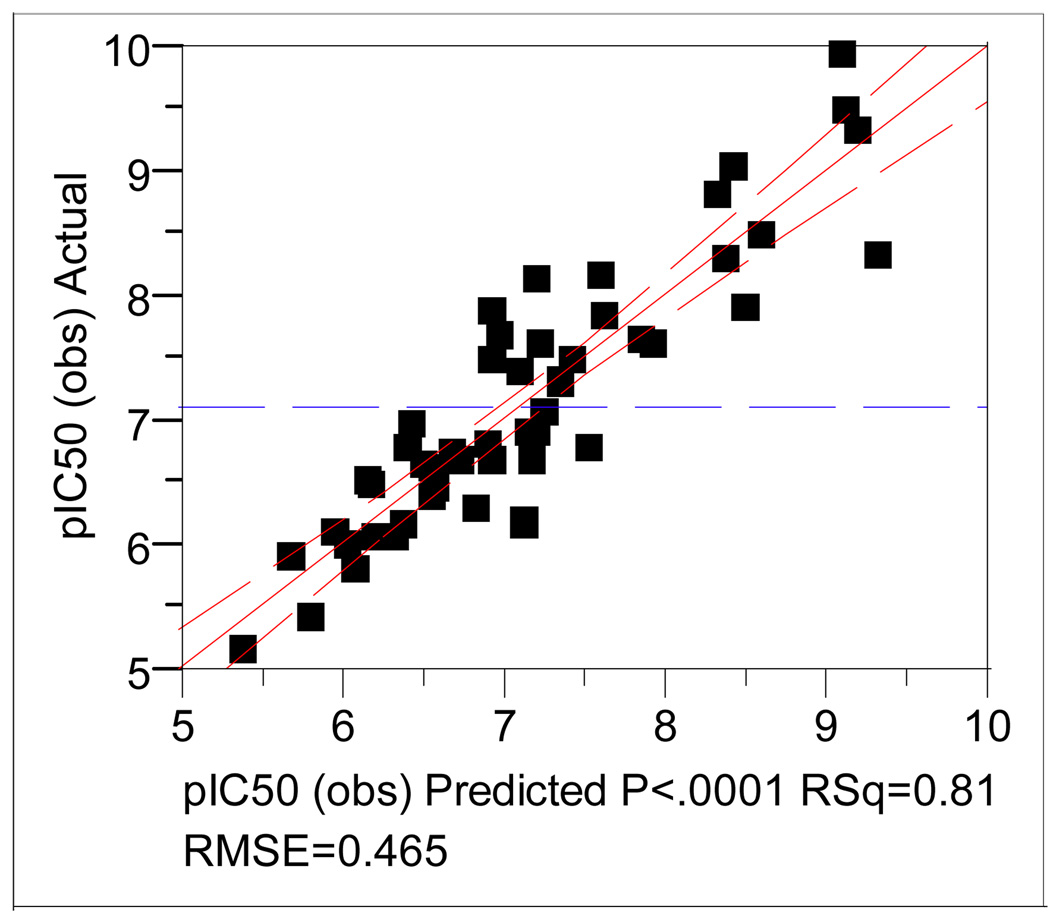
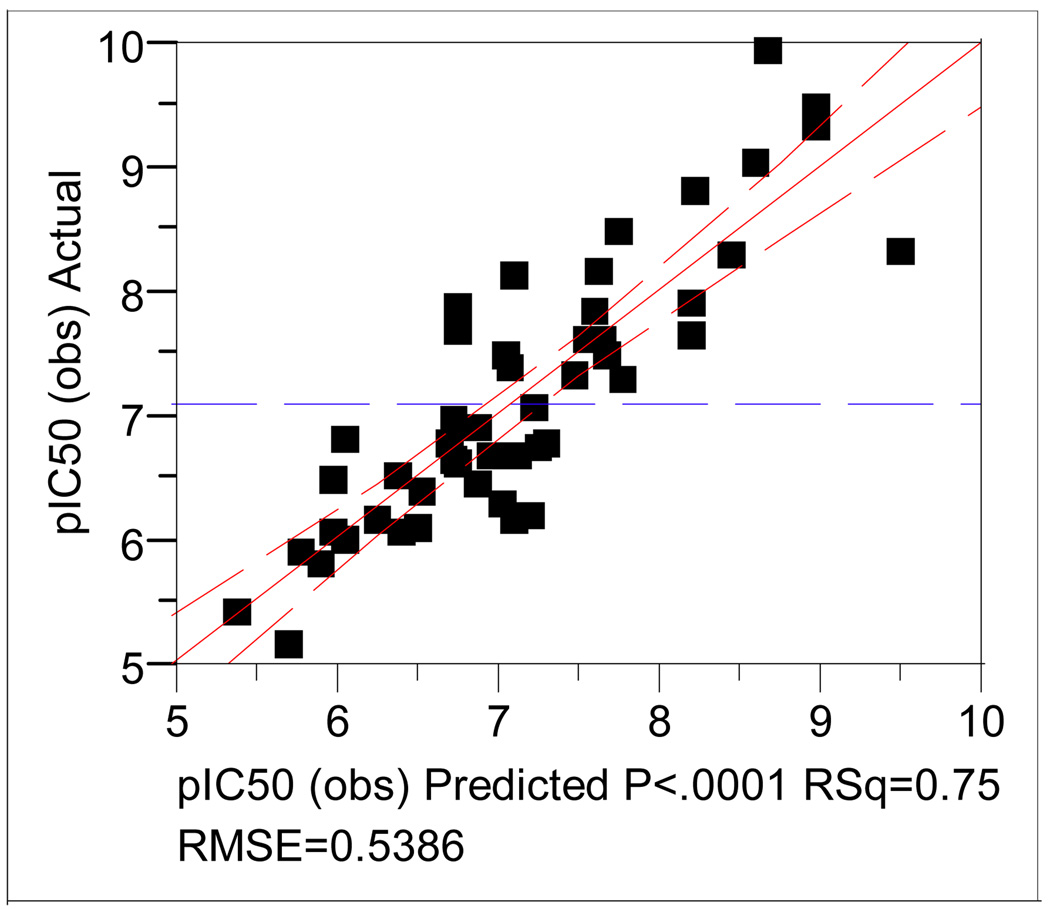
Figure 6 is a plot of the observed and the calculated pIC50 values from the three-component CoMFA model (eq. 1 in Table 3) from the steric and electrostatic fields.
Figure 6.
A plot between the observed and the calculated pIC50 values from the three-component CoMFA model of the ligand-binding mode 1.
II. CoMFA for Binding Model 2
Figure 7 shows all the compounds superimposed in the docked conformation of binding mode 2. Binding mode 2 corresponds to the conformation 2b shown in Figure 2. The best CoMFA model for the 51 substituted maleimides obtained is a three-component model from the steric and electrostatic fields with the following statistics (see Table 3): R2(cv) = 0.296 and SE(cv) = 0.915 for the cross-validation, and R2 = 0.784 and SE = 0.507 for the fitted. F (3,47) = 56.784, and Prob. of R2 = 0 (3,47) = 0.000. The steric contribution of these maleimide analogs toward their inhibitory potency as described by this model is 49%, whereas the electrostatic portion is 51%. The first component explains 51% of the variance in the pIC50, and the second and the third components account for additional 16% and 12% of the variance, respectively. An essentially identical CoMFA model was obtained when the steric and the electrostatic fields were considered separately.
Figure 7.
Superposition of 51 substituted maleimides as GSK-3β inhibitors obtained from docking into the binding site of GSK-3β (1P0E) structure in binding mode 2.
Comparison of Two Binding Modes with CoMFA results
It is interesting to examine the CoMFA results from the two different binding modes of the compounds listed in Table 6: whether the CoMFA results can be used to distinguish the two different binding modes and select the correct binding mode from the wrong one. In general, the two binding modes of the inhibitors alone are not significantly different; the entire molecules are rotated about 180° in the binding pocket as can be seen in Figure 3 and 7, especially for the study of CoMFA. The differences between the two superpositions are due to the different X- and Y-substituents and the environment of the GSK-3β binding site.
The CoMFA results summarized in Table 3 show that the superposition from the binding mode 1 (eq 1) accounts for the variation in the pIC50 values of the compounds studied more than the superposition from the binding mode 2 (eq 2): while eq 1 explains 81% of the variance, eq 2 explains 78%. Even though the difference between the two results (percent of the variance explained by the two models) is not large, the statistics (SE and R2 for both the cross-validation and the fitted) indicate that the binding mode 1 is better than the binding mode 2. The relatively similar results are due to the similar superposition of the two binding modes. It is interesting, in this case, that the correct and the incorrect binding modes (or superpositions) yielded similar CoMFA results, even though the correct binding mode gave the better results. Usually in CoMFA, the model having R2(cv) > 0.3 is considered significant. Therefore, the CoMFA model for binding mode 2 is at the borderline of statistical significance. Therefore, the results overall indicate that the binding mode 1 would be favored over binding mode 2 for the benzofuran-3-yl-(indol-3-yl)maleimides examined in this study.
Further Validation of CoMFA Model from the Binding Mode 1
In order to further validate the CoMFA model derived from the binding mode 1, the inhibitory potency values pIC50 were scrambled and used as such to develop a CoMFA model. The resulting CoMFA model was then compared with the CoMFA model developed using the correct pIC50 values. Such procedures have been used to prove the robustness of the derived CoMFA model. Five different scrambled pIC50 data sets were used in this validation procedure (See Table 4 for further details). The statistics for the CoMFA models developed using the scrambled pIC50 values for the 51 substituted maleimides are summarized in Table 4. The results show that none of the scrambled pIC50 data sets yielded a statistically significant CoMFA model. The results provide additional support for the CoMFA model derived from the binding mode 1 of these compounds.
Table 4.
Validation of CoMFA and CoMSIA Models for 51 Substituted Maleimides from the Binding Modes 1 using Scrambling pIC50 values.
| Scramble | Cross-validation | Fitted | |||||
|---|---|---|---|---|---|---|---|
| Runa | Fields | N | L | SE(cv) | R2(cv) | SE | R2 |
| CoMFA (Eq 1) | |||||||
| 1 | Steric+Electrostatic | 51 | 1 | 1.102 | −0.065 | 0.902 | 0.287 |
| 2 | Steric+Electrostatic | 51 | 1 | 1.057 | 0.021 | 0.836 | 0.387 |
| 3 | Steric+Electrostatic | 51 | 1 | 1.042 | 0.048 | 0.807 | 0.430 |
| 4 | Steric+Electrostatic | 51 | 1 | 1.098 | −0.056 | 0.828 | 0.399 |
| 5 | Steric+Electrostatic | 51 | 1 | 1.116 | −0.090 | 0.890 | 0.306 |
| CoMSIA (Eq 3) | |||||||
| 6 | Steric+Electrostatic | 51 | 1 | 1.194 | −0.249 | 0.928 | 0.249 |
| 7 | Steric+Electrostatic | 51 | 1 | 1.077 | −0.016 | 0.918 | 0.262 |
| 8 | Steric+Electrostatic | 51 | 1 | 1.047 | 0.040 | 0.877 | 0.326 |
| 9 | Steric+Electrostatic | 51 | 1 | 1.093 | −0.047 | 0.913 | 0.270 |
| 10 | Steric+Electrostatic | 51 | 1 | 1.130 | −0.119 | 0.923 | 0.253 |
The scrambling of the inhibitory potency value (pIC50) values used in this validation were generated as follows: The original pIC50 values in the data table were initially sorted by the compounds id (not shown). For scramble run 1, the pIC50 values of the entire set of compounds were resorted in ascending order and then the resulting pIC50 values in the newly sorted order were assigned to the compounds in the initial order as the scrambled pIC50 values. For scramble run 2, the compound id of the entire set of compounds were resorted in ascending order and then the resulting pIC50 values in the newly sorted order were assigned to the compounds sorted in the descending order as the scrambled pIC50 values. For scramble run 3, the pIC50 values of the entire set of compounds were resorted in descending order and then the resulting pIC50 values in the newly sorted order were assigned to the compounds in the initial order as the scrambled pIC50 values. For scramble run 4, the pIC50 values of the entire set of compounds were resorted in ascending order and then the resulting pIC50 values in the newly sorted order were assigned to the compounds in the initial order as the scrambled pIC50 values. For scramble run 5, the pIC50 values of the entire set compounds were resorted in ascending order and then the resulting pIC50 values in the newly sorted order were assigned to the compounds sorted in the descending order as the scrambled pIC50 values
Comparative Molecular Similarity Index Analysis
The two binding modes of the compounds (Figure 2 and 5) in Table 6 used in the CoMFA studies described above were also used to study 3D-QSAR using the CoMSIA approach. The CoMSIA results obtained from the binding mode 1 and 2 are summarized in Table 5.
Table 5.
CoMSIA Models for 51 Substituted Maleimides from the Binding Modes 1 and 2.
| Cross- validation |
Fitted | Contribution | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Eq | Fields | Na | Lb | SE(cv) | R2(cv) | SE | R2 | Ster | Elec | Hydrophob |
| Binding Mode 1 | ||||||||||
| 3 | Steric+Electrostatic | 51 | 3 | 0.835 | 0.414 | 0.550 | 0.746 | 25% | 75% | |
| 51 | 2 | 0.853 | 0.375 | 0.621 | 0.668 | 25% | 75% | |||
| 51 | 1 | 0.912 | 0.271 | 0.735 | 0.527 | 26% | 74% | |||
| 4 | Ster+Elec+Hydrophob | 51 | 3 | 0.839 | 0.408 | 0.557 | 0.739 | 18% | 52% | 30% |
| 51 | 2 | 0.864 | 0.360 | 0.622 | 0.668 | 17% | 51% | 32% | ||
| 51 | 1 | 0.913 | 0.270 | 0.755 | 0.500 | 16% | 46% | 38% | ||
| Binding Mode 2 | ||||||||||
| 5 | Steric+Electrostatic | 51 | 2 | 0.915 | 0.281 | 0.638 | 0.650 | 22% | 78% | |
| 51 | 1 | 0.971 | 0.174 | 0.737 | 0.525 | 22% | 78% | |||
| 6 | Ster+Elec+Hydrophob | 51 | 2 | 0.921 | 0.272 | 0.603 | 0.688 | 14% | 54% | 31% |
| 51 | 1 | 0.951 | 0.208 | 0.758 | 0.496 | 13% | 47% | 40% | ||
Number of compounds used in the model.
Number of components in the model.
I. CoMSIA for Binding Model 1
The best CoMSIA model obtained from 51 substituted maleimides in Table 6 is a three-component model from the steric and electrostatic fields with the following statistics (see Table 5): R2(cv) = 0.414 and SE(cv) = 0.835 for the cross-validation, and R2 = 0.746 and SE = 0.550 for the fitted. F (3,47) = 45.952, and Prob. of R2 = 0 (3,47) = 0.000. The steric portion of the influences of maleimide analogs for the inhibitory potency described by this model is 25%, whereas the electrostatic portion is 75%. The first component explains 53% of the variance in the pIC50, and the second and the third component account for additional 14% and 8% of the variance. An essentially identical CoMFA model was obtained when the steric and the electrostatic fields were considered separately. Addition of hydrophobic and/or hydrogen donor or acceptor components did not improve the correlation already obtained (However, see further discussion below). It is interesting to note that the calculated pIC50 value of Compound 1 has the largest deviation (1.258) from the observed pIC50 value. Compound 1 was also found to be an outlier in the classical QSAR discussed above. In this aspect, the CoMSIA results are similar to the classical QSAR.
As the validation process for the CoMFA model derived from the binding mode 1, the inhibitory potency values pIC50 were scrambled and used as such to develop a CoMSIA model. The same five different scrambled pIC50 data sets used for the validation of the CoMFA model were used in this validation procedure. The results are also summarized in Table 4. The results show that none of the scrambled pIC50 data sets yielded a statistically significant CoMSIA model.
Figure 8 is the coefficient contour map of the three-component CoMSIA model derived from 51 compounds. In this contour map, the sterically favored regions are shown in green. The positive electrostatic contours are shown in blue. The model indicated that there are no sterically disfavored regions and no electrostatically negative regions at this contour level. Table 6 shows the observed and the calculated pIC50 values from this three-component CoMSIA model along their observed values. Figure 9 is a plot of the observed and the calculated pIC50 values from this model.
Figure 8.
(a) Steric contour map (70% level) of the three-component CoMSIA model. (b) Electrostatic contour map (30% level) of the corresponding three-component CoMSIA model in the ligand-binding mode 1.
Figure 9.
A plot between the observed and the calculated pIC50 values from the three-component CoMSIA model from the steric and electrostatic fields of the ligand-binding mode 1.
II. CoMSIA for Binding Model 2
The best CoMSIA model obtained from 51 substituted maleimides in Table 6 is a two-component model from the steric and the electrostatic fields with the following statistics (see Table 5): R2(cv) = 0.281 and SE(cv) = 0.915 for the cross-validation, and R2 = 0.650 and SE = 0.638 for the fitted. F (2,48) = 44.89, and Prob. of R2 = 0 (2,48) = 0.000. The steric portion of the influences of maleimide analogs for the inhibitory potency described by this model is 22%, whereas the electrostatic portion is 78%. The first component explains 53% of the variance in the pIC50, and the second component accounts for additional 13% of the variance. An essentially identical CoMSIA model was obtained when the steric and the electrostatic fields were considered separately. Addition of hydrophobic and/or hydrogen donor or acceptor components did not improve the correlation already obtained.
Comparison of Two Binding Modes with CoMSIA results
In light of the similar CoMFA results from the two different binding modes of the compounds studied, it is interesting to examine the CoMSIA results with respect to the two different binding modes.
The CoMSIA results (eq 3, 4, 5, and 6) summarized in Table 5 show that the superposition from the binding mode 1 (eq 3 and 4) accounts for the variation in the pIC50 values of the compounds studied more than the superposition from the binding mode 2 (eq 5 and 6). These results are consistent with those of the CoMFA discussed above. The differences in statistics between the two CoMSIA results are larger than the corresponding CoMFA results. The statistics of the CoMSIA analyses (SE and R2 for both the cross-validation and the fitted) also indicate that the binding mode 1 explains the variation in pIC50 better than the binding mode 2. The results further support the binding mode 1 as the correct binding modes of the compounds studied. Another interesting aspect of the CoMSIA results is that eq. 4 and 6 indicate some possible contributions of hydrophobic contribution of the substituents toward the observed pIC50 values as in the classical QSAR discussed above. It was previously studied that the steric contribution of CoMFA may include the hydrophobic contribution.38–43 The present CoMSIA results indicate separate contributions from the hydrophobic contributions even though the steric contributions may include the hydrophobic contributions of the substituents as in the case of CoMFA.
As in CoMFA, the model having R2(cv) > 0.3 may be considered significant in CoMSIA. Therefore, the CoMSIA model for binding mode 2 may not be statistically significant in this standard. Therefore, the results of CoMSIA are consistent with those of CoMFA, and both results indicate that the binding mode 1 would be favored over the binding mode 2 for the benzofuran-3-yl-(indol-3-yl)maleimides examined in this study.
Comparison of 3D-QSAR Results with the Current X-ray Structures
While this manuscript was in preparation, the preliminary results of ligand-bound X-ray crystal structures of GSK-3β became available. The present 3D-QSAR results are interesting to compare with the preliminary X-ray crystal structures of two GSK-3β inhibitors, namely compound 5 and compound 14. Compound 5 has Br as X5 and (CH2)3OH as R. Compound 14 has 5-cyclopropylethynyl as X5, F as Y5, and CH3 as R. Even though there are clear differences in the substituent pattern of these two compounds, the initial X-ray crystal structures could not readily discern the relative positions of the indole ring and the benzofuran ring. In fact, the positions of these two rings were thought to be switched and be similar to the binding mode 1 in the initial preliminary X-ray results. This initial observation was not consistent with the 3D-QSAR results. In the updated X-ray crystal structures of these two compounds, however, it was determined that the positions of the two rings are consistent with the binding mode 1.
The CoMFA results indicate that the binding mode 1 is preferred over the binding mode 2. The same is true for the CoMSIA results. The CoMSIA results indicate more clearly that the binding mode 1 better accounts for the variance in pIC50 than does binding mode 2, even though their statistics are inferior to those of CoMFA.
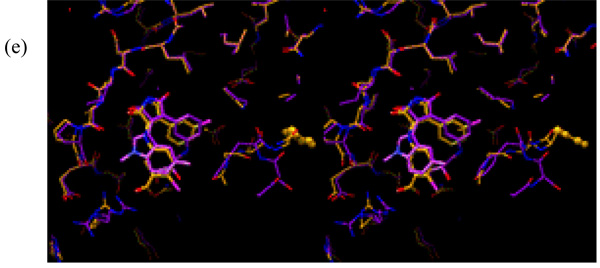
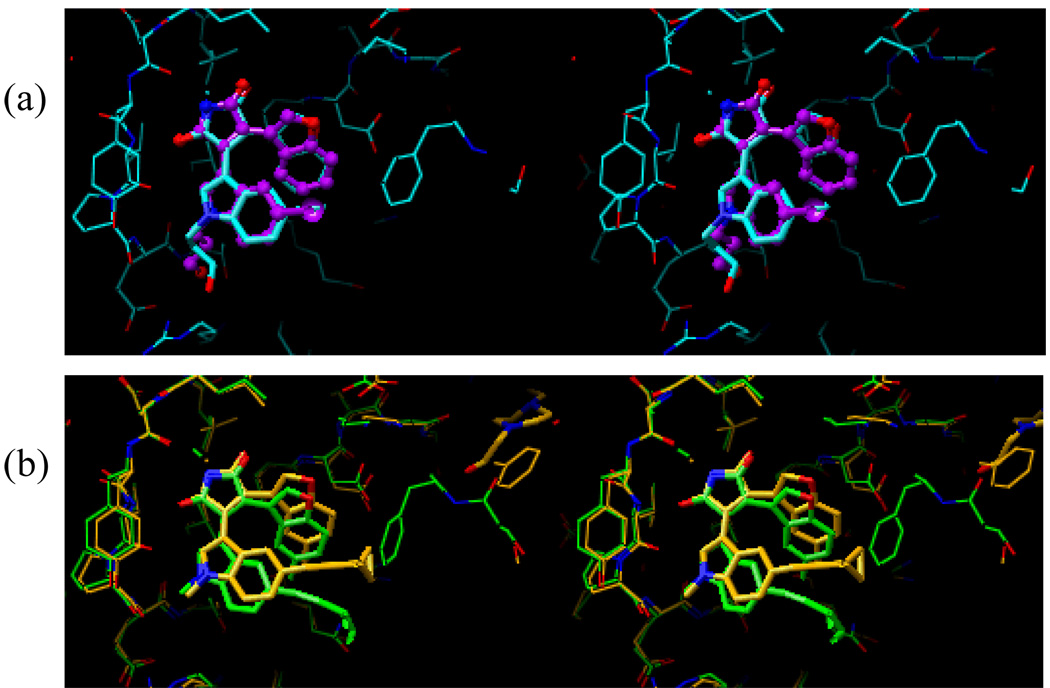
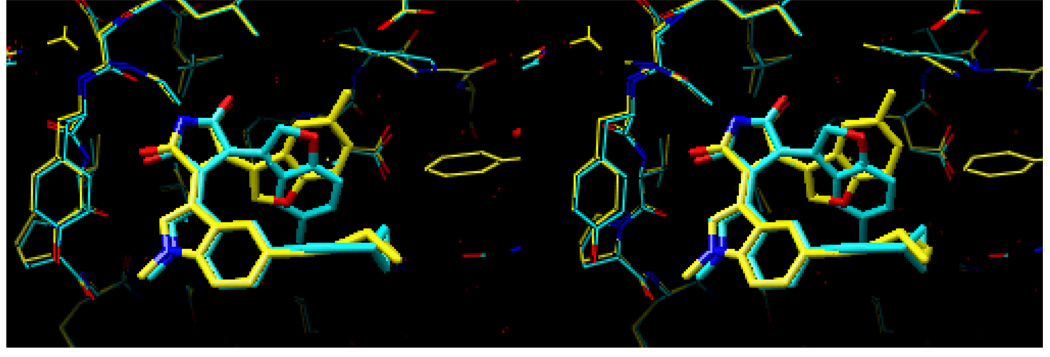
Figure 10 shows a superposition of the docked conformation and the current X-ray crystal structures of compounds 5 and 14. Figure 10a shows that the docked conformation (shown in purple) and the current X-ray crystal structure (shown in cyan) of compound 5 are essentially identical. Although the superposition of the docked conformation (shown in green) and the X-ray crystal structure (shown in orange) of compound 14 are very similar, Figure 10b shows that there is some movement in the binding pocket of GSK-3β (See further discussion below about the flexible binding pocket relating to the binding mode of compound 14).
Figure 10.
Superposition of the docked and the preliminary X-ray crystal structures of compounds 5 (a) and 14 (b). In Figure 10a, the docked structure of compound 5 is shown in purple, while the X-ray crystal structure of the same compound is shown in cyan. In Figure 10b, the docked structure of compound 14 is shown in green, while the X-ray crystal structure of the same compound is shown in orange.
Unexplained pIC50 Portions by the 3D-QSAR results
The current CoMFA and CoMSIA models account for the variance of pIC50 values about 80% and 75%, (R2 = 0.81 for CoMFA and 0.75 for CoMSIA), and the corresponding SE values are 0.48 and 0.55, respectively. Unexplained portion or outliers of QSARs can be very important and interesting, especially when the observed biological activity is higher than that predicted by the QSAR model. Unexplained portions or outliers by the QSAR model may imply several possibilities in addition to experimental errors. They may imply that the QSAR may lack certain descriptors to describe the QSAR of the entire group of compounds studied, or that the mathematical model or approach may not be appropriate. The outliers or unexplained portions may also be due to the inappropriate calculation of the parameter values used, indicate a different mechanism of action, or may result from a different binding mode or a flexible binding site.44, 45
One possible source of the unexplained component of the pIC50 values in the present case, which is about 20% of the variance in pIC50 values, is likely due to the flexible binding pocket as shown in Figure 1b. Depending on the size of the ligand, the flexible glycine-rich loop of GSK-3β may change its loop conformation as indicated by the different position of Phe67 in the various X-ray crystal structures. (Also see the discussion below for Figure 10b and compound 14.) Another possible source is the flexible side chain conformation as observed by the different side chain conformations of Arg141 (see the lower left corner of Figure 1b). The third possibility is the binding mode of the benzofuran ring. The size of the binding pocket where the benzofuran ring binds is large enough to accommodate the unsubstituted benzofuran ring in two different positions. One possibility is the one shown by the binding of the corresponding indole ring in staurosporine (Figure 1c, 1Q3D) or in the bis-(indole)maleimide pyridinophane (Figure 1d, 2OW3), and the other is the one seen in the conformation used in the current CoMFA or CoMSIA studies. Interestingly, two different benzofuran binding conformations were observed in the Compound 14-bound GSK-3β X-ray crystal structure which is shown in Figure 11.
Figure 11.
Two different binding modes of the benzofuran ring of compound 14 observed in the X-ray crystal structure of the ligand-bound GSK-3β.
In QSAR, if the difference between the observed and the calculated activity values is greater than twice the standard error of the model, such compounds are normally considered as outliers. Table 6 shows that one compound (compound 7; X = 7-CH2OH, Y = 6-CH2OH, and R = CH3) is an outlier of the CoMFA model derived from the binding mode 1. The calculated pIC50 value of this compound is 9.31, whereas the observed pIC50 value is 8.29. The difference is larger than 1.0 in logarithmic unit. Thus, the compound is calculated to be more a potent binder than observed. Possible reasons for the discrepancy between the calculated and the observed inhibitory potencies of this compound could be due to various reasons as described above, as well as to possible experimental error. This discrepancy suggests that this compound might be studied further.
Unlike the CoMFA model, the corresponding CoMSIA model from the binding mode 1 yielded five outliers (compounds 1, 7, 10, 12, and 41). The larger number of outliers from the CoMSIA model shows that the CoMSIA model does not explain the observed pIC50 values as well as the CoMFA model, and is not as good as the corresponding CoMFA model to describe the 3D-QSAR for the set studied. However, it is interesting to see that the CoMSIA model also suggests that compound 7 would be a more potent binder than observed. The difference between the two values is greater than 1.0 in logarithmic unit. These results are consistent with those of CoMFA.
Further utilization of the 3D-QSAR results
The aim of this study was to identify the binding mode of the substituted maleimides (I) to the binding site of GSK-3β. Understanding the binding modes of compounds under study is critical in drug discovery research. Utilizing the 3D-QSAR methodologies of both CoMFA and CoMSIA, the possible binding mode of the maleimides of interest was determined in this study. The CoMFA model was further validated statistically using the scrambled pIC50 values. The suggested binding modes of these compounds were further supported by the two preliminary X-ray crystal structures of inhibitor-bound GSK-3β.
Even though the major aim of our study was accomplished, it was interesting to test the CoMFA model developed from the binding mode 1 for its predictability. Two compounds were synthesized and their pIC50 values were estimated using the final CoMFA and CoMSIA models while they were being tested for their biological activity. One compound (52) is X5 = Cl, X6 = OMe, R = CH3, and the other compound (53) is X7 = CH2OMe, Y6 = CH2OH, R = CH3. The calculated pIC50 values from the CoMFA model are 6.13 for compound 52 and 7.67 for compound 53. The calculated pIC50 values from the CoMSIA model are 5.95 for compound 52 and 5.77 for compound 53. The experimentally determined pIC50 values are 7.09 (81.4 nM) and 9.14 (0.73 nM) for compound 52 and 53, respectively. The experimentally determined pIC50 values are higher than the calculated values for both compounds from both CoMFA and CoMSIA methods. Although compound 53 is not the most potent inhibitor in this series, it is still significantly potent. It provides an example of utilizing the binding mode and 3D-QSAR of these GSK-3β inhibitors in our drug discovery research. The results of CoMFA fit better than the results of CoMSIA.
Although we were delighted to see the higher inhibitory potencies of the newly synthesized compounds, it was also puzzling to see the poor predictabilities of both 3D-QSAR models, at least for these two compounds. For compound 52 (5-Cl, 6-OCH3), one can compare the results with those of compound 39 (5-OCH3, 6-Cl). The observed pIC50 of compound 39 is 6.36, whereas the calculated values are 6.54 and 6.51 from CoMFA and CoMSIA, respectively. One can see that while both 3D-QSAR models predicted well the pIC50 value of compound 39, those models did not predict well the pIC50 value of compound 52. The results indicate that the current models do not describe the effects of these structural changes on the inhibitory potencies. However, inclusion of these two compounds in the further development of the CoMFA and CoMSIA models could improve the predictabilities of both models for future compounds containing such structural modifications. If a 3D-QSAR model does not contain certain structural information, it is not surprising to find that the model is generally unable to predict the activity of a compound embodying such structural modifications.30
Conclusion and Summary
The binding modes of GSK-3β inhibitors have been studied with molecular modelling and docking methods along with 3D-QSAR approaches. The approaches of CoMFA and CoMSIA were used for 3D-QSAR with 51 substituted 3-benzofuranyl-4-indolyl-maleimides as GSK-3β inhibitors.
Two binding modes of our inhibitors to the binding pocket of GSK-3β were investigated. The binding mode 1 yielded better CoMFA and CoMSIA correlations. The binding mode determined by the results of this study is consistent with the preliminary results of an X-ray crystal structure analysis of inhibitor-bound GSK-3β. This study shows that the 3D-QSAR methodologies are useful in identifying the correct binding modes of the substituted 3-benzofuranyl-4-indolyl-maleimides to GSK-3β. These models will be updated with additional compounds and used in our continued work to estimate the inhibitory potency of other novel GSK-3β inhibitors of this structural class.
Several possible sources of the unexplained component of the pIC50 values by the 3D-QSAR models are discussed.
The present study provides the first example of identifying the correct binding mode of GSK-3β inhibitors using the molecular modelling, docking, and 3D-QSAR approaches.
Experimental Section
The publicly available protein structures used in this study and listed in Table 1 were obtained from the RSCS protein data bank.34
Two binding modes of the 51 compounds in the GSK-3β binding site were obtained by docking each compound into the binding site (see the discussions in the text) starting from the initial 2a and 2b conformations in Table 2.
The ligands were manually docked into the binding site of GSK-3β (1R0E). The initial binding position of the ligands was set by superimposing the pyrrolidin-2-one ring of the inhibitors over the corresponding ring of the ligand in the ligand-bound X-ray crystal structure of GSK-3β, 1R0E. Initial docking conformations of the substituted indole ring and the substituted benzofuran ring of the inhibitors were set to be similar to the two conformations (2a and 2b) shown in Figure 2. The orientation of each side chain of the molecules were set in such a way that the substituents would exhibit minimal steric clashes with any amino acid residues of the protein, but would be able to engage in possible hydrogen bonding interactions with nearby amino acid residues.
The geometry optimizations of the ligand-bound GSK-3β complexes were then performed using the molecular modelling software Sybyl version 7.3 of Tripos. The optimization of the protein-ligand complex was done by the Powell method without any initial optimization using the MMFF94 force field, the Gasteiger-Marsili charges, constant dielectric function, NB cut-off of 8.0, and dielectric constant of 1.0. The default settings were used for others with termination when the gradient reaches 0.05 kcal/mol. The maximum iteration for the geometry optimizations was set to be 1000.
The 3D-QSAR models were developed using the techniques of CoMFA and CoMSIA available in the Sybyl software package. The superpositions of the inhibitors used for each CoMFA and CoMSIA models were those of the docked positions and conformations obtained as described above. The CoMFA and CoMSIA modules of the molecular modelling software Sybyl version 7.3 of Tripos were used for these 3D-QSAR analyses. Default settings for all parameters were used using CH3+ as the probe, a 2-angstrom lattice box, and the Gasteiger-Marsili charges. Leave-one-out method was used for the cross-validation step. The PLS analysis was done using the SAMPLS method available through Sybyl. The selections of the final CoMFA and CoMSIA models were based on the results of the cross-validation, where are summarized in Table 3 (CoMFA) and Table 5 (CoMSIA). Further validation of the CoMFA and CoMSIA models from the binding mode 1 using scrambling pIC50 values are summarized in Table 4.
All the figures were generated using the UCSF chimera molecular modelling program production version 1.46
Acknowledgements
Ki Kim thanks Dr. Pavel Petukhov for the use of molecular modeling software. Andrew Mesecar is supported by grants from the National Cancer Institute of the National Institutes of Health (CA92744 and CA48112). This work was supported in part by NIH grant (1R01 MH072940-01, Grantee Alan P. Kozikowski)
References
- 1.Martinez A. Preclinical Efficacy on GSK-3 Inhibitors:Towards a Future Generation of Powerful Drugs. Med. Res. Rev. 2008;5:773–796. doi: 10.1002/med.20119. [DOI] [PubMed] [Google Scholar]
- 2.Jope RS, Yuskaitis CJ, Beurel E. Glycogen Synthase Kinase-3 (GSK3): Inflammation, Diseases, and Therapeutics. Neurochem. Res. 2007;32:577–595. doi: 10.1007/s11064-006-9128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat R, Haeberlein SLB, Avila J. Glycogen synthase kinase 3: a drug target for CNS therapies. J. Neurochem. 2004;89:1313–1317. doi: 10.1111/j.1471-4159.2004.02422.x. [DOI] [PubMed] [Google Scholar]
- 4.Martinez A, Castro A, Alonso M. Glycogen Synthase Kinase 3 (GSK-3) Inhibitors as New Promising Drugs for Diabetes, Neurodegeneration, Cancer, and Inflammation. Med. Res. Rev. 2002;22:373–384. doi: 10.1002/med.10011. [DOI] [PubMed] [Google Scholar]
- 5.Martinez A, Alonso M, Castro A, Pérez C, Moreno FJ. First Non-ATP Competitive Glycogen Synthase Kinase 3 (GSK-3) Inhibitors: Thiadiazolidinones (TDZD) as Potential Drugs for the Treatment of Alzheimer's Disease. J. Med. Chem. 2002;45:1292–1299. doi: 10.1021/jm011020u. [DOI] [PubMed] [Google Scholar]
- 6.Meijer L, Skaltsounis AL, Magiatis P, Polychronopoulos P, Knockaert M, Leost M, Ryan XP, Vonica CA, Brivanlou A, Dajani R, Crovace C, Tarricone C, Musacchio A, Roe SM, Pearl L, Greengard P. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem. Biol. 2003;10:1255–1266. doi: 10.1016/j.chembiol.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Kuo G-H, Prouty C, DeAngelis A, Shen LQ, O'Neill DJ, Shah C, Connolly PJ, Murray WV, Conway BR, Cheung P, Westover L, Xu JZ, Look RA, Demarest KT, Emanuel S, Middleton SA, Jolliffe L, Beavers MP, Chen X. Synthesis and Discovery of Macrocyclic Polyoxygenated Bis-7-azaindolylmaleimides as a Novel Series of Potent and Highly Selective Glycogen Synthase Kinase-3â Inhibitors. J. Med. Chem. 2003;46:4021–4031. doi: 10.1021/jm030115o. [DOI] [PubMed] [Google Scholar]
- 8.Witherington J, Bordas V, Gaiba A, Garton NS, Naylor A, Rawlings AD, Slingsby BP, Smith DG, Takle AK, Ward RW. 6-Aryl-pyrazolo[3,4-b]pyridines: potent inhibitors of glycogen synthase kinase-3 (GSK-3) Bioorg. Med. Chem. Lett. 2003;13:3055–3057. doi: 10.1016/s0960-894x(03)00645-0. [DOI] [PubMed] [Google Scholar]
- 9.Witherington J, Bordas V, Gaiba A, Naylor A, Rawlings AD, Slingsby BP, Smith DG, Takle AK, Ward RW. 6-Heteroaryl-pyrazolo[3,4-b]pyridines: Potent and Selective Inhibitors of Glycogen Synthase Kinase-3 (GSK-3) Bioorg. Med. Chem. Lett. 2003;13:3059–3062. doi: 10.1016/s0960-894x(03)00646-2. [DOI] [PubMed] [Google Scholar]
- 10.Bregman H, Williams DS, Atilla GE, Carroll PJ, Meggers E. An Organometallic Inhibitor for Glycogen Synthase Kinase 3. J. Am. Chem. Soc. 2004;126:13594–13595. doi: 10.1021/ja046049c. [DOI] [PubMed] [Google Scholar]
- 11.Engler TA, Henry JR, Malhotra S, Cunningham B, Furness K, Brozinick J, Burkholder TP, Clay MP, Clayton J, Diefenbacher C, Hawkins E, Iversen PW, Li Y, Lindstrom TD, Marquart AL, McLean J, Mendel D, Misener E, Briere D, O'Toole JC, Porter WJ, Queener S, Reel JK, Owens RA, Brier RA, Eessalu TE, Wagner JR, Campbell RM, Vaughn R. Substituted 3-Imidazo[1,2-a]pyridin-3-yl-4-(1,2,3,4-tetrahydro-[1,4]diazepino-[6,7,1-hi]indol-7-yl)pyrrole-2,5-diones as Highly Selective and Potent Inhibitors of Glycogen Synthase Kinase-3. J. Med. Chem. 2004;47:3934–3937. doi: 10.1021/jm049768a. [DOI] [PubMed] [Google Scholar]
- 12.Kunick C, Laurenroth K, Wieking K, Xie X, Schultz C, Gussio R, Zaharevitz D, Leost M, Meijer L, Weber A, Jorgensen FS, Lemcke T. Evaluation and Comparison of 3D-QSAR CoMSIA Models for CDK1, CDK5, and GSK-3 Inhibition by Paullones. J. Med. Chem. 2004;47:22–36. doi: 10.1021/jm0308904. [DOI] [PubMed] [Google Scholar]
- 13.Peat AJ, Garrido D, Boucheron JA, Schweiker SL, Dickerson SH, Wilson JR, Wang TY, Thomson SA. Novel GSK-3 inhibitors with improved cellular activity. Bioorg. Med. Chem. Lett. 2004;14:2127–2130. doi: 10.1016/j.bmcl.2004.02.037. [DOI] [PubMed] [Google Scholar]
- 14.Meijer L, Flajolet M, Greengard P. Pharmacological inhibitors of glycogen synthase kinase 3. TRENDS in Pharmacological Sciences. 2004;9:471–480. doi: 10.1016/j.tips.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H-C, Ye H, Conway BR, Derian CK, Addo MF, Kuo G-H, Hecker LR, Croll DR, Li J, Westover L, Xu JZ, Look R, Demarest KT, Andrade-Gordon P, Damiano BP, Maryanoff BE. 3-(7-Azaindolyl)-4-arylmaleimides as potent, selective inhibitors of glycogen synthase kinase-3. Bioorg. Med. Chem. Lett. 2004;14:3245–3250. doi: 10.1016/j.bmcl.2004.03.090. [DOI] [PubMed] [Google Scholar]
- 16.Polychronopoulos P, Magiatis P, Skaltsounis A-L, Myrianthopoulos V, Mikros E, Tarricone A, Musacchio A, Roe SM, Pearl L, Leost M, Greengard P, Meijer L. Structural Basis for the Synthesis of Indirubins as Potent and Selective Inhibitors of Glycogen Synthase Kinase-3 and Cyclin-Dependent Kinases. J. Med. Chem. 2004;47:935–946. doi: 10.1021/jm031016d. [DOI] [PubMed] [Google Scholar]
- 17.Martinez A, Alonso M, Castro A, Dorronsoro I, Gelpi JL, Luque FJ, Perez C, Moreno FJ. SAR and 3D-QSAR Studies on Thiadiazolidinone Derivatives: Exploration of Structural Requirements for Glycogen Synthase Kinase 3 Inhibitors. J. Med. Chem. 2005;48:7103–7112. doi: 10.1021/jm040895g. [DOI] [PubMed] [Google Scholar]
- 18.Zeng M, Jiang Y, Zhang B, Zheng K, Zhang N, Yu Q. 3D QSAR studies on GSK-3 inhibition by aloisines. Bioorg. Med. Chem. Lett. 2005;15:395–399. doi: 10.1016/j.bmcl.2004.10.060. [DOI] [PubMed] [Google Scholar]
- 19.Kozikowski AP, Gaisina IN, Petukhov PA, Sridhar J, King LT, Blond SY, Duka T, Rusnak M, Sidhu A. Highly Potent and Specific GSK-3b Inhibitors That Block Tau Phosphorylation and Decrease a-Synuclein Protein Expression in a Cellular Model of Parkinson's Disease. ChemMedChem. 2006;1:256–266. doi: 10.1002/cmdc.200500039. [DOI] [PubMed] [Google Scholar]
- 20.Dessalew N, Bharatam PV. 3D-QSAR and molecular docking study on bisarylmaleimide series as glycogen synthase kinase 3, cyclin dependent kinase 2 and cyclin dependent kinase 4 inhibitors: An insight into the criteria for selectivity. Eur. J. Med. Chem. 2007;42:1014–1027. doi: 10.1016/j.ejmech.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Dessalew N, Patel DS, Bharatam PV. 3D-QSAR and molecular docking studies on pyrazolopyrimidine derivatives as glycogen synthase kinase-3beta inhibitors. J. Mol. Graph. Model. 2007;25:885–895. doi: 10.1016/j.jmgm.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Hamann M, Alonso D, Martin-Aparicio E, Fuertes A, Perez-Puerto MJ, Castro A, Morales S, Navarro ML, del Monte-Millan M, Medina M, Pennaka H, Balaiah A, Peng J, Cook J, Wahyuono S, Martinez A. Glycogen Synthase Kinase-3 (GSK-3) Inhibitory Activity and Structure-Activity Relationship (SAR) Studies of the Manzamine Alkaloids. Potential for Alzheimer’s Disease. J. Nat. Prod. 2007;70:1397–1405. doi: 10.1021/np060092r. [DOI] [PubMed] [Google Scholar]
- 23.Shin D, Lee SC, Heo YS, Lee WY, M CJ, Lee YS, Ro S. Design and synthesis of 7-hydroxy-1H-benzoimidazole derivatives as novel inhibitors of glycogen synthase kinase-3beta. Bioorg. Med. Chem. Lett. 2007;17:5686–5689. doi: 10.1016/j.bmcl.2007.07.056. [DOI] [PubMed] [Google Scholar]
- 24.Kozikowski AP, Gaisina IN, Yuan H, Petukhov PA, Blond SY, Fedolak A, Caldarone B, McGonigle P. Structure-Based Design Leads to the Identification of Lithium Mimetics That Block Mania-like Effects in Rodents. Possible New GSK-3β Therapies for Bipolar Disorders. J. Am. Chem. Soc. 2007;129:8328–8332. doi: 10.1021/ja068969w. [DOI] [PubMed] [Google Scholar]
- 25.Stukenbrock H, Mussmann R, Geese M, Ferandin Y, Lozach O, Lemcke T, Kegel S, Lomow A, Burk U, Dohrmann C, Meijer L, Austen M, Kunick C. 9-Cyano-1-azapaullone (Cazpaullone), a Glycogen Synthase Kinase-3 (GSK-3) Inhibitor Activating Pancreatic β Cell Protection and Replication. J. Med. Chem. 2008;51:2196–2207. doi: 10.1021/jm701582f. [DOI] [PubMed] [Google Scholar]
- 26.Voigt B, Krug M, Schachtele C, Totzke F, Hilgeroth A. Probing Novel 1-Aza-9-oxafluorenes as Selective GSK-3b Inhibitors. ChemMedChem. 2008;3:120–126. doi: 10.1002/cmdc.200700175. [DOI] [PubMed] [Google Scholar]
- 27.Taha MO, Bustanji Y, Al-Ghussein MAS, Mohammad M, Zalloum H, Al-Masri IM, Atallah N. Pharmacophore Modeling, Quantitative Structure-Activity Relationship Analysis, and in Silico Screening Reveal Potent Glycogen Synthase Kinase-3 Inhibitory Activities for Cimetidine, Hydroxychloroquine, and Gemifloxacin. J. Med. Chem. 2008;51:2062–2077. doi: 10.1021/jm7009765. [DOI] [PubMed] [Google Scholar]
- 28.Gaisina IN, Gallier F, Kim KH, Ougolkov AV, Guo S, Holzle D, Luchini DN, Kurome T, Blond SY, Billadeau D, Kozikowski AP. Identification of Potent and Selective GSK-3β Inhibitors that Suppress Proliferation and Survival of Pancreatic Cancer Cells. J. Med. Chem. doi: 10.1021/jm801317h. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim KH. Comparative Molecular Field Analysis. In: Dean PM, editor. Molecular Similarity in Drug Design. London: Chapman & Hall; 1995. pp. 291–331. [Google Scholar]
- 30.Kim KH, Greco G, Novellino E. A critical review of recent CoMFA applications. Perspectives in Drug Discovery and Design. 1998;12–14:233–255. [Google Scholar]
- 31.Martin YC, Kim KH, Lin CT. Comparative Molecular Field Analysis: CoMFA. Vol. 1. JAI Press; 1996. pp. 1–52. [Google Scholar]
- 32.Klebe G. Comparative Molecular Similarity Indices Analysis: CoMSIA. In: Kubinyi HFG, Martin YC, editors. Three-Dimensional Quantitative Structure Activity Relationships. 3D QSAR in Drug Design. Recent Advances. Vol. 3. Netherlands: Springer; 1998. pp. 87–104. [Google Scholar]
- 33.Bertrand JA, Thieffine S, Vulpetti A, Cristiani C, Valsasina B, Knapp S, Kalisz HM, Flocco M. Structural Characterization of the GSK-3β Active Site Using Selective and Non-selective ATP-mimetic Inhibitors. J. Mol. Biol. 2003;333:393–407. doi: 10.1016/j.jmb.2003.08.031. [DOI] [PubMed] [Google Scholar]
- 34.Berman HM, Westbrook J, Feng Z, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hubbard SR. Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- 36.Huse M. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 37.Johnson LN. Active and inactive protein kinases: Structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 38.Kim KH. A Novel Method of Describing Hydrophobic Effects Directly From 3D Structures in 3D-Quantitative Structure-Activity Relationships Study. Med. Chem. Res. 1991;1:259–264. [Google Scholar]
- 39.Kim KH. Description of Nonlinear Dependence Directly from 3D Structures in 3D-Quantitative Structure-Activity Relationships. Med. Chem. Res. 1992;2:22–27. [Google Scholar]
- 40.Kim KH. Quantitative Structure-Activity Relationships: Nonlinear Dependence Described Directly from 3D Structures Using Comparative Molecular Field Analysis (CoMFA) Quant. Struct.-Act. Relat. 1992;11:309–317. [Google Scholar]
- 41.Kim KH. Nonlinear Dependence in GRID-Comparative Molecular Field Analysis (CoMFA) J. Comput.-Aid. Mol. Design. 1993;7:71–82. doi: 10.1007/BF00141576. [DOI] [PubMed] [Google Scholar]
- 42.Kim KH. Quantitative Structure-Activity Relationships: Describing Hydrophobic Interactions Directly from 3D Structures Using a Comparative Molecular Field Analysis (CoMFA) Approach. Quant. Struct.-Act. Relat. 1993;12:232–238. [Google Scholar]
- 43.Kim KH. Calculation of Hydrophobic Parameters Directly from Three-Dimensional Structures Using Comparative Molecular Field Analysis (CoMFA) J. Comput.-Aid. Mol. Design. 1995;9:308–318. doi: 10.1007/BF00125172. [DOI] [PubMed] [Google Scholar]
- 44.Kim KH. Outliers in SAR and QSAR: 2. Is flexible binding site a possible source of outliers? Journal of Computer-Aided Mol. Design. 2007;21:421–435. doi: 10.1007/s10822-007-9126-y. [DOI] [PubMed] [Google Scholar]
- 45.Kim KH. Outliers in SAR and QSAR: Is unusual binding mode a possible source of outliers? Journal of Computer-Aided Mol. Design. 2007;21:63–86. doi: 10.1007/s10822-007-9106-2. [DOI] [PubMed] [Google Scholar]
- 46.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 47.Allard J, Nikolcheva T, Gong L, Wang J, Dunten P, Avnur Z, Waters R, Sun Q, Skinner B. From genetics to therapeutics: the Wnt pathway and osteoporosis. To be Published. [Google Scholar]
- 48.Zhang HC, Bonaga LV, Ye H, Derian CK, Damiano BP, Maryanoff BE. Novel bis(indolyl)maleimide pyridinophanes that are potent, selective inhibitors of glycogen synthase kinase-3. Bioorg. Med. Chem. Lett. 2007;17:2863–2868. doi: 10.1016/j.bmcl.2007.02.059. [DOI] [PubMed] [Google Scholar]
- 49.Bax B, Carter PS, Lewis C, Guy AR, Bridges A, Tanner R, Pettman G, Mannix C, Culbert AA, Brown MJ, Smith DG, Reith AD. The structure of phosphorylated GSK-3beta complexed with a peptide, FRATtide, that inhibits beta-catenin phosphorylation. Structure. 2001;9:1143–1152. doi: 10.1016/s0969-2126(01)00679-7. [DOI] [PubMed] [Google Scholar]
- 50.Dajani R, Fraser E, Roe SM, Yeo M, Good VM, Thompson V, Dale TC, Pearl LH. Structural basis for recruitment of glycogen synthase kinase 3beta to the axin-APC scaffold complex. Embo J. 2003;22:494–501. doi: 10.1093/emboj/cdg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertrand JA, Thieffine S, Vulpetti A, Cristiani C, Valsasina B, Knapp S, Kalisz HM, Flocco M. Structural characterization of the GSK-3beta active site using selective and non-selective ATP-mimetic inhibitors. J. Mol. Biol. 2003;333:393–407. doi: 10.1016/j.jmb.2003.08.031. [DOI] [PubMed] [Google Scholar]
- 52.Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, Pearl LH. Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105:721–732. doi: 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 53.ter Haar E, Coll JT, Austen DA, Hsiao HM, Swenson L, Jain J. Structure of GSK3beta reveals a primed phosphorylation mechanism. Nat. Struct. Biol. 2001;8:593–596. doi: 10.1038/89624. [DOI] [PubMed] [Google Scholar]
- 54.Aoki M, Yokota T, Sugiura I, Sasaki C, Hasegawa T, Okumura C, Ishiguro K, Kohno T, Sugio S, Matsuzaki T. Structural insight into nucleotide recognition in tau-protein kinase I/glycogen synthase kinase 3 beta. Acta Crystallogr.,Sect.D. 2004;60:439–446. doi: 10.1107/S090744490302938X. [DOI] [PubMed] [Google Scholar]
- 55.Bhat R, Xue Y, Berg S, Hellberg S, Ormo M, Nilsson Y, Radesater AC, Jerning E, Markgren PO, Borgegard T, Nylof M, Gimenez-Cassina A, Hernandez F, Lucas JJ, Diaz-Nido J, Avila J. Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418. J. Biol. Chem. 2003;278:45937–45945. doi: 10.1074/jbc.M306268200. [DOI] [PubMed] [Google Scholar]
- 56.Kim H-J, Choo H, Cho YS, No KT, Pae AN. Novel GSK-3b inhibitors from sequential virtual screening. Bioorg. Med. Chem. 2008;16:636–643. doi: 10.1016/j.bmc.2007.10.047. [DOI] [PubMed] [Google Scholar]
- 57.GSK-3β in vitro kinase assay was done as described previously: The in vitro kinase assay was performed in a 40 µL reaction volume containing 250 ng GSK-3β Calbiochem, 10 µM pGS peptide (RRRPASVPPSPSLRHSS(P)HQRR), 10 µM ATP plus 1 µCi γ32P ATP, and increasing concentrations of the inhibitors in kinase buffer (20 mM MOPS pH 7.2, 15 mM MgCl2, 2 mM EGTA, 25 mM β-glycerol phosphate, 1 mM Na3VO4, 1mM DTT). The inhibitors were allowed to bind GSK-3β for 10 min prior to the addition of peptide. The reaction was started by the addition of the ATP solution and was carried out at 30 °C for 30 min after which 25 µL of the reaction mixture was spotted on P81 Whatman filters, dried, washed twice with 0.75% H3PO4, and rinsed with acetone. The dried filters were placed in scintillation vials containing 2 ml of Scinti-safe Econo 2 solution and read in a Beckman Coulter LS 600IC scintillation counter. IC50 determination as below: The data were plotted as average percent activity versus the log of the concentration of inhibitor. The IC50 was determined according to NIH guidelines and the 4-parameter logistic plot (4PL), which calculates the relative IC50 based on the maximum (top) and minimum (bottom) percent activity as well as the Hill slope. In some cases the 4PL did not give an accurate fitting of the data (error fit above 40%) and thus a 3-parameter logistic fit top (3PLFT) or 3-parameter logistic fit bottom (3PLFB) were used to obtain the IC50 using the equation y = Bottom + (Top-Bottom)/(1+10^((LogIC50-x)*HillSlope)) from Graphpad Prism 5. This program displays the LogIC50 as well as the IC50 and gives the standard error of the LogIC50, this was converted back to the % fitting error of the IC50 by using the equation %FE(IC50)=FE(LogIC50)*Ln(10)*100.